Efficient Reprogramming of Mouse Embryonic Stem Cells into Trophoblast Stem-like Cells via Lats Kinase Inhibition
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods and Materials
2.1. Animal Care and Use
2.2. Mouse Embryo Culture
2.3. Stem Cells and Culture Conditions
2.4. Immunofluorescence of Embryos or Cells
2.5. Quantitative RT-PCR
2.6. Flow Cytometry
2.7. Chimeric Embryos and Implantation
2.8. Quantification and Statistical Analysis
3. Results
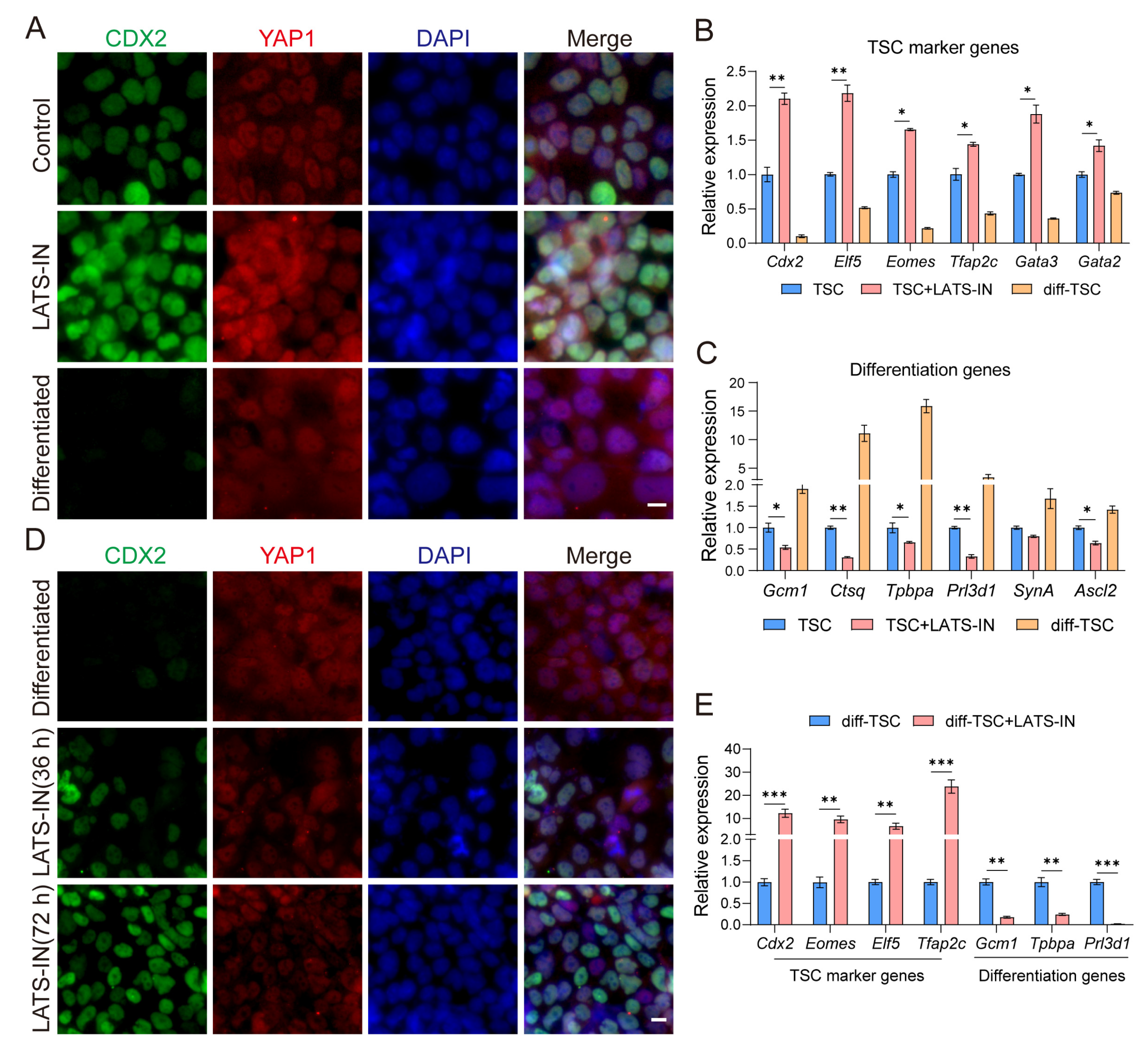
3.1. The Cellular Localization of YAP Regulates the Differentiation State of TSCs
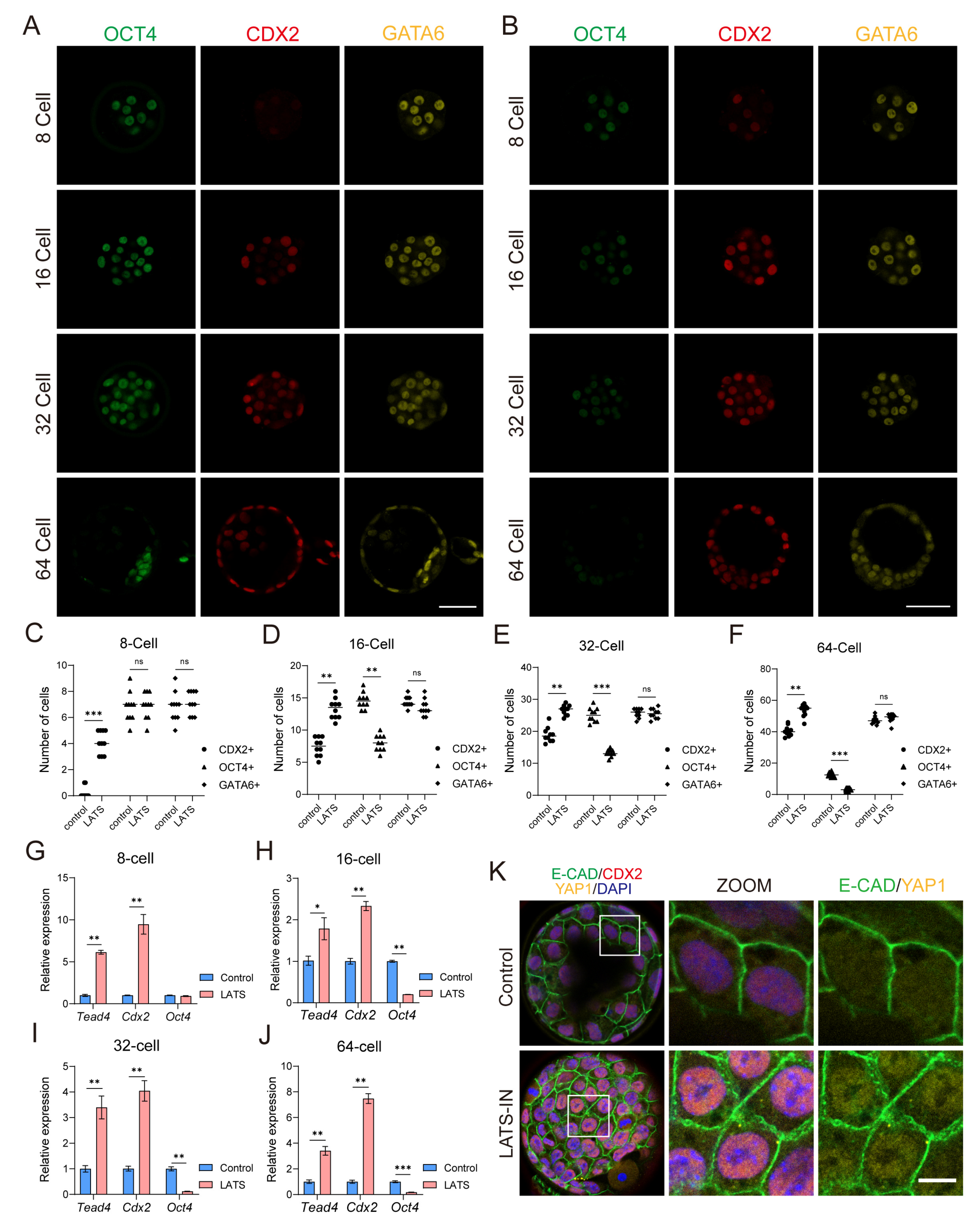
3.2. Inhibition of LATS Kinase Promotes the Specialization of Embryos into the TE Lineage
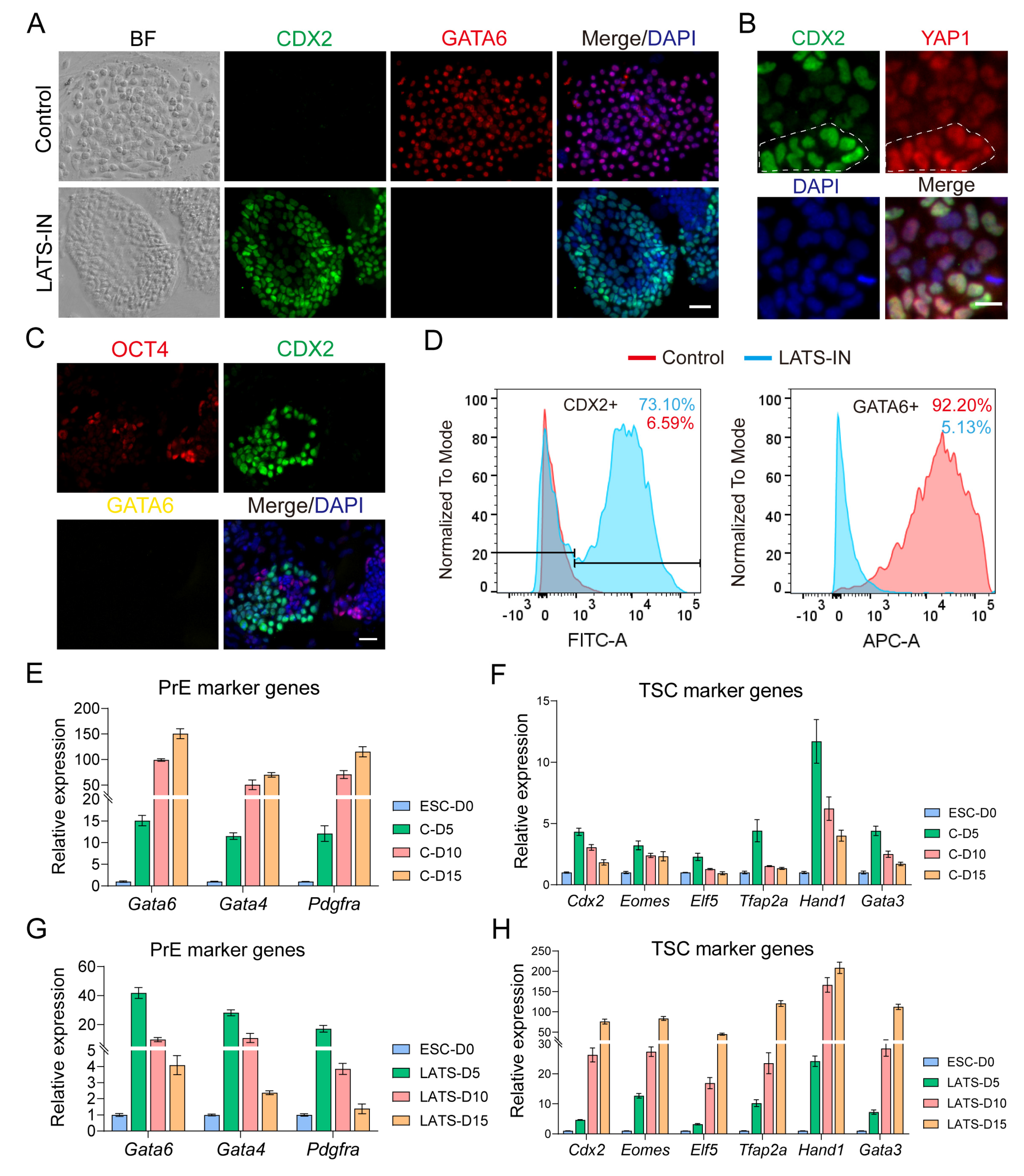
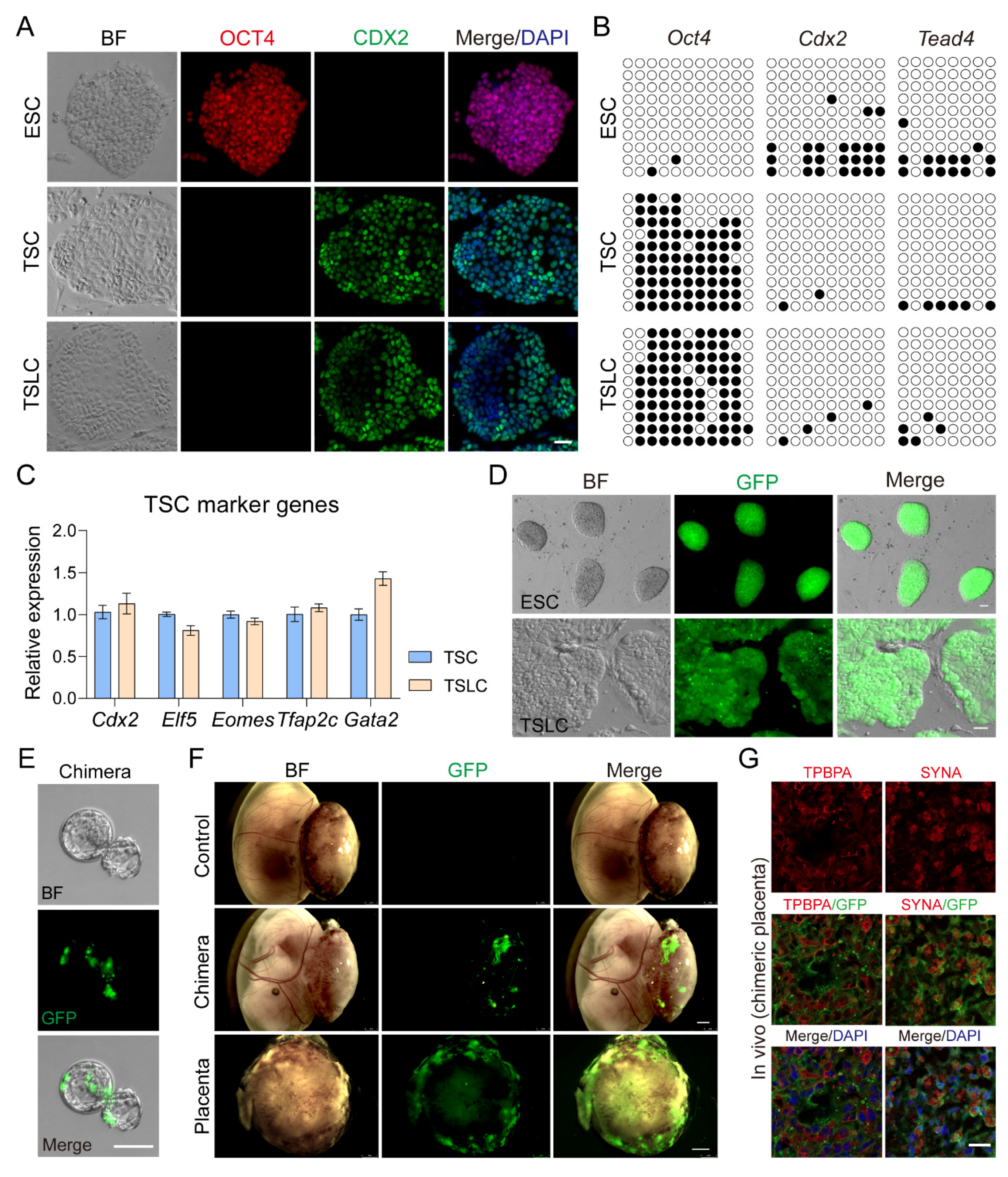
3.3. Inhibition of LATS Kinase Promotes ESCs Transformation to TSC-like Cells
3.4. TSLCs Can Differentiate into Placental Terminal Trophoblast Cells In Vivo
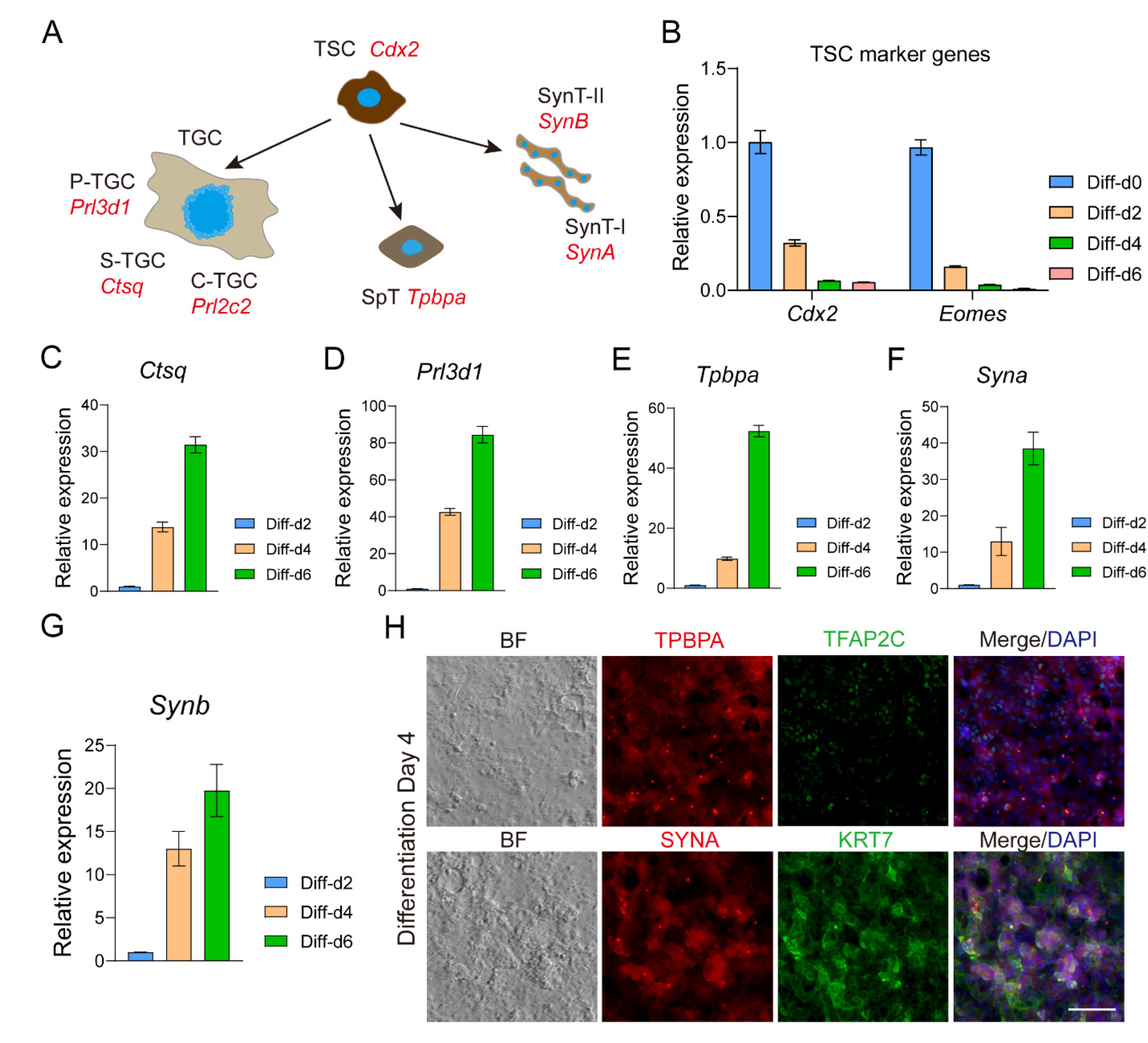
3.5. TSLCs Can Differentiate into Mature Trophoblast Cells In Vitro
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fierro-González, J.C.; White, M.D.; Silva, J.C.; Plachta, N. Cadherin-dependent filopodia control preimplantation embryo compaction. Nat. Cell Biol. 2013, 15, 1424–1433. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.A.; Teo, R.T.Y.; Li, H.L.; Robson, P.; Glover, D.M.; Zernicka-Goetz, M. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc. Natl. Acad. Sci. USA 2010, 107, 6364–6369. [Google Scholar] [CrossRef]
- Nishioka, N.; Inoue, K.; Adachi, K.; Kiyonari, H.; Ota, M.; Ralston, A.; Yabuta, N.; Hirahara, S.; Stephenson, R.O.; Ogonuki, N.; et al. The hippo signaling pathway components lats and yap pattern tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 2009, 16, 398–410. [Google Scholar] [CrossRef] [PubMed]
- Posfai, E.; Rovic, I.; Jurisicova, A. The mammalian embryo's first agenda: Making trophectoderm. Int. J. Dev. Biol. 2019, 63, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Rossant, J. Genetic control of early cell lineages in the mammalian embryo. Annu. Rev. Genet. 2018, 52, 185–201. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.H.; McConnell, J.M. Lineage allocation and cell polarity during mouse embryogenesis. Semin Cell Dev Biol 2004, 15, 583–597. [Google Scholar] [CrossRef]
- Yamanaka, Y.; Lanner, F.; Rossant, J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 2010, 137, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.S.; Rahman, S.; Raina, D.; Schröter, C.; Hadjantonakis, A.K. Live visualization of ERK activity in the mouse blastocyst reveals lineage-specific signaling dynamics. Dev. Cell 2020, 55, 341–353.e5. [Google Scholar] [CrossRef]
- Frum, T.; Ralston, A. Cell signaling and transcription factors regulating cell fate during formation of the mouse blastocyst. Trends Genet. 2015, 31, 402–410. [Google Scholar] [CrossRef]
- Ohinata, Y.; Endo, T.A.; Sugishita, H.; Watanabe, T.; Iizuka, Y.; Kawamoto, Y.; Saraya, A.; Kumon, M.; Koseki, Y.; Kondo, T.; et al. Establishment of mouse stem cells that can recapitulate the developmental potential of primitive endoderm. Science 2022, 375, 574–578. [Google Scholar] [CrossRef]
- Yang, J.; Ryan, D.J.; Wang, W.; Tsang, J.C.H.; Lan, G.C.; Masaki, H.; Gao, X.F.; Antunes, L.; Yu, Y.; Zhu, Z.X.; et al. Establishment of mouse expanded potential stem cells. Nature 2017, 550, 393–397. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, B.; Xu, J.; Wang, J.L.; Wu, J.; Shi, C.; Xu, Y.X.; Dong, J.B.; Wang, C.Y.; Lai, W.F.; et al. Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell 2017, 169, 243–257. [Google Scholar] [CrossRef]
- Kunath, T.; Arnaud, D.; Uy, G.D.; Okamoto, L.; Chureau, C.; Yamanaka, Y.; Heard, E.; Gardner, R.L.; Avner, P.; Rossant, J. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 2005, 132, 1649–1661. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Kunath, T.; Hadjantonakis, A.K.; Nagy, A.; Rossant, J. Promotion of trophoblast stem cell proliferation by FGF4. Science 1998, 282, 2072–2075. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Rayon, T.; Menchero, S.; Nieto, A.; Xenopoulos, P.; Crespo, M.; Cockburn, K.; Cañon, S.; Sasaki, H.; Hadjantonakis, A.K.; de la Pompa, J.L.; et al. Notch and hippo converge on to specify the trophectoderm lineage in the mouse blastocyst. Dev. Cell 2014, 30, 410–422. [Google Scholar] [CrossRef]
- Stephenson, R.O.; Yamanaka, Y.; Rossant, J. Disorganized epithelial polarity and excess trophectoderm cell fate in preimplantation embryos lacking E-cadherin. Development 2010, 137, 3383–3391. [Google Scholar] [CrossRef] [PubMed]
- Vinot, S.; Le, T.; Ohno, S.; Pawson, T.; Maro, B.; Louvet-Vallée, S. Asymmetric distribution of PAR proteins in the mouse embryo begins at the 8-cell stage during compaction. Dev. Biol. 2005, 282, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Leung, C.Y.; Shahbazi, M.N.; Zernicka-Goetz, M. Actomyosin polarisation through PLC-PKC triggers symmetry breaking of the mouse embryo. Nat. Commun. 2017, 8, 921. [Google Scholar] [CrossRef]
- Fleming, T.P.; Javed, Q.; Collins, J.; Hay, W. Biogenesis of structural intercellular-junctions during cleavage in the mouse embryo. J. Cell Sci. 1993, 17, 119–125. [Google Scholar] [CrossRef]
- Maître, J.L.; Berthoumieux, H.; Krens, S.F.G.; Salbreux, G.; Jülicher, F.; Paluch, E.; Heisenberg, C.P. Adhesion Functions in Cell Sorting by Mechanically Coupling the Cortices of Adhering Cells. Science 2012, 338, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Anani, S.; Bhat, S.; Honma-Yamanaka, N.; Krawchuk, D.; Yamanaka, Y. Initiation of Hippo signaling is linked to polarity rather than to cell position in the pre-implantation mouse embryo. Development 2014, 141, 2813–2824. [Google Scholar] [CrossRef] [PubMed]
- Hirate, Y.; Hirahara, S.; Inoue, K.; Suzuki, A.; Alarcon, V.B.; Akimoto, K.; Hirai, T.; Hara, T.; Adachi, M.; Chida, K.; et al. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr. Biol. 2013, 23, 1181–1194. [Google Scholar] [CrossRef] [PubMed]
- Home, P.; Saha, B.; Ray, S.; Dutta, D.; Gunewardena, S.; Yoo, B.; Pal, A.; Vivian, J.L.; Larson, M.; Petroff, M.; et al. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc. Natl. Acad. Sci. USA 2012, 109, 7362–7367. [Google Scholar] [CrossRef] [PubMed]
- Gerri, C.; McCarthy, A.; Scott, G.M.; Regin, M.; Stamatiadis, P.; Brumm, S.; Simon, C.S.; Lee, J.; Montesinos, C.; Hassitt, C.; et al. A conserved role of the Hippo signalling pathway in initiation of the first lineage specification event across mammals. Development 2023, 150, dev201112. [Google Scholar] [CrossRef] [PubMed]
- Yagi, R.; Kohn, M.J.; Karavanova, I.; Kaneko, K.J.; Vullhorst, D.; DePamphilis, M.L.; Buonanno, A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 2007, 134, 3827–3836. [Google Scholar] [CrossRef]
- Nishioka, N.; Yamamoto, S.; Kiyonari, H.; Sato, H.; Sawada, A.; Ota, M.; Nakao, K.; Sasaki, H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 2008, 125, 270–283. [Google Scholar] [CrossRef]
- Hao, Y.W.; Chun, A.; Cheung, K.; Rashidi, B.; Yang, X.L. Tumor suppressor LATS1 is a negative regulator of oncogene. J. Biol. Chem. 2008, 283, 5496–5509. [Google Scholar] [CrossRef]
- Gu, B.; Bradshaw, B.; Zhu, M.; Sun, Y.; Hopyan, S.; Rossant, J. Live imaging YAP signalling in mouse embryo development. Open Biol. 2022, 12, 210335. [Google Scholar] [CrossRef]
- Hirate, Y.; Cockburn, K.; Rossant, J.; Sasaki, H. Tead4 is constitutively nuclear, while nuclear vs. cytoplasmic Yap distribution is regulated in preimplantation mouse embryos. Proc. Natl. Acad. Sci. USA 2012, 109, E3389–E3390. [Google Scholar] [CrossRef]
- Niwa, H.; Toyooka, T.; Shimosato, D.; Strumpf, D.; Takahashi, K.; Yagi, R.; Rossant, J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 2005, 123, 917–929. [Google Scholar] [CrossRef]
- Strumpf, D.; Mao, C.A.; Yamanaka, Y.; Ralston, A.; Chawengsaksophak, K.; Beck, F.; Rossant, J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 2005, 132, 2093–2102. [Google Scholar] [CrossRef] [PubMed]
- Cockburn, K.; Biechele, S.; Garner, J.; Rossant, J. The Hippo pathway member Nf2 is required for inner cell mass specification. Curr. Biol. 2013, 23, 1195–1201. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.Y.; Zernicka-Goetz, M. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nat. Commun. 2013, 4, 2251. [Google Scholar] [CrossRef] [PubMed]
- Lorthongpanich, C.; Messerschmidt, D.M.; Chan, S.W.; Hong, W.J.; Knowles, B.B.; Solter, D. Temporal reduction of LATS kinases in the early preimplantation embryo prevents ICM lineage differentiation. Genes Dev. 2013, 27, 1441–1446. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H. Mouse ES cell culture system as a model of development. Dev. Growth Differ. 2010, 52, 275–283. [Google Scholar] [CrossRef]
- Wang, H.Y.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef]
- Martello, G.; Smith, A. The nature of embryonic stem cells. Annu. Rev. Cell Dev. Biol. 2014, 30, 647–675. [Google Scholar] [CrossRef]
- Nagy, A.; Gocza, E.; Diaz, E.M.; Prideaux, V.R.; Ivanyi, E.; Markkula, M.; Rossant, J. Embryonic stem-cells alone are able to support fetal development in the mouse. Development 1990, 110, 815–821. [Google Scholar] [CrossRef]
- Denker, H.W. Autonomy in the development of stem cell-derived embryoids: Sprouting blastocyst-like cysts, and ethical implications. Cells 2021, 10, 1461. [Google Scholar] [CrossRef]
- Amadei, G.; Handford, C.E.; Qiu, C.X.; De Jonghe, J.; Greenfeld, H.; Tran, M.; Martin, B.K.; Chen, D.Y.; Aguilera-Castrejon, A.; Hanna, J.H.; et al. Embryo model completes gastrulation to neurulation and organogenesis. Nature 2022, 610, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.Y.C.; Rubinstein, H.; Gantner, C.W.; Hadas, R.; Amadei, G.; Stelzer, Y.; Zernicka-Goetz, M. Mouse embryo model derived exclusively from embryonic stem cells undergoes neurulation and heart development. Cell Stem Cell 2022, 29, 1445–1458.e8. [Google Scholar] [CrossRef]
- Li, R.H.; Zhong, C.Q.; Yu, Y.; Liu, H.S.; Sakurai, M.; Yu, L.Q.; Min, Z.Y.; Shi, L.; Wei, Y.L.; Takahashi, Y.; et al. Generation of blastocyst-like structures from mouse embryonic and adult cell cultures. Cell 2019, 179, 687–702.e18. [Google Scholar] [CrossRef]
- Rivron, N.C.; Frias-Aldeguer, J.; Vrij, E.J.; Boisset, J.C.; Korving, J.; Vivié, J.; Truckenmüller, R.K.; van Oudenaarden, A.; van Blitterswijk, C.A.; Geijsen, N. Blastocyst-like structures generated solely from stem cells. Nature 2018, 557, 106–111. [Google Scholar] [CrossRef] [PubMed]
- van den Brink, S.C.; Baillie-Johnson, P.; Balayo, T.; Hadjantonakis, A.-K.; Nowotschin, S.; Turner, D.A.; Martinez Arias, A. Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development 2014, 141, 4231–4242. [Google Scholar] [CrossRef] [PubMed]
- Morgani, S.M.; Canham, M.A.; Nichols, J.; Sharov, A.A.; Migueles, R.P.; Ko, M.S.H.; Brickman, J.M. Totipotent embryonic stem cells arise in ground-state culture conditions. Cell Rep. 2013, 3, 1945–1957. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.; Smith, A. Pluripotency in the Embryo and in Culture. CSH Perspect. Biol. 2012, 4, a008128. [Google Scholar] [CrossRef] [PubMed]
- Brons, I.G.M.; Smithers, L.E.; Trotter, M.W.B.; Rugg-Gunn, P.; Sun, B.W.; Lopes, S.M.C.D.; Howlett, S.K.; Clarkson, A.; Ahrlund-Richter, L.; Pedersen, R.A.; et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 2007, 448, 191–195. [Google Scholar] [CrossRef]
- Tesar, P.J.; Chenoweth, J.G.; Brook, F.A.; Davies, T.J.; Evans, E.P.; Mack, D.L.; Gardner, R.L.; McKay, R.D.G. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007, 448, 196–199. [Google Scholar] [CrossRef]
- Kojima, Y.; Kaufman-Francis, K.; Studdert, J.B.; Steiner, K.A.; Power, M.D.; Loebel, D.A.F.; Jones, V.; Hor, A.; de Alencastro, G.; Logan, G.J.; et al. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell 2014, 14, 107–120. [Google Scholar] [CrossRef]
- Smith, A. Formative pluripotency: The executive phase in a developmental continuum. Development 2017, 144, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M.; Barber, M.; Mansfield, W.; Cui, Y.; Spindlow, D.; Stirparo, G.G.; Dietmann, S.; Nichols, J.; Smith, A. Capture of mouse and human stem cells with features of formative pluripotency. Cell Stem Cell 2021, 28, 453–471.e458. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.; Kunath, T.; Rossant, J. Mouse trophoblast stem cells. Methods Mol. Med. 2006, 121, 125–148. [Google Scholar] [CrossRef] [PubMed]
- Kubaczka, C.; Senner, C.; Araúzo-Bravo, M.J.; Sharma, N.; Kuckenberg, P.; Becker, A.; Zimmer, A.; Brüstle, O.; Peitz, M.; Hemberger, M.; et al. Derivation and maintenance of murine trophoblast stem cells under defined conditions. Stem Cell Rep. 2014, 2, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, K.; Himeno, E.; Tanaka, S.; Kunath, T. Isolation and manipulation of mouse trophoblast stem cells. Curr. Protoc. Stem Cell Bio. 2015, 32, 1E.4.1–1E.4.32. [Google Scholar] [CrossRef] [PubMed]
- Azami, T.; Bassalert, C.; Allègre, N.; Estrella, L.V.; Pouchin, P.; Ema, M.; Chazaud, C. Regulation of the ERK signalling pathway in the developing mouse blastocyst. Development 2019, 146, dev177139. [Google Scholar] [CrossRef]
- Krawczyk, K.; Wilczak, K.; Szczepanska, K.; Maleszewski, M.; Suwinska, A. Paracrine interactions through FGFR1 and FGFR2 receptors regulate the development of preimplantation mouse chimaeric embryo. Open Biol. 2022, 12, 220193. [Google Scholar] [CrossRef]
- Chai, N.; Patel, Y.; Jacobson, K.; McMahon, J.; McMahon, A.; Rappolee, D.A. FGF is an essential regulator of the fifth cell division in preimplantation mouse embryos. Dev. Biol. 1998, 198, 105–115. [Google Scholar] [CrossRef]
- Singh, V.P.; Gerton, J.L. Protocol for mouse trophoblast stem cell isolation, differentiation, and cytokine detection. STAR Protoc. 2021, 2, 100242. [Google Scholar] [CrossRef]
- Zhu, D.M.; Gong, X.; Miao, L.Y.; Fang, J.S.; Zhang, J. Efficient induction of syncytiotrophoblast layer II cells from trophoblast stem cells by canonical wnt signaling activation. Stem Cell Rep. 2017, 9, 2034–2049. [Google Scholar] [CrossRef]
- Seong, J.; Frias-Aldeguer, J.; Holzmann, V.; Kagawa, H.; Sestini, G.; Heidari Khoei, H.; Scholte Op Reimer, Y.; Kip, M.; Pradhan, S.J.; Verwegen, L.; et al. Epiblast inducers capture mouse trophectoderm stem cells in vitro and pattern blastoids for implantation in utero. Cell Stem Cell 2022, 29, 1102–1118.e1108. [Google Scholar] [CrossRef]
- Wu, T.; Wang, H.; He, J.; Kang, L.; Jiang, Y.H.; Liu, J.C.; Zhang, Y.; Kou, Z.H.; Liu, L.J.; Zhang, X.H.; et al. Reprogramming of trophoblast stem cells into pluripotent stem cells by oct4. Stem Cells 2011, 29, 755–763. [Google Scholar] [CrossRef]
- Ohinata, Y.; Tsukiyama, T. Establishment of trophoblast stem cells under defined culture conditions in mice. PLoS ONE 2014, 9, e107308. [Google Scholar] [CrossRef]
- Perez-Garcia, V.; Lea, G.; Lopez-Jimenez, P.; Okkenhaug, H.; Burton, G.J.; Moffett, A.; Turco, M.Y.; Hemberger, M. BAP1/ASXL complex modulation regulates epithelial-mesenchymal transition during trophoblast differentiation and invasion. Elife 2021, 10, e63254. [Google Scholar] [CrossRef] [PubMed]
- Motomura, K.; Oikawa, M.; Hirose, M.; Honda, A.; Togayachi, S.; Miyoshi, H.; Ohinata, Y.; Sugimoto, M.; Abe, K.; Inoue, K.; et al. Cellular dynamics of mouse trophoblast stem cells: Identification of a persistent stem cell type. Biol. Reprod. 2016, 94, 122. [Google Scholar] [CrossRef] [PubMed]
- Seong, J.; Rivron, N.C. Protocol for capturing trophectoderm stem cells reflecting the blastocyst stage. STAR Protoc. 2023, 4, 102151. [Google Scholar] [CrossRef] [PubMed]
- Beddington, R.S.P.; Robertson, E.J. An assessment of the developmental potential of embryonic stem-cells in the midgestation mouse embryo. Development 1989, 105, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H.; Miyazaki, J.; Smith, A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000, 24, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H.; Ogawa, K.; Shimosato, D.; Adachi, K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 2009, 460, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Blij, S.; Parenti, A.; Tabatabai-Yazdi, N.; Ralston, A. Cdx2 efficiently induces trophoblast stem-like cells in naïve, but not primed, pluripotent stem cells. Stem Cells Dev. 2015, 24, 1352–1365. [Google Scholar] [CrossRef]
- Kuckenberg, P.; Buhl, S.; Woynecki, T.; van Füerden, B.; Tolkunova, E.; Seiffe, F.; Moser, M.; Tomilin, A.; Winterhager, E.; Schorle, H. The Transcription Factor TCFAP2C/AP-2γ Cooperates with CDX2 To Maintain Trophectoderm Formation. Mol. Cell. Biol. 2010, 30, 3310–3320. [Google Scholar] [CrossRef]
- Gao, H.B.; Gao, R.; Zhang, L.F.; Xiu, W.C.; Zang, R.G.; Wang, H.; Zhang, Y.; Chen, J.Y.; Gao, Y.W.; Gao, S.R. Esrrb plays important roles in maintaining self-renewal of trophoblast stem cells (TSCs) and reprogramming somatic cells to induced TSCs. J. Mol. Cell Biol. 2019, 11, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Ralston, A.; Cox, B.J.; Nishioka, N.; Sasaki, H.; Chea, E.; Rugg-Gunn, P.; Guo, G.J.; Robson, P.; Draper, J.S.; Rossant, J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 2010, 137, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Rhee, C.; Lee, B.K.; Beck, S.; Anjum, A.; Cook, K.R.; Popowski, M.; Tucker, H.O.; Kim, J. Arid3a is essential to execution of the first cell fate decision via direct embryonic and extraembryonic transcriptional regulation. Genes Dev. 2014, 28, 2219–2232. [Google Scholar] [CrossRef]
- Rhee, C.; Lee, B.K.; Beck, S.; LeBlanc, L.; Tucker, H.O.; Kim, J. Mechanisms of transcription factor-mediated direct reprogramming of mouse embryonic stem cells to trophoblast stem-like cells. Nucleic Acids Res. 2017, 45, 10103–10114. [Google Scholar] [CrossRef] [PubMed]
- Basak, T.; Ain, R. Molecular regulation of trophoblast stem cell self-renewal and giant cell differentiation by the Hippo components yap and lats1. Stem Cell Res. Ther. 2022, 13, 189. [Google Scholar] [CrossRef] [PubMed]
- Russ, A.P.; Wattler, S.; Colledge, W.H.; Aparicio, S.A.J.R.; Carlton, M.B.L.; Pearce, J.J.; Barton, S.C.; Surani, M.A.; Ryan, K.; Nehls, M.C.; et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 2000, 404, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Ying, Q.L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The ground state of embryonic stem cell self-renewal. Nature 2008, 453, 519–523. [Google Scholar] [CrossRef]
- Kastan, N.; Gnedeva, K.; Alisch, T.; Petelski, A.A.; Huggins, D.J.; Chiaravalli, J.; Aharanov, A.; Shakked, A.; Tzahor, E.; Nagiel, A.; et al. Small-molecule inhibition of Lats kinases may promote Yap-dependent proliferation in postmitotic mammalian tissues. Nat. Commun. 2021, 12, 3100. [Google Scholar] [CrossRef]
- Hayashi, Y.; Furue, M.K.; Tanaka, S.; Hirose, M.; Wakisaka, N.; Danno, H.; Ohnuma, K.; Oeda, S.; Aihara, Y.; Shiota, K.; et al. BMP4 induction of trophoblast from mouse embryonic stem cells in defined culture conditions on laminin. In Vitro Cell. Dev. An. 2010, 46, 416–430. [Google Scholar] [CrossRef]
- Ullah, Z.; Kohn, M.J.; Yagi, R.; Vassilev, L.T.; DePamphilis, M.L. Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes Dev. 2008, 22, 3024–3036. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, D.; Morimoto, H.; Yoshimura, K.; Kono, T.; Ogawa, H. The differentiation potency of trophoblast stem cells from mouse androgenetic embryos. Stem Cells Dev. 2019, 28, 290–302. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Li, W.; Lv, Z.; Liu, L.; Tong, M.; Hai, T.; Hao, J.; Guo, C.L.; Ma, Q.W.; Wang, L.; et al. iPS cells produce viable mice through tetraploid complementation. Nature 2009, 461, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Wang, J.L.; Zhang, Y.; Kou, Z.H.; Gao, S.R. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell 2009, 5, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Latos, P.A.; Hemberger, M. From the stem of the placental tree: Trophoblast stem cells and their progeny. Development 2016, 143, 3650–3660. [Google Scholar] [CrossRef] [PubMed]
- Posfai, E.; Schell, J.P.; Janiszewski, A.; Rovic, I.; Murray, A.; Bradshaw, B.; Yamakawa, T.; Pardon, T.; El Bakkali, M.; Talon, I.; et al. Evaluating totipotency using criteria of increasing stringency. Nat. Cell Biol. 2021, 23, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chang, L.; Wu, J.; Huang, J.; Guan, W.; Bates, L.E.; Stuart, H.T.; Guo, M.; Zhang, P.; Huang, B.; et al. In vitro generation of mouse morula-like cells. Dev. Cell 2023, 58, 2510–2527.e2517. [Google Scholar] [CrossRef]
- Yang, M.Z.; Yu, H.W.; Yu, X.; Liang, S.Q.; Hu, Y.L.; Luo, Y.X.; Izsvák, Z.; Sun, C.B.; Wang, J.C. Chemical-induced chromatin remodeling reprograms mouse ESCs to totipotent-like stem cells. Cell Stem Cell 2022, 29, 400–418.e13. [Google Scholar] [CrossRef]
- Shen, H.; Yang, M.; Li, S.Y.; Zhang, J.; Peng, B.; Wang, C.H.; Chang, Z.; Ong, J.N.; Du, P. Mouse totipotent stem cells captured and maintained through spliceosomal repression. Cell 2021, 184, 2843–2859.e20. [Google Scholar] [CrossRef]
- Sakashita, A.; Kitano, T.; Ishizu, H.; Guo, Y.; Masuda, H.; Ariura, M.; Murano, K.; Siomi, H. Transcription of MERVL retrotransposons is required for preimplantation embryo development. Nat. Genet. 2023, 55, 484–495. [Google Scholar] [CrossRef]
- Suzuki, D.; Okura, K.; Nagakura, S.; Ogawa, H. CDX2 downregulation in mouse mural trophectoderm during peri-implantation is heteronomous, dependent on the YAP-TEAD pathway and controlled by estrogen-induced factors. Rep. Med. Biol. 2022, 21, e12446. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Han, W.; Dong, R.; Wei, S.; Chen, L.; Gu, Z.; Liu, Y.; Guo, W.; Yan, F. Efficient Reprogramming of Mouse Embryonic Stem Cells into Trophoblast Stem-like Cells via Lats Kinase Inhibition. Biology 2024, 13, 71. https://doi.org/10.3390/biology13020071
Gao Y, Han W, Dong R, Wei S, Chen L, Gu Z, Liu Y, Guo W, Yan F. Efficient Reprogramming of Mouse Embryonic Stem Cells into Trophoblast Stem-like Cells via Lats Kinase Inhibition. Biology. 2024; 13(2):71. https://doi.org/10.3390/biology13020071
Chicago/Turabian StyleGao, Yake, Wenrui Han, Rui Dong, Shu Wei, Lu Chen, Zhaolei Gu, Yiming Liu, Wei Guo, and Fang Yan. 2024. "Efficient Reprogramming of Mouse Embryonic Stem Cells into Trophoblast Stem-like Cells via Lats Kinase Inhibition" Biology 13, no. 2: 71. https://doi.org/10.3390/biology13020071
APA StyleGao, Y., Han, W., Dong, R., Wei, S., Chen, L., Gu, Z., Liu, Y., Guo, W., & Yan, F. (2024). Efficient Reprogramming of Mouse Embryonic Stem Cells into Trophoblast Stem-like Cells via Lats Kinase Inhibition. Biology, 13(2), 71. https://doi.org/10.3390/biology13020071





