A Comprehensive Assessment of Nutritional Value, Antioxidant Potential, and Genetic Diversity in Metapenaeus ensis from Three Different Populations
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Sample Collection
2.3. General Nutrition Analysis
2.4. Physiological Analysis
2.5. Genetic Diversity
2.6. Data Analysis
3. Results
3.1. General Nutrition
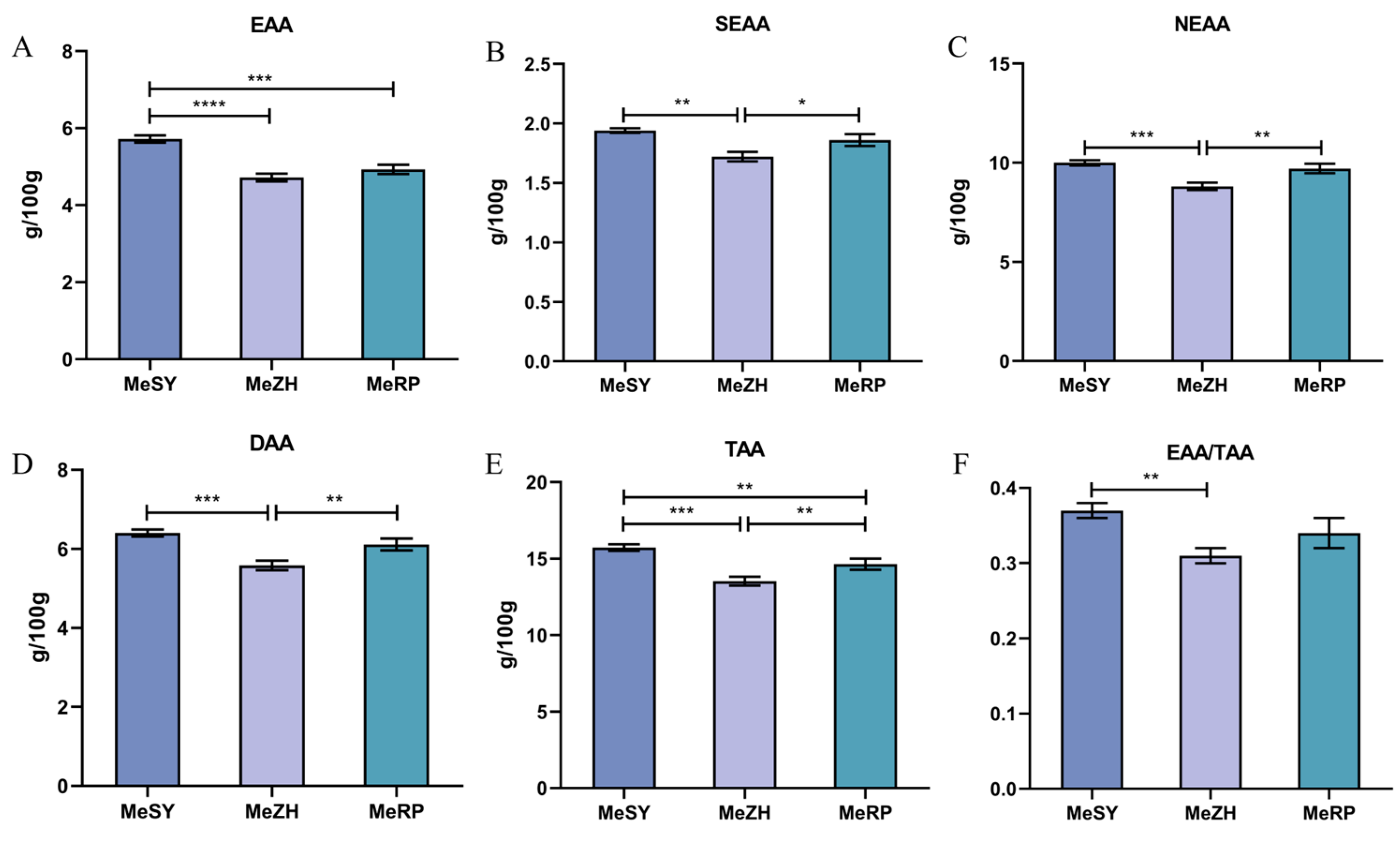
3.2. Amino Acids
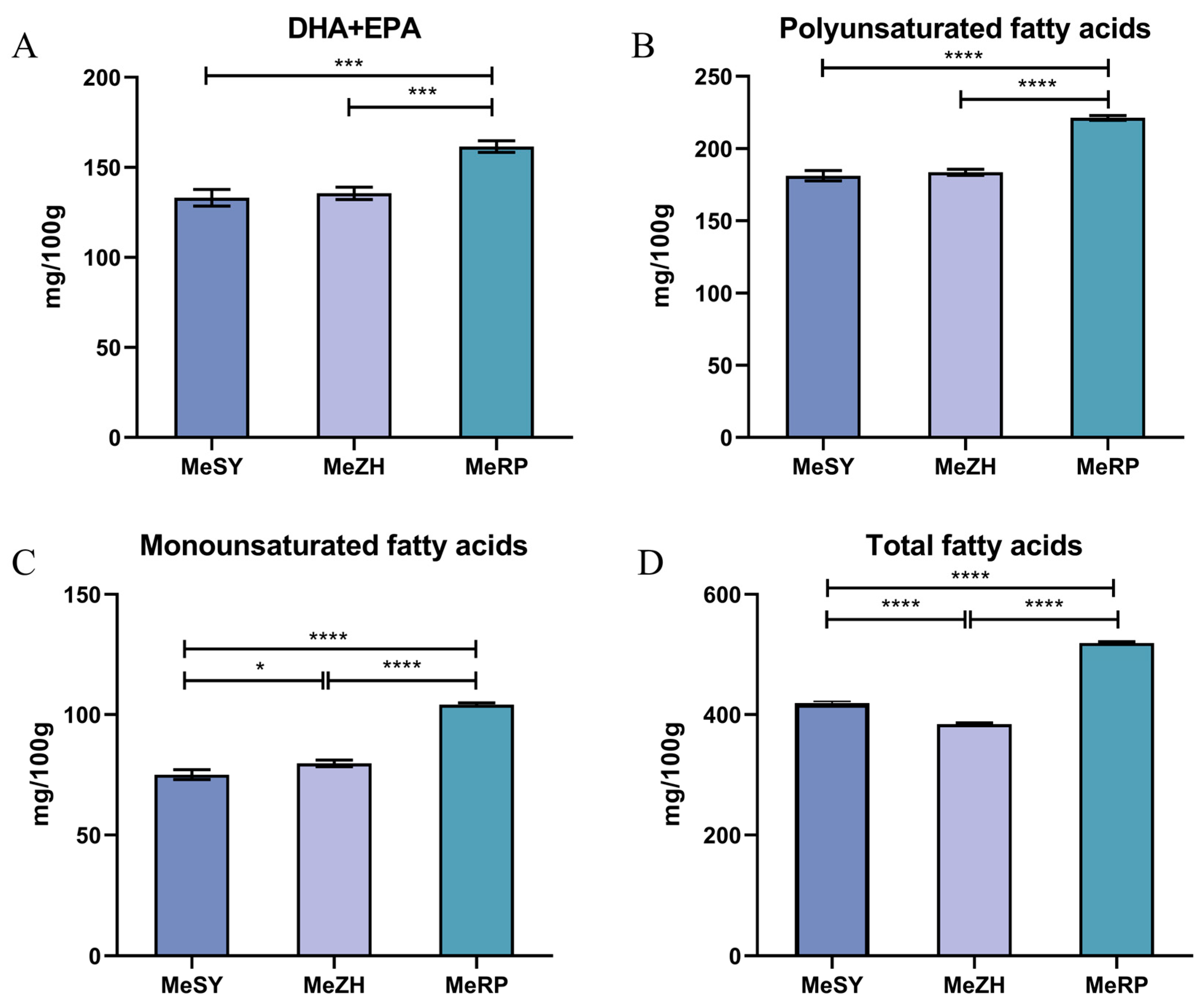
3.3. Fatty Acids
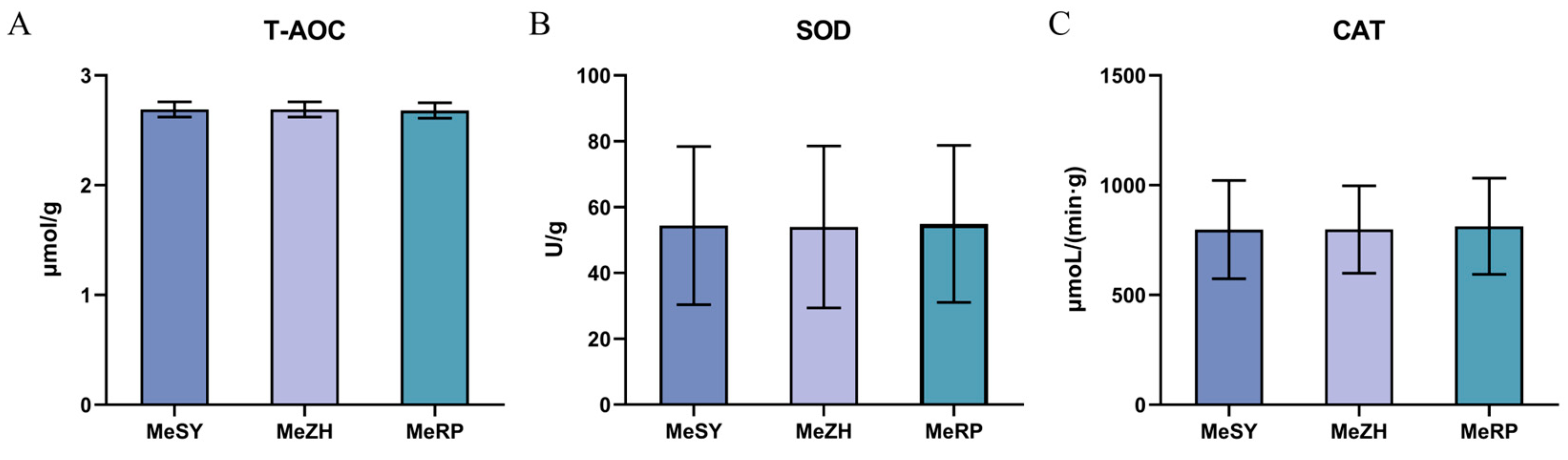
3.4. Oxidative Stress Indicators
3.5. Genetic Diversity
4. Discussion
4.1. Analysis of Basic Nutritional Components in Different Populations of M. ensis
4.2. Analysis of Amino Acid Content in Different Populations of M. ensis
4.3. Analysis of Fatty Acid Composition in Different Populations of M. ensis
4.4. Analysis of Antioxidant Capacity in Different Populations of M. ensis
4.5. Analysis of Genetic Diversity in Different Populations of M. ensis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holthuis, L.B. FAO Species Catalogue: Vol. 1. Shrimps and Prawns of the World. An Annotated Catalogue of Species of Interest to Fisheries; FAO: Rome, Italy, 1980. [Google Scholar]
- Chu, K.H.; Chen, Q.C.; Huang, L.M.; Wong, C.K. Morphometric Analysis of Commercially Important Penaeid Shrimps from the Zhujiang Estuary, China. Fish. Res. 1995, 23, 83–93. [Google Scholar] [CrossRef]
- Liu, L.; Liu, C.C.; Li, J.L. Comparison of Biochemical Composition and Nutritional Value of Antarctic Krill (Euphausia superb) with Several Species of Shrimps. Adv. Mater. Res. 2012, 361, 799–803. [Google Scholar] [CrossRef]
- Li, Y.; Chen, J.; Jiang, S.; Huang, J.; Jiang, S.; Yang, Q.; Yang, L.; Shi, J.; Zhou, F. A Comprehensive Study on Nutritional Quality, Physiological Enzyme Activity and Genetic Diversity in Six Populations of Penaeus monodon. Aquac. Int. 2024, 1–17. [Google Scholar] [CrossRef]
- Gonçalves, G.R.L.; Dos Santos, P.V.M.; Negreiros-Fransozo, M.L.; Castilho, A.L.; De Troch, M. Environmental Factors Modulated the Fatty Acid Profile of the Shrimp Xiphopenaeus Spp. in Cananéia and Ubatuba Southeast Brazilian Coast. Environ. Sci. Pollut. Res. 2023, 30, 76936–76949. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, G.R.L.; Denadai, A.C.; Sousa, A.N.; Castilho, A.L.; De Troch, M. Fatty Acid Profiles of Three Commercial Shrimp from Southeastern Brazil. Reg. Stud. Mar. Sci. 2021, 48, 102032. [Google Scholar] [CrossRef]
- Liao, M.; Liao, X.; Long, X.; Zhao, J.; He, Z.; Zhang, J.; Wu, T.; Sun, C. Host-Microbiota Interactions and Responses of Metapenaeus ensis Infected with Decapod Iridescent Virus 1. Front. Microbiol. 2023, 13, 1097931. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-D.; Si, M.-R.; Jiang, S.-G.; Yang, Q.-B.; Jiang, S.; Yang, L.-S.; Huang, J.-H.; Zhou, F.-L. First Transcriptome Profiling in Gill and Hepatopancrease Tissues of Metapenaeus ensis in Response to Acute Ammonia-N Stress. Fish Shellfish. Immunol. 2023, 139, 108926. [Google Scholar] [CrossRef]
- Ji, D.; Yan, M.; Hu, L.; Chen, C.; Zhang, M. Analysis of Nutritional Compositions in Muscle of Wild Sword Prawn Metapenaeus ensis with Different Sizes. Fish. Sci. 2022, 41, 1045–1051. [Google Scholar]
- Xiao, F.; Wang, J.; Liu, H.; Zhuang, M.; Wen, X.; Zhao, H.; Wu, K. Effects of Dietary Protein Levels on Growth, Digestive Enzyme Activity, Antioxidant Capacity, and Gene Expression Related to Muscle Growth and Protein Synthesis of Juvenile Greasyback Shrimp (Metapenaeus ensis). Animals 2023, 13, 3886. [Google Scholar] [CrossRef]
- Huy, N.X.; Ty, N.; Van Giang, T.; Phuong, T.V. A First Look at Genetic Diversity of Metapenaeus ensis Populations in Tam Giang–Cau Hai Lagoon, Vietnam. Isr. J. Aquac.-Bamidgeh 2024, 76, 158–167. [Google Scholar]
- Li, Y.; Zhou, F.; Yang, O.; Jiang, S.; Huang, J.; Yang, L.; Jiang, S. Complete Mitochondrial Genome of Metapenaeus affinis (H. Milne Edwards, 1837) and Metapenaeus ensis (De Haan, 1844). Isr. J. Aquac.-Bamidgeh 2022. [Google Scholar] [CrossRef]
- GB 5009.3–2016; Determination of Moisture in Food. National Standards of the People’s Republic of China: Beijing, China, 2016.
- GB 5009.5-2016; National standards for food safety-Determination of proteins in foods. National Standards of the People’s Republic of China: Beijing, China, 2016.
- GB 5009.6–2016; National Food Safety Standard-Determination of Crude Fat in Food. National Standards of the People’s Republic of China: Beijing, China, 2016.
- GB 5009.4–2016; National Food Safety Standard - Determination of Ash in Food. National Standards of the People’s Republic of China: Beijing, China, 2016.
- GB/T 15672–2009; Determination of Total Sugar Content in Edible Fungi. National Standards of the People’s Republic of China: Beijing, China, 2009.
- GB 5009.124-2016; National Food Safety Standard-Determination of Amino Acids in Foods by Hydrochloric Acid Hydrolysis. National Standards of the People’s Republic of China: Beijing, China, 2016.
- GB 5009.168-2016; National Food Safety Standard-Determination of Fatty Acids in Foods by Gas Chromatography. National Standards of the People’s Republic of China: Beijing, China, 2016.
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Freed, D.; Aldana, R.; Weber, J.A.; Edwards, J.S. The Sentieon Genomics Tools–A Fast and Accurate Solution to Variant Calling from next-Generation Sequence Data. BioRxiv 2017. [Google Scholar] [CrossRef]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Kendig, K.I.; Baheti, S.; Bockol, M.A.; Drucker, T.M.; Hart, S.N.; Heldenbrand, J.R.; Hernaez, M.; Hudson, M.E.; Kalmbach, M.T.; Klee, E.W. Sentieon DNASeq Variant Calling Workflow Demonstrates Strong Computational Performance and Accuracy. Front. Genet. 2019, 10, 736. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z. SOAPnuke: A MapReduce Acceleration-Supported Software for Integrated Quality Control and Preprocessing of High-Throughput Sequencing Data. Gigascience 2018, 7, gix120. [Google Scholar] [CrossRef]
- Wang, J. Comparision of Nutritional Composition in Muscle of Penaeus Chinensis, Penaeus Vannamei Boone and Penaeus Japonicuss Bate. Food Sci. Technol. 2013, 38, 145–150. [Google Scholar]
- Liu, Z.; Liu, Q.; Zhang, D.; Wei, S.; Sun, Q.; Xia, Q.; Shi, W.; Ji, H.; Liu, S. Comparison of the Proximate Composition and Nutritional Profile of Byproducts and Edible Parts of Five Species of Shrimp. Foods 2021, 10, 2603. [Google Scholar] [CrossRef]
- Xu, X.; Liu, H.; Yan, B. Nutritional Component Analysis and Quality Evaluation of Penaeus Japonicus. Food Sci. 2011, 32, 297–301. [Google Scholar]
- Li, X.; Wang, Y.; Li, H.; Jiang, X.; Ji, L.; Liu, T.; Sun, Y. Chemical and Quality Evaluation of Pacific White Shrimp Litopenaeus vannamei: Influence of Strains on Flesh Nutrition. Food Sci. Nutr. 2021, 9, 5352–5360. [Google Scholar] [CrossRef]
- Fafournoux, P.; Bruhat, A.; Jousse, C. Amino Acid Regulation of Gene Expression. Biochem. J. 2000, 351, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Church, D.D.; Hirsch, K.R.; Park, S.; Kim, I.-Y.; Gwin, J.A.; Pasiakos, S.M.; Wolfe, R.R.; Ferrando, A.A. Essential Amino Acids and Protein Synthesis: Insights into Maximizing the Muscle and Whole-Body Response to Feeding. Nutrients 2020, 12, 3717. [Google Scholar] [CrossRef]
- Shi, L.; Hao, G.; Chen, J.; Ma, S.; Weng, W. Nutritional Evaluation of Japanese Abalone (Haliotis Discus Hannai Ino) Muscle: Mineral Content, Amino Acid Profile and Protein Digestibility. Food Res. Int. 2020, 129, 108876. [Google Scholar] [CrossRef] [PubMed]
- Tan DeQing, T.D.; Wang JianWei, W.J.; Dan ShengGuo, D.S. The Ratio of Flesh to Body and Analysis on Nutritive Composition of Muscle in Ancherythroculter nigrocauda. Acta Hydrobiol. Sin. 2004, 28, 240–246. [Google Scholar]
- Yuan JuLin, Y.J.; Liu Mei, L.M.; Ni Meng, N.M.; Mi GuoQiang, M.G.; Zhang Chao, Z.C.; Gu ZhiMin, G.Z. Effects of Different Culture Models on Growth Performances, Morphological Traits and Nutritional Quality in Muscles of Micropterus salmoides. Acta Agric. Univ. Jiangxiensis 2018, 40, 1276–1285. [Google Scholar]
- Cao, X.; Xia, J.; Zhou, Y.; Wang, Y.; Xia, H.; Wang, S.; Liao, W.; Sun, G. The Effect of Mufa-Rich Food on Lipid Profile: A Meta-Analysis of Randomized and Controlled-Feeding Trials. Foods 2022, 11, 1982. [Google Scholar] [CrossRef]
- Cui, G.; Jiang, Z.; Wang, J.; Hu, Z.; Wei, W. Comparison of Nutritional Components of Macrobrachium rosenbergii under Two Cultivation Modes. Jiangsu Agric. Sci. 2018, 46, 212–214. [Google Scholar]
- Chen, J.; Liu, H. Nutritional Indices for Assessing Fatty Acids: A Mini-Review. Int. J. Mol. Sci. 2020, 21, 5695. [Google Scholar] [CrossRef]
- Araujo, P.; Truzzi, C.; Belghit, I.; Antonucci, M. The Impact of Seawater Warming on Fatty Acid Composition and Nutritional Quality Indices of Trematomus bernacchii from the Antarctic Region. Food Chem. 2021, 365, 130500. [Google Scholar] [CrossRef]
- Kang, Y.J.; Lang, M.Y.; Zhao, W. Advance in Antioxidant Enzymes and Its Effect Factors in Aquatic Organisms. J. Microbiol. 2013, 33, 75–80. [Google Scholar]
- Zhao, H.; Zhang, R.; Yan, X.; Fan, K. Superoxide dismutase nanozymes: An emerging star for anti-oxidation. J. Mater. Chem. B 2021, 9, 6939–6957. [Google Scholar] [CrossRef] [PubMed]
- Carmo De Carvalho E Martins, M.D.; Martins; Da Silva Santos Oliveira, A.S.; Da Silva, L.A.A.; Primo, M.G.S.; De Carvalho Lira, V.B. Biological Indicators of Oxidative Stress [Malondialdehyde, Catalase, Glutathione Peroxidase, and Superoxide Dismutase] and Their Application in Nutrition. In Biomarkers in Nutrition; Patel, V.B., Preedy, V.R., Eds.; Biomarkers in Disease: Methods, Discoveries and Applications; Springer International Publishing: Cham, Switzerland, 2022; pp. 1–25. ISBN 978-3-030-81304-8. [Google Scholar]
- Silvestrini, A.; Meucci, E.; Ricerca, B.M.; Mancini, A. Total Antioxidant Capacity: Biochemical Aspects and Clinical Significance. Int. J. Mol. Sci. 2023, 24, 10978. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.H.; Deng, X.Y.; Jiang, W.M.; He, F.L. Effects of High Level Chromium on Antioxidant Enzyme System in Gill and Hepatopancreas of Procambarus clarkii. J. Agro-Environ. Sci. 2007, 23, 24–34. [Google Scholar]
- Tang, Q.-Y.; Xie, J.-H.; Xia, Z.-L.; Cai, M.-Y.; Wu, Y.-M.; Bai, L.-H.; Du, H.-K.; Li, J.-F.; Yang, G.-L. Genetic Diversity of the Breeding Populations of Giant Freshwater Prawn Macrobrachium rosenbergii. Acta Hydrobiol. Sin. 2020, 44, 1097–1104. [Google Scholar]
- Matukumalli, L.K.; Lawley, C.T.; Schnabel, R.D.; Taylor, J.F.; Allan, M.F.; Heaton, M.P.; O’Connell, J.; Moore, S.S.; Smith, T.P.; Sonstegard, T.S. Development and Characterization of a High Density SNP Genotyping Assay for Cattle. PLoS ONE 2009, 4, e5350. [Google Scholar] [CrossRef]
- Luo, W.; Luo, C.; Wang, M.; Guo, L.; Chen, X.; Li, Z.; Zheng, M.; Folaniyi, B.S.; Luo, W.; Shu, D. Genome Diversity of Chinese Indigenous Chicken and the Selective Signatures in Chinese Gamecock Chicken. Sci. Rep. 2020, 10, 14532. [Google Scholar] [CrossRef]
- Holsinger, K.E.; Weir, B.S. Genetics in Geographically Structured Populations: Defining, Estimating and Interpreting F ST. Nat. Rev. Genet. 2009, 10, 639–650. [Google Scholar] [CrossRef]
| Population | Body Length (mm) | Weight (g) | Location |
|---|---|---|---|
| Sanya (MeSY) | 112.00 ± 12.00 | 29.60 ± 17.82 | 18°30′ N, 110°10′ E |
| Zhuhai (MeZH) | 124.00 ± 18.00 | 18.95 ± 13.75 | 22°14′ N, 113°36′ E |
| Raoping (MeRP) | 121.00 ± 18.00 | 19.20 ± 8.90 | 23°66′ N, 117°00′ E |
| Nutritional Components | MeSY | MeZH | MeRP |
|---|---|---|---|
| Ash (g/100 g) | 1.40 ± 0.10 a | 1.47 ± 0.07 a | 1.40 ± 0.00 a |
| Moisture (g/100 g) | 74.50 ± 0.90 a | 75.30 ± 1.20 a | 74.90 ± 0.50 a |
| Crude fat (g/100 g) | 0.97 ± 0.13 a | 0.83 ± 0.07 b | 1.03 ± 0.07 a |
| Crude protein (g/100 g) | 22.10 ± 0.80 a | 20.80 ± 0.60 a | 21.17 ± 0.17 a |
| Total sugar (%) | 0.34 ± 0.03 a | 0.36 ± 0.03 a | 0.29 ± 0.04 b |
| Amino Acid (g/100 g) | MeSY | MeZH | MeRP |
|---|---|---|---|
| Aspartic acid @ | 1.59 ± 0.02 a | 1.37 ± 0.03 b | 1.53 ± 0.01 a |
| Threonine * | 0.61 ± 0.0 a | 0.54 ± 0.02 b | 0.59 ± 0.01 a |
| Serine | 0.50 ± 0.0 a | 0.49 ± 0.01 a | 0.54 ± 0.01 a |
| Glutamic acid @ | 2.31 ± 0.03 a | 2.02 ± 0.04 b | 2.26 ± 0.01 a |
| Glycine @ | 1.56 ± 0.02 a | 1.35 ± 0.03 b | 1.42 ± 0.02 b |
| Alanine @ | 0.94 ± 0.02 a | 0.84 ± 0.02 b | 0.90 ± 0.01 a |
| Cystine | 0.15 ± 0.00 a | 0.13 ± 0.00 a | 0.09 ± 0.01 b |
| Valine * | 0.72 ± 0.01 a | 0.59 ± 0.01 b | 0.55 ± 0.01 b |
| Methionine * | 0.34 ± 0.01 a | 0.30 ± 0.01 a | 0.31 ± 0.03 a |
| Isoleucine * | 0.66 ± 0.01 a | 0.53 ± 0.01 b | 0.48 ± 0.01 b |
| Leucine * | 1.26 ± 0.02 a | 1.06 ± 0.02 b | 1.16 ± 0.02 c |
| Tyrosine | 0.33 ± 0.0 a | 0.34 ± 0.01 a | 0.44 ± 0.01 b |
| Phenylalanine * | 0.64 ± 0.00 a | 0.54 ± 0.01 b | 0.58 ± 0.02 b |
| Lysine * | 1.49 ± 0.02 a | 1.16 ± 0.02 b | 1.26 ± 0.02 c |
| Histidine & | 0.30 ± 0.00 a | 0.25 ± 0.01 a | 0.27 ± 0.01 a |
| Arginine & | 1.64 ± 0.02 a | 1.47 ± 0.03 b | 1.59 ± 0.04 a |
| Proline | 0.68 ± 0.00 a | 0.55 ± 0.01 b | 0.67 ± 0.01 a |
| Fatty Acid(mg/100 g) | MeSY | MeZH | MeRP |
|---|---|---|---|
| C16:0 | 68.40 ± 4.80 a | 62.00 ± 3.20 b | 89.00 ± 3.70 c |
| C16:1 | 16.40 ± 1.20 a | 14.50 ± 0.70 a | 23.90 ± 1.00 b |
| C17:0 | 13.30 ± 0.90 a | 12.30 ± 0.70 a | 16.80 ± 0.70 b |
| C18:0 | 79.30 ± 5.00 a | 75.40 ± 3.70 b | 94.10 ± 3.80 c |
| C18:1n9c | 45.40 ± 3.20 a | 53.00 ± 3.20 b | 63.50 ± 2.60 c |
| C18:2n6c | 6.00 ± 0.40 a | 5.20 ± 0.20 a | 8.70 ± 0.40 b |
| C20:2 | 4.40 ± 0.40 a | 5.45 ± 0.25 a | 4.65 ± 0.25 a |
| C22:0 | 9.55 ± 0.75 a | 9.60 ± 0.40 a | 11.05 ± 0.45 b |
| C20:4n6 | 37.60 ± 2.60 a | 37.35 ± 1.35 a | 46.30 ± 1.90 b |
| C22:1n9 | 13.95 ± 0.85 a | 25.75 ± 0.85 b | 13.85 ± 0.55 a |
| C20:5n3(EPA) | 68.60 ± 4.90 a | 69.50 ± 3.50 a | 82.40 ± 3.50 b |
| C22:6n3(DHA) | 64.55 ± 4.35 a | 66.10 ± 3.30 a | 79.15 ± 2.85 b |
| Population | MeSY | MeZH | MeRP |
|---|---|---|---|
| SNP density (SNP/Kb) | 5.39 | 2.79 | 2.62 |
| Nucleotide diversity (π) | 8.70 × 10−4 ± 1.16 × 10−3 | 5.34 × 10−4 ± 7.59 × 10−4 | 5.52 × 10 −4 ± 7.79 × 10−4 |
| Polymorphism information content (PIC) | 1.23 × 10−1 ± 8.44 × 10−2 | 0.14 ± 0.11 | 0.15 ± 0.11 |
| Observed heterozygosity (Ho) | 6.99 × 10−2 ± 1.30 × 10−1 | 0.14 ± 0.16 | 0.16 ± 0.17 |
| Inbreeding coefficient (FHOM) | 4.83 × 10−1 ± 6.09 × 10−2 | 8.31 × 10−2 ± 6.97 × 10 −2 | 5.93 × 10−2 ± 2.48 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Chen, J.; Jiang, S.; Yang, Q.; Yang, L.; Huang, J.; Shi, J.; Zhang, Y.; Lu, Z.; Zhou, F. A Comprehensive Assessment of Nutritional Value, Antioxidant Potential, and Genetic Diversity in Metapenaeus ensis from Three Different Populations. Biology 2024, 13, 838. https://doi.org/10.3390/biology13100838
Li Y, Chen J, Jiang S, Yang Q, Yang L, Huang J, Shi J, Zhang Y, Lu Z, Zhou F. A Comprehensive Assessment of Nutritional Value, Antioxidant Potential, and Genetic Diversity in Metapenaeus ensis from Three Different Populations. Biology. 2024; 13(10):838. https://doi.org/10.3390/biology13100838
Chicago/Turabian StyleLi, Yundong, Juan Chen, Song Jiang, Qibin Yang, Lishi Yang, Jianhua Huang, Jianzhi Shi, Yan Zhang, Zhibin Lu, and Falin Zhou. 2024. "A Comprehensive Assessment of Nutritional Value, Antioxidant Potential, and Genetic Diversity in Metapenaeus ensis from Three Different Populations" Biology 13, no. 10: 838. https://doi.org/10.3390/biology13100838
APA StyleLi, Y., Chen, J., Jiang, S., Yang, Q., Yang, L., Huang, J., Shi, J., Zhang, Y., Lu, Z., & Zhou, F. (2024). A Comprehensive Assessment of Nutritional Value, Antioxidant Potential, and Genetic Diversity in Metapenaeus ensis from Three Different Populations. Biology, 13(10), 838. https://doi.org/10.3390/biology13100838






