Research Progress on Viruses of Passiflora edulis
Simple Summary
Abstract
1. Introduction
2. Classification of Passion Fruit Viruses
2.1. Potyviruses Infecting Passion Fruit
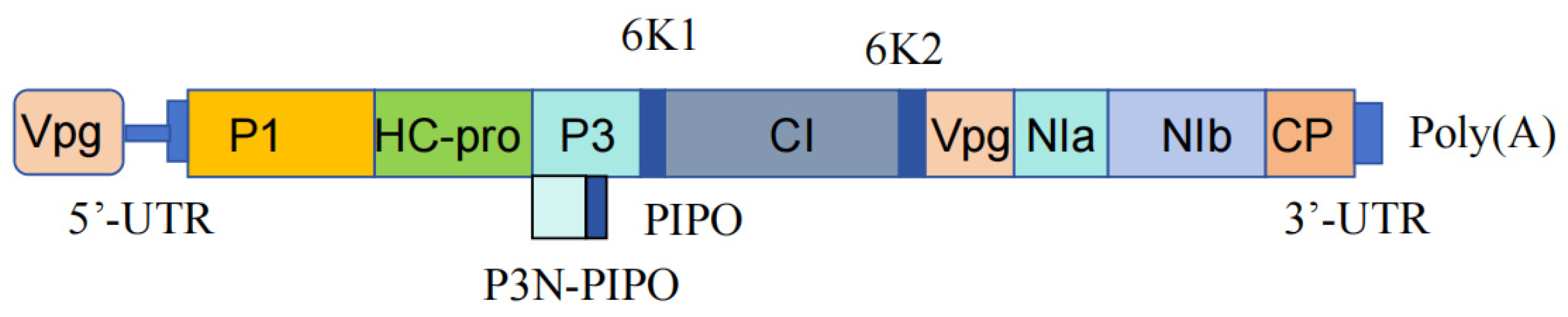
2.1.1. Proteins Encoded by Potyviruses
2.1.2. Potyvirus Species Infecting Passion Fruit
2.2. Begomoviruses Infecting Passion Fruit
2.2.1. Begomovirus Genome and Encoded Proteins
2.2.2. Begomoviruses Infecting Passion Fruit
2.3. Viruses of Other Genera Infecting Passion Fruit
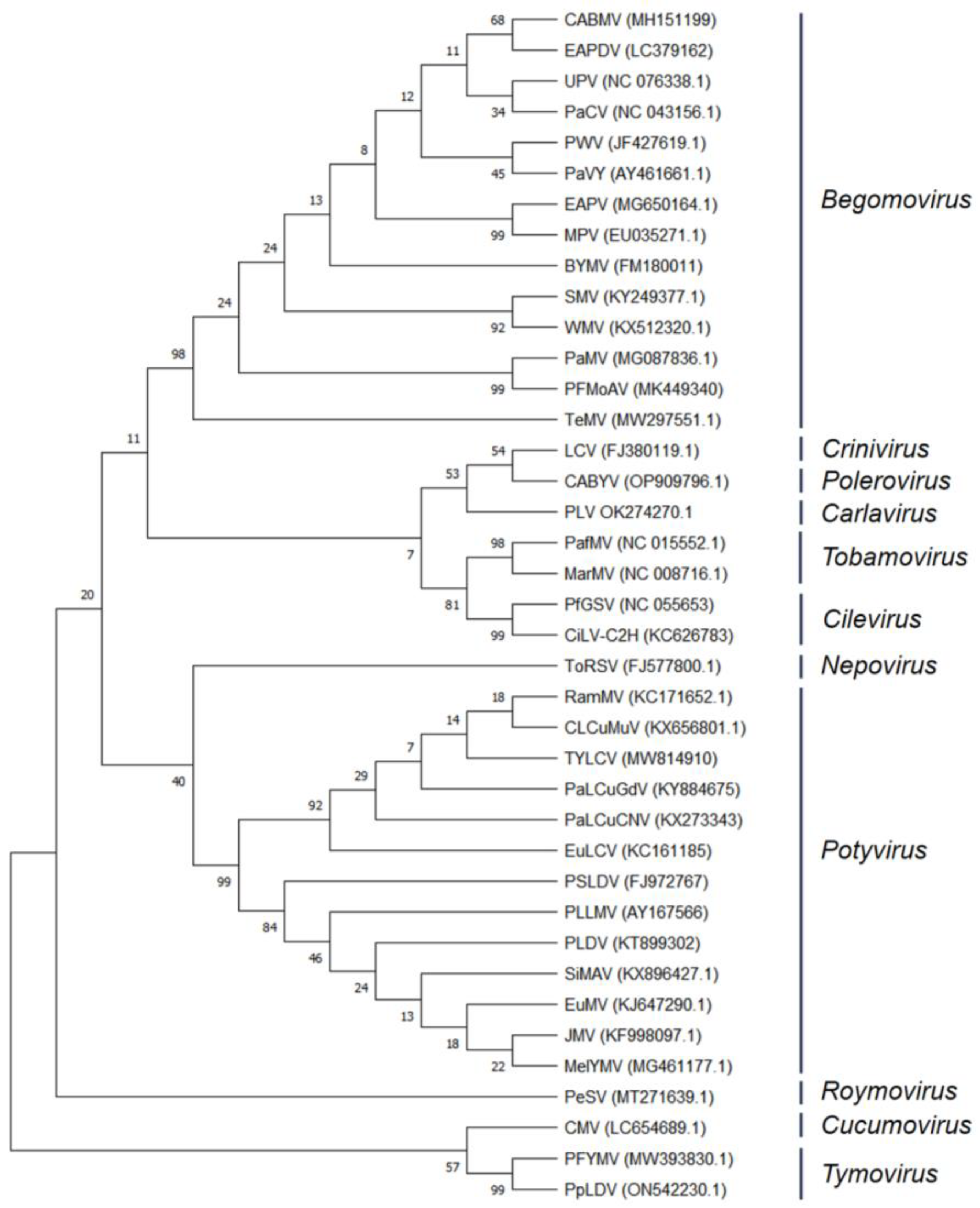
2.4. Evolutionary Relationships of Taxa
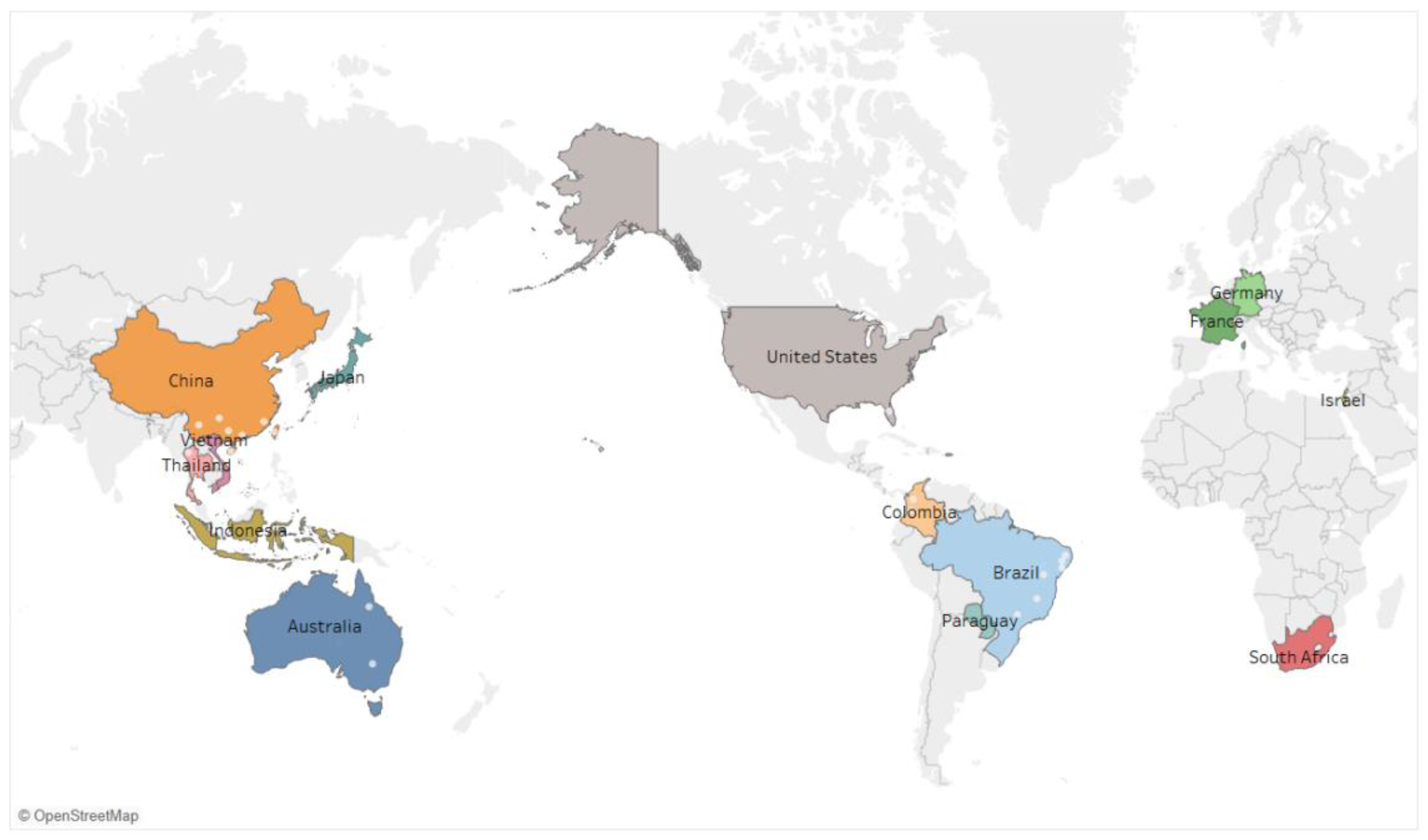
2.5. Viruses Infecting Passion Fruit Around the World
3. Modes of Transmission of Passion Fruit Viral Diseases
4. Biological Characteristics of Viral Diseases Affecting Passion Fruit
5. Plant Immune Mechanisms and the Mechanisms of Disease Symptom Development
6. Methods of Controlling Viral Diseases in Passion Fruit
6.1. Selecting Disease-Resistant Varieties
6.2. Agrobacterium-Mediated Genetic Transformation
6.2.1. Against Viruses
6.2.2. Against Vector Insects
6.3. RNAi in Antiviral Applications
6.4. Physical Control
6.5. Chemical Control of Viruses
6.6. Mild Strain Cross-Protection
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Santos, E.A.; Souza, M.M.; Abreu, P.P.; Da Conceição, L.D.H.C.S.; Araújo, I.S.; Viana, A.P.; De Almeida, A.-A.F.; Freitas, J.C.D.O. Confirmation and characterization of interspecific hybrids of Passiflora L. (Passifloraceae) for ornamental use. Euphytica 2012, 184, 389. [Google Scholar] [CrossRef]
- Xia, Z.; Huang, D.; Zhang, S.; Wang, W.; Ma, F.; Wu, B.; Xu, Y.; Xu, B.; Chen, D.; Zou, M.; et al. Chromosome-scale genome assembly provides insights into the evolution and flavor synthesis of passion fruit (Passiflora edulis Sims). Hortic. Res. 2021, 8, 14. [Google Scholar] [CrossRef]
- Song, S.; Zhang, D.; Ma, F.; Xing, W.; Huang, D.; Wu, B.; Chen, J.; Chen, D.; Xu, B.; Xu, Y. Genome-Wide Identification and Expression Analyses of the Aquaporin Gene Family in Passion Fruit (Passiflora edulis), Revealing PeTIP3-2 to Be Involved in Drought Stress. Int. J. Mol. Sci. 2022, 23, 5720. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.L.; Jesus, O.N.D.; Oliveira, G.A.F.; Oliveira, E.J.D. Effect of selection on genetic variability in yellow passion fruit. Crop Breed. Appl. Biotechnol. 2012, 12, 253–260. [Google Scholar] [CrossRef]
- Li, W.; Li, C.J.; Zhang, S.F.; Huang, B. Research progress on the nutritional quality and functional substances of passionflower and its application. J. China Agric. Univ. 2022, 27, 79–92. [Google Scholar] [CrossRef]
- Shi, B.B.; Yuan, Q.F.; Li, S.P. Research Progress on Nutrition and Functional Components of Passiflora coerulea. Guizhou Agric. Sci. 2019, 47, 95–98. [Google Scholar]
- Fonseca, A.M.A.; Geraldi, M.R.; Junior, M.R.M.; Silvestre, A.J.D.; Rocha, S.M. Purple passion fruit (Passiflora edulis f. edulis): A comprehensive review on the nutritional value, phytochemical profile and associated health effects. Food Res. Int. 2022, 160, 111665. [Google Scholar] [PubMed]
- Kim, M.; Lim, H.S.; Lee, H.H.; Kim, T.H. Role Identification of Passiflora Incarnata Linnaeus: A Mini Review. J. Menopausal Med. 2017, 23, 156–159. [Google Scholar] [CrossRef]
- Chen, G.L. Production status and cultivation management of passionflower in Taiwan. Fujian Fruit 1991, 4, 26–27. [Google Scholar]
- Zhang, C.; Jiang, J.; Chen, S.; Wang, F. Telosma mosaic virus: An emerging plant RNA virus causing production loss in passion fruit across Asia. Plant Pathol. 2023, 73. [Google Scholar] [CrossRef]
- Fischer, I.H.; Rezende, J.A. Diseases of passion flower (Passiflora spp.). Pest Technol. 2008, 2, 1–19. [Google Scholar]
- Luo, J.S.; Zhou, Z.E.; Wang, L.S.; Chen, Z.D.; Wu, Z.J.; Lin, R.Y. Occurrence and Prevention and Control Suggestion of Virus Diseases of Passion Fruit. Southeast Hortic. 2019, 7, 36–40. [Google Scholar]
- Fukumoto, T.; Nakamura, M.; Wylie, S.J.; Chiaki, Y.; Iwai, H. Complete nucleotide sequence of a new isolate of passion fruit woodiness virus from Western Australia. Arch. Virol. 2013, 158, 1821–1824. [Google Scholar] [CrossRef]
- Barros, D.R.; Alfenas-Zerbini, P.; Beserra, J.E., Jr.; Antunes, T.F.; Zerbini, F.M. Comparative analysis of the genomes of two isolates of cowpea aphid-borne mosaic virus (CABMV) obtained from different hosts. Arch. Virol. 2011, 156, 1085–1091. [Google Scholar] [CrossRef]
- Yan, J.W.; Yuan, Q.F.; Peng, Z.J.; Wang, L.J.; Xie, P.; Chen, N.; Ma, Y.H. Research Progress on Viruses of Passionfruit. Chin. J. Trop. Agric. 2018, 38, 85–94. [Google Scholar]
- De Wijs, J.J. A virus causing ringspot of Passiflora edulis in the Ivory Coast. Ann. Appl. Biol. 1974, 77, 33–40. [Google Scholar] [CrossRef]
- Song, R.N.; Hou, Y.X.; Cui, M.D.; Feng, Y.; Han, Q.X.; Rao, X.Q. Detection of virus infecting passion fruit in Guangdong. J. Zhongkai Univ. Agric. Eng. 2020, 33, 11–15. [Google Scholar] [CrossRef]
- Camelo-Garcia, V.M.; Esquivel-Fariña, A.; Ferro, C.G.; Kitajima, E.W.; Rezende, J.A.M. Strongylodon macrobotrys: New host of soybean mosaic virus in Brazil. Plant Dis. 2021, 105, 1573. [Google Scholar] [CrossRef]
- Chen, B.; Wu, D.; Zheng, H.; Li, G.; Cao, Y.; Chen, J.; Yan, F.; Song, X.; Lin, L. Complete genome sequence of Passiflora virus Y infecting passion fruit in China. Arch. Virol. 2021, 166, 1489–1493. [Google Scholar] [CrossRef]
- Iwai, H.; Terahara, R.; Yamashita, Y.; Ueda, S.; Nakamura, M. Complete nucleotide sequence of the genomic RNA of an Amami-O-shima strain of East Asian Passiflora potyvirus. Arch. Virol. 2006, 151, 1457–1460. [Google Scholar] [CrossRef]
- Mbeyagala, E.K.; Maina, S.; Macharia, M.W.; Mukasa, S.B.; Holton, T. Illumina sequencing reveals the first near-complete genome sequence of Ugandan Passiflora virus. Microbiol. Resour. Announc. 2019, 8, e00358-19. [Google Scholar] [CrossRef]
- Kehoe, M.A.; Coutts, B.A.; Buirchell, B.J.; Jones, R.A. Plant virology and next generation sequencing: Experiences with a Potyvirus. PLoS ONE 2014, 9, e104580. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Yan, H.; Song, L.; Jin, P.; Miao, W.; Cui, H. Analysis of the complete genome sequence of a potyvirus from passion fruit suggests its taxonomic classification as a member of a new species. Arch. Virol. 2018, 163, 2583–2586. [Google Scholar] [CrossRef]
- Fresnillo, P.; Jover-Gil, S.; Samach, A.; Candela, H. Complete genome sequence of an isolate of Passiflora chlorosis virus from passion fruit (Passiflora edulis Sims). Plants 2022, 11, 1838. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Gao, F.; Zheng, S.; Zhang, X.; Zhang, L.; Li, T. Molecular characterization of a new potyvirus infecting passion fruit. Arch. Virol. 2019, 164, 1903–1906. [Google Scholar] [CrossRef]
- Do, D.H.; Chong, Y.H.; Ha, V.C.; Cheng, H.W.; Chen, Y.K.; Bui, T.N.; Nguyen, T.B.; Yeh, S.D. Characterization and Detection of Passiflora Mottle Virus and Two Other Potyviruses Causing Passionfruit Woodiness Disease in Vietnam. Phytopathology 2021, 111, 1675–1685. [Google Scholar] [CrossRef] [PubMed]
- Riska, Y.; Inudo, K.; Nakamura, M.; Fukumoto, T.; Takushi, T.; Fuji, S.-I.; Iwai, H. East Asian Passiflora distortion virus: A novel potyvirus species causing deformation of passion fruits in Japan. J. Gen. Plant Pathol. 2019, 85, 221–231. [Google Scholar] [CrossRef]
- Novaes, Q.S.; Freitas-Astua, J.; Yuki, V.A.; Kitajima, E.W.; Camargo, L.E.A.; Rezende, J.A.M. Partial characterization of a bipartite begomovirus infecting yellow passion flower in Brazil. Plant Pathol. 2003, 52, 648–654. [Google Scholar] [CrossRef]
- Vaca-Vaca, J.C.; Carrasco-Lozano, E.C.; López-López, K. Molecular identification of a new begomovirus infecting yellow passion fruit (Passiflora edulis) in Colombia. Arch. Virol. 2017, 162, 573–576. [Google Scholar] [CrossRef]
- Polston, J.E.; Londoño, M.A.; Cohen, A.L.; Padilla-Rodriguez, M.; Rosario, K.; Breitbart, M. Genome sequence of Euphorbia mosaic virus from passionfruit and Euphorbia heterophylla in Florida. Genome Announc. 2017, 5, e01714-16. [Google Scholar] [CrossRef]
- Cheng, Y.H.; Deng, T.C.; Chen, C.C.; Chiang, C.H.; Chang, C.A. First report of Euphorbia leaf curl virus and Papaya leaf curl Guangdong virus on Passion Fruit in Taiwan. Plant Dis. 2014, 98, 1746. [Google Scholar] [CrossRef]
- Chen, L.J.; Sun, D.L.; Lu, Y.L.; An, Y.X. First report of ramie mosaic virus on passion fruit in Guangdong, southern China. J. Plant Pathol. 2020, 102, 1305. [Google Scholar] [CrossRef]
- Li, J.Y. Pathogenicity Analysis of Papaya Leaf Curl Guangdong Virus and Euphorbia Leaf Curl Virus. Master’s Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, 2022. [Google Scholar] [CrossRef]
- Ferreira, S.S.; Barros, D.R.; De Almeida, M.R.; Zerbini, F.M. Characterization of Passionfruit severe leaf distortion virus, a novel begomovirus infecting passionfruit in Brazil, reveals a close relationship with tomato-infecting begomoviruses. Plant Pathol. 2010, 59, 221–230. [Google Scholar] [CrossRef]
- Fontenele, R.S.; Abreu, R.A.; Lamas, N.S.; Alves-Freitas, D.M.T.; Vidal, A.H.; Poppiel, R.R.; Melo, F.L.; Lacorte, C.; Martin, D.P.; Campos, M.A.; et al. Passion fruit chlorotic mottle virus: Molecular characterization of a new divergent geminivirus in Brazil. Viruses 2018, 10, 169. [Google Scholar] [CrossRef] [PubMed]
- Spadotti, D.M.A.; Bello, V.H.; Favara, G.M.; Stangarlin, O.S.; Krause-Sakate, R.; Rezende, A.M. Passiflora edulis: New natural host of Melochia yellow mosaic virus in Brazil. Australas. Plant Dis. 2019, 14, 23. [Google Scholar] [CrossRef]
- Tang, Y.; He, Z.; Zhou, G. Passiflora edulis is a new host of Cotton leaf curl Multan virus-betasatellite complex in China. Can. J. Plant Pathol. 2020, 42, 493–498. [Google Scholar] [CrossRef]
- Mituti, T.; Spadotti, D.M.A.; Narita, N.; Rezende, J.A.M. First report of Sida mottle Alagoas virus infecting Passiflora edulis in Brazil. Plant Dis. 2019, 103, 169. [Google Scholar] [CrossRef]
- Chen, L.; Sun, D.; Zhang, X.; Shao, D.; Lu, Y.; An, Y. Transcriptome analysis of yellow passion fruit in response to cucumber mosaic virus infection. PLoS ONE 2021, 16, e0247127. [Google Scholar] [CrossRef]
- Song, Y.S.; Ryu, K.H. The complete genome sequence and genome structure of passion fruit mosaic virus. Arch. Virol. 2011, 156, 1093–1095. [Google Scholar] [CrossRef]
- Song, Y.S.; Min, B.E.; Hong, J.S.; Rhie, M.J.; Kim, M.J.; Ryu, K.H. Molecular evidence supporting the confirmation of maracuja mosaic virus as a species of the genus Tobamovirus and production of an infectious cDNA transcript. Arch. Virol. 2006, 151, 2337–2348. [Google Scholar] [CrossRef]
- Kitajima, E.W.; Rezende, J.A.; Rodrigues, J.C. Passion fruit green spot virus vectored by Brevipalpus phoenicis (Acari: Tenuipalpidae) on passion fruit in Brazil. Exp. Appl. Acarol. 2003, 30, 225–231. [Google Scholar] [CrossRef]
- Olmedo Velarde, A.; Roy, A.; Larrea-Sarmiento, A.; Wang, X.; Padmanabhan, C.; Nunziata, S.; Nakhla, M.K.; Hu, J.; Melzer, M. First report of the hibiscus strain of citrus leprosis virus C2 infecting passionfruit (Passiflora edulis). Plant Dis. 2022, 106, 2539. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, S.; Zeidan, M.; Sobolev, I.; Beckelman, Y.; Holdengreber, V.; Tam, Y.; Bar Joseph, M.; Lipsker, Z.; Gera, A. The complete nucleotide sequence of Passiflora latent virus and its phylogenetic relationship to other carlaviruses. Arch. Virol. 2007, 152, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.H.; Lacorte, C.; Sanches, M.M.; Alves-Freitas, D.M.T.; Abreu, E.F.M.; Pinheiro-Lima, B.; Rosa, R.C.C.; Jesus, O.N.; Campos, M.A.; Felix, G.P.; et al. Occurrence of lettuce chlorosis virus in Passiflora spp. in Brazil. J. Plant Pathol. 2021, 103, 443–447. [Google Scholar] [CrossRef]
- Crestani, O.A.; Kitajima, E.W.; Lin, M.T.; Marinho, V.L. Passion fruit yellow mosaic virus, a new tymovirus found in Brazil. Phytopathology 1986, 76, 951–955. [Google Scholar] [CrossRef]
- Cardona, D.; Restrepo, A.; Higuita, M.; Gallo, Y.; Marin, M.; Gutiérrez, P. Natural infection of purple passion fruit (Passiflora edulis f. edulis) by a novel member of the family Tymoviridae in Colombia. Acta Virol. 2022, 66, 254–262. [Google Scholar] [CrossRef]
- Pares, R.D.; Martin, A.B.; Morrison, W. Rhabdovirus-like particles in passion fruit. Australas. Plant Pathol. 1983, 12, 51–52. [Google Scholar] [CrossRef]
- Zhang, S.; Huang, A.; Zhou, X.; Li, Z.; Dietzgen, R.G.; Zhou, C.; Cao, M. Natural Defect of a Plant Rhabdovirus Glycoprotein Gene: A Case Study of Virus-Plant Coevolution. Phytopathology 2021, 111, 227–236. [Google Scholar] [CrossRef]
- Vidal, A.H.; Lacorte, C.; Sanches, M.M.; Alves-Freitas, D.M.T.; Abreu, E.F.M.; Pinheiro-Lima, B.; Rosa, R.C.C.; Jesus, O.N.; Campos, M.A.; Felix, G.P.; et al. Characterization of Cucurbit Aphid-Borne Yellows Virus (CABYV) from Passion Fruit in Brazil: Evidence of a Complex of Species within CABYV Isolates. Viruses 2023, 15, 410. [Google Scholar] [CrossRef] [PubMed]
- Jover-Gil, S.; Beeri, A.; Fresnillo, P.; Samach, A.; Candela, H. Complete genome sequence of a novel virus, classifiable within the Potyviridae family, which infects passion fruit (Passiflora edulis). Arch. Virol. 2018, 163, 3191–3194. [Google Scholar] [CrossRef]
- Gu, P.P. Biological Characteristics of TeMV and Omics Analysis of Passiflora edulis Infected by TeMV. Master’s Thesis, Gannan Normal University, Ganzhou, China, 2023. Available online: https://d.wanfangdata.com.cn/thesis/D03261157 (accessed on 1 March 2024).
- Adams, M.J.; Antoniw, J.F.; Fauquet, C.M. Molecular criteria for genus and species discrimination within the family Potyviridae. Arch. Virol. 2005, 150, 459–479. [Google Scholar] [CrossRef]
- Adams, M.J.; Antoniw, J.F.; Beaudoin, F. Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol. Plant Pathol. 2005, 6, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Gao, F.; Shen, J.; Zhang, X.; Zheng, S.; Zhang, L.; Li, T. Molecular characterization of two recombinant isolates of telosma mosaic virus infecting Passiflora edulis from Fujian Province in China. PeerJ 2020, 8, e8576. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Zhang, X.; Zheng, S.; Zhang, L.; Li, T. Molecular Identification and Specific Detection of Telosma mosaic virus Infecting Passion Fruit. Scientia Agricultura Sinica. 2017, 50, 4725–4734. [Google Scholar]
- Rohozková, J.; Navrátil, M. P1 peptidase—A mysterious protein of family Potyviridae. J. Biosci. 2011, 36, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Hajimorad, M.R.; Domier, L.L.; Tolin, S.A.; Whitham, S.A.; Saghai Maroof, M.A. Soybean mosaic virus: A successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 2018, 19, 1563–1579. [Google Scholar] [CrossRef]
- Fukuzawa, N.; Itchoda, N.; Ishihara, T.; Goto, K.; Masuta, C.; Matsumura, T. HC-Pro, a potyvirus RNA silencing suppressor, cancels cycling of Cucumber mosaic virus in Nicotiana benthamiana plants. Virus Genes 2010, 40, 440–446. [Google Scholar] [CrossRef]
- Tu, Y.; Jin, Y.; Ma, D.; Li, H.; Zhang, Z.; Dong, J.; Wang, T. Interaction between PVY HC-Pro and the NtCF1β-subunit reduces the amount of chloroplast ATP synthase in virus-infected tobacco. Sci. Rep. 2015, 5, 15605. [Google Scholar] [CrossRef]
- Deng, P.; Wu, Z.; Wang, A. The multifunctional protein CI of potyviruses plays interlinked and distinct roles in viral genome replication and intercellular movement. Virol. J. 2015, 12, 141. [Google Scholar] [CrossRef]
- Geng, C.; Cong, Q.Q.; Li, X.D.; Mou, A.L.; Gao, R.; Liu, J.L.; Tian, Y.P. Developmentally regulated plasma membrane protein of Nicotiana benthamiana contributes to potyvirus movement and transports to plasmodesmata via the early secretory pathway and the actomyosin system. Plant Physiol. 2015, 167, 394–410. [Google Scholar] [CrossRef]
- Luan, H.; Shine, M.B.; Cui, X.; Chen, X.; Ma, N.; Kachroo, P.; Zhi, H.; Kachroo, A. The Potyviral P3 Protein Targets Eukaryotic Elongation Factor 1A to Promote the Unfolded Protein Response and Viral Pathogenesis. Plant Physiol. 2016, 172, 221–234. [Google Scholar] [CrossRef]
- Zhang, L.; Ren, T.L.; Liu, Y.; Sun, P.P.; Ma, J.; Narisu, L.I.Z.N. Research progress of melon virus. China Fruits 2023, 4, 24–30. [Google Scholar] [CrossRef]
- Bera, S.; Arena, G.D.; Ray, S.; Flannigan, S.; Casteel, C.L. The Potyviral Protein 6K1 Reduces Plant Proteases Activity during Turnip mosaic virus Infection. Viruses 2022, 14, 1341. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.; Jean-François, L. The genome-linked protein VPg of plant viruses—a protein with many partners. Curr. Opin. Virol. 2011, 1, 347–354. [Google Scholar]
- Sabharwal, P.; Srinivas, S.; Savithri, H.S. Mapping the domain of interaction of PVBV VPg with NIa-Pro: Role of N-terminal disordered region of VPg in the modulation of structure and function. Virology 2018, 524, 18–31. [Google Scholar] [CrossRef]
- Léonard, S.; Plante, D.; Wittmann, S.; Daigneault, N.; Fortin, M.G.; Laliberté, J.F. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 2000, 74, 7730–7737. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Shi, Y.; Dai, Z.; Wang, A. The RNA-Dependent RNA Polymerase NIb of Potyviruses Plays Multifunctional, Contrasting Roles during Viral Infection. Viruses 2020, 12, 77. [Google Scholar] [CrossRef]
- Liu, J.; Wu, X.; Fang, Y.; Liu, Y.; Bello, E.O.; Li, Y.; Xiong, R.; Li, Y.; Fu, Z.Q.; Wang, A.; et al. A plant RNA virus inhibits NPR1 sumoylation and subverts NPR1-mediated plant immunity. Nat. Commun. 2023, 14, 3580. [Google Scholar] [CrossRef]
- Xiao, H.; Lord, E.; Sanfaçon, H. Proteolytic Processing of Plant Proteins by Potyvirus NIa Proteases. J. Virol. 2022, 96, e0144421. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Turiño, S.; García, J.A. Potyviral coat protein and genomic RNA: A striking partnership leading virion assembly and more. Adv. Virus Res. 2020, 108, 165–211. [Google Scholar] [CrossRef]
- Parrella, G.; Lanave, C. Identification of a new pathotype of Bean yellow mosaic virus (BYMV) infecting blue passion flower and some evolutionary characteristics of BYMV. Arch. Virol. 2009, 154, 1689–1694. [Google Scholar] [CrossRef]
- Kumar, R.; Dasgupta, I. Geminiviral C4/AC4 proteins: An emerging component of the viral arsenal against plant defence. Virology 2023, 579, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Mubin, M.; Ijaz, S.; Nahid, N.; Hassan, M.; Younus, A.; Qazi, J.; Nawaz-Ul-Rehman, M.S. Journey of begomovirus betasatellite molecules: From satellites to indispensable partners. Virus Genes 2020, 56, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.R.; Hagen, C.; Lucas, W.J.; Gilbertson, R.L. Exploiting chinks in the plant’s armor: Evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 2005, 43, 361–394. [Google Scholar] [CrossRef]
- Breves, S.S.; Silva, F.A.; Euclydes, N.C.; Saia, T.F.F.; Jean-Baptiste, J.; Andrade Neto, E.R.; Fontes, E.P.B. Begomovirus-Host Interactions: Viral Proteins Orchestrating Intra and Intercellular Transport of Viral DNA While Suppressing Host Defense Mechanisms. Viruses 2023, 15, 1593. [Google Scholar] [CrossRef]
- Gong, P.; Tan, H.; Zhao, S.; Li, H.; Liu, H.; Ma, Y.; Zhang, X.; Rong, J.; Fu, X.; Lozano-Durán, R.; et al. Geminiviruses encode additional small proteins with specific subcellular localizations and virulence function. Nat. Commun. 2021, 12, 4278. [Google Scholar] [CrossRef] [PubMed]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef]
- Lozano-Durán, R.; Rosas-Díaz, T.; Gusmaroli, G.; Luna, A.P.; Taconnat, L.; Deng, X.W.; Bejarano, E.R. Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell 2011, 23, 1014–1032. [Google Scholar] [CrossRef]
- Dai, K.W.; Tsai, Y.T.; Wu, C.Y.; Lai, Y.C.; Lin, N.S.; Hu, C.C. Identification of Crucial Amino Acids in begomovirus C4 Proteins Involved in the Modulation of the Severity of Leaf Curling Symptoms. Viruses 2022, 14, 499. [Google Scholar] [CrossRef]
- Li, P.; Su, F.; Meng, Q.; Yu, H.; Wu, G.; Li, M.; Qing, L. The C5 protein encoded by Ageratum leaf curl Sichuan virus is a virulence factor and contributes to the virus infection. Mol. Plant Pathol. 2021, 22, 1149–1158. [Google Scholar] [CrossRef]
- Wu, H.; Liu, M.; Kang, B.; Liu, L.; Hong, N.; Peng, B.; Gu, Q. AC5 protein encoded by squash leaf curl China virus is an RNA silencing suppressor and a virulence determinant. Front. Microbiol. 2022, 13, 980147. [Google Scholar] [CrossRef]
- Li, F.; Xu, X.; Huang, C.; Gu, Z.; Cao, L.; Hu, T.; Ding, M.; Li, Z.; Zhou, X. The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol. 2015, 208, 555–569. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olivé, E.; Navas-Castillo, J. The role of extensive recombination in the evolution of geminiviruses. Curr. Top. Microbiol. Immunol. 2023, 439, 139–166. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Yang, T.Z.; Cai, R.P.; Lin, X.M.; Deng, N.K.; Che, H.Y.; Lin, Y.T.; Kong, X.Y. Molecular Detection and Identification of Viruses from Passiflora edulis in Hainan. Acta Hortic. Sin. 2022, 49, 1785–1794. [Google Scholar] [CrossRef]
- Qiu, H.Y. Study of the Interaction between the Coat Protein of Cucumber Mosaic Virus and Host Factors. Ph.D. Dissertation, China Agricultural University, Beijing, China, 2015. [Google Scholar]
- Wang, R.L. The Subcellular Distribution of Cucumber Mosaic Virus LS2b Protein, and the Interaction with Endogenous 30S Ribosomal Protein Subunit S11 Affects Viral Replication, Infection and Gene Silencing Suppressor Activity. Ph.D. Dissertation, Northwest A&F University, Xianyang, China, 2017. Available online: https://kns.cnki.net/kcms2/article/abstract?v=SY7jeTtuViJMdSFIXfrztFqAQE3E4j-kUuzmIgDrUYt0I8QvJrzoaImu7dYo0D7kb-Pv2LlOSpBLzNc0sJ4OnuQeEA0JiWv88nv4G-8brmScJez9kmHIkgQ33yCaB7X79KFlqQYG4ImLe7mG5_V4r4udFLkSqo7IH5BAqgXAdSSquiXpeN9rSFQBuR4g_PAKc7_6-TGe968=&uniplatform=NZKPT&language=CHS (accessed on 1 June 2024).
- Lu, J.D. Heterogenous Combination of Cucumovirus Replicases Regulates Virus Genomic and Subgenomic RNA Replication. Master’s Thesis, Zhejiang Sci-Tech University, Hangzhou, China, 2016. Available online: https://kns.cnki.net/kcms2/article/abstract?v=SY7jeTtuViJlxpSS5zD4mSO2NwWGn0fkBgWE3a6RT-WP7Jc4BkKwSnN5Sdqhc1G3UyVkDJnVPZDkRPGLX2A20y2xxw7CL0kcSuP6AYV-vVDgbX9ioeg7w6cdbByXtNn7tqcIVFeGilwrrI6j2-S16b4NPhPniISP3aB9mmhsSGMQaiaBYMAMBgX_Bb6KNeUCLUuBIKX5FvU=&uniplatform=NZKPT&language=CHS (accessed on 1 May 2024).
- Stobbe, A.H.; Melcher, U.; Palmer, M.W.; Roossinck, M.J.; Shen, G. Co-divergence and host-switching in the evolution of tobamoviruses. J. Gen. Virol. 2012, 93, 408–418. [Google Scholar] [CrossRef]
- Conti, G.; Rodriguez, M.C.; Venturuzzi, A.L.; Asurmendi, S. Modulation of host plant immunity by Tobamovirus proteins. Ann. Bot. 2017, 119, 737–747. [Google Scholar] [CrossRef]
- Ishibashi, K.; Ishikawa, M. Replication of Tobamovirus RNA. Annu. Rev. Phytopathol. 2016, 54, 55–78. [Google Scholar] [CrossRef]
- Ramos-González, P.L.; Dos Santos, G.F.; Chabi-Jesus, C.; Harakava, R.; Kitajima, E.W.; Freitas-Astúa, J. Passion Fruit Green Spot Virus Genome Harbors a New Orphan ORF and Highlights the Flexibility of the 5′-End of the RNA2 Segment Across Cileviruses. Front. Microbiol. 2020, 11, 206. [Google Scholar] [CrossRef]
- Cho, I.S.; Yang, C.Y.; Yoon, J.Y.; Kwon, T.R.; Hammond, J.; Lim, H.S. First report of Passiflora latent virus infecting persimmon in Korea. Plant Dis. 2020, 105, 1236. [Google Scholar] [CrossRef]
- Choi, M.K.; Ju, H.J. First report of Passiflora latent virus infecting passion fruit (Passiflora edulis) in South Korea. Plant Dis. 2023, 107, 2893. [Google Scholar] [CrossRef]
- Bao, S.; Ge, F.; Li, X.; Bo, B. First report of Passiflora latent virus in Passion fruit (Passiflora edulis) in China. Plant Dis. 2023, 107, 3326. [Google Scholar] [CrossRef] [PubMed]
- Morales, F.J.; Lozano, I.; Castaño, M.; Arroyave, J.; Velasco, A.C.; Varon, F.J. Partial characterization of a Tymovirus infecting passion fruit in Colombia, South America. J. Phytopathol. 2002, 150, 292–296. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Gonçalves, Z.S.; Jesus, O.N.; Lima, L.K.S.; Corrêa, R.X. Responses of Passiflora spp. to cowpea aphid-borne mosaic virus reveal infection in asymptomatic plants and new species with probable immunity. Arch. Virol. 2021, 166, 2419–2434. [Google Scholar] [CrossRef]
- Roy, A.; Guillermo, L.M.; Nunziata, S.; Padmanabhan, C.; Rivera, Y.; Brlansky, R.H.; Hartung, J. First report of Passion fruit green spot virus in yellow Passion fruit (Passiflora edulis f. flavicarpa) in Casanare, Colombia. Plant Dis. 2023, 107, 2270. [Google Scholar] [CrossRef]
- Ren, Y.Y.; Wang, L.J.; Chen, N.; Yuan, Q.F.; Ma, Y.H. Identification of Viruses Pathogen Infecting Passion Fruit in Guizhou. Guizhou Agric. Sci. 2023, 51, 35–41. [Google Scholar] [CrossRef]
- Liu, Q.H.; Lan, P.X.; Lu, X.; Li, F.; Tan, G.L. Identification of the viruses infecting Passiflora edulis in Yunnan and sequenceanalysis of the cp gene of CMV infecting P. edulis. Acta Phytopathol. Sin. 2022, 52, 1021–1024. [Google Scholar] [CrossRef]
- Xing, B.; Yang, L.; Gulinuer, A.; Ye, G. Research progress on horizontal gene transfer and its functions in insects. Trop. Plants 2023, 2, 3. [Google Scholar] [CrossRef]
- Rosen, R.; Kanakala, S.; Kliot, A.; Pakkianathan, B.C.; Farich, B.A.; Santana-Magal, N.; Elimelech, M.; Kontsedalov, S.; Lebedev, G.; Cilia, M.; et al. Persistent, circulative transmission of begomoviruses by whitefly vectors. Curr. Opin. Virol. 2015, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Munguti, F.; Maina, S.; Nyaboga, E.N.; Kilalo, D.; Kimani, E.; Macharia, M.; Holton, T. Transcriptome Sequencing Reveals a Complete Genome Sequence of Cowpea Aphid-Borne Mosaic Virus from Passion Fruit in Kenya. Microbiol. Resour. Announc. 2019, 10, e01607-18. [Google Scholar] [CrossRef]
- Huang, C.M.; Hu, C.J.; Shi, G.Y.; Luo, H.B.; Cao, H.Q.; Wu, X.J.; Jiang, S.L.; Ye, L.P.; Wei, Y.W. Effects of TeMV and PWV infection on metabolic physiology and tissue microstructure of passion fruit. Acta Phytopathol. Sin. 2023, 53, 22–30. [Google Scholar]
- Chen, S.; Yu, N.; Yang, S.; Zhong, B.; Lan, H. Identification of Telosma mosaic virus infection in Passiflora edulis and its impact on phytochemical contents. Virol. J. 2018, 15, 168. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.M.; He, S.Y.; Xin, X.F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Mandadi, K.K.; Scholthof, K.B. Plant immune responses against viruses: How does a virus cause disease? Plant Cell 2013, 25, 1489–1505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhao, F.; Jiang, L.; Chen, C.; Wu, L.; Liu, Z. Different pathogen defense strategies in Arabidopsis: More than pathogen recognition. Cells 2018, 7, 252. [Google Scholar] [CrossRef]
- Na, C.; Zhou, J.R.; Rott, P.C.; Li, J.; Fu, H.Y.; Huang, M.T.; Zhang, H.L.; Gao, S.J. ScPR1 plays a positive role in the regulation of resistance to diverse stresses in sugarcane (Saccharum spp.) and Arabidopsis thaliana. Ind. Crops Prod. 2022, 180, 114736. [Google Scholar]
- Sett, S.; Prasad, A.; Prasad, M. Resistance genes on the verge of plant-virus interaction. Trends Plant Sci. 2022, 27, 1242–1252. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z. The Effect of Cucumber Mosaic Virus (CMV) on Passion Fruit Quality and Research on Control Strategies. Master’s Thesis, Guizhou University, Guiyang, China, 2021. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Chakraborty, S. Chloroplast: The Trojan horse in plant-virus interaction. Mol. Plant Pathol. 2018, 19, 504–518. [Google Scholar] [CrossRef] [PubMed]
- Jiao, N. Establishment of Passiflora caerulea L. Rapid Propagation System and Research on Detoxification Technology. Master’s Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, 2019. [Google Scholar] [CrossRef]
- Preisigke, S.C.; Viana, A.P.; Santos, E.A.; Santos, P.R.D.; AmbrÓsio, M.; Santos, V.O.D.; Silva, F.A.D. Individual selection of the first backcross generation of passion fruit potentially resistant to the fruit woodiness disease. An. Acad. Bras. Cienc. 2020, 92 (Suppl. S1), e20180797. [Google Scholar] [CrossRef] [PubMed]
- Freitas, J.C.O.; Viana, A.P.; Santos, E.A.; Silva, F.H.L.; Paiva, C.L.; Rodrigues, R.; Souza, M.M.; Eiras, M. Genetic basis of the resistance of a passion fruit segregant population to Cowpea aphid-borne mosaic virus (CABMV). Trop. Plant Pathol. 2015, 40, 291–297. [Google Scholar] [CrossRef]
- Santos, E.A.; Viana, A.P.; de Oliveira Freitas, J.C.; Silva, F.H.L.; Rodrigues, R.; Eiras, M. Resistance to Cowpea aphid-borne mosaic virus in species and hybrids of Passiflora: Advances for the control of the passion fruit woodiness disease in Brazil. Eur. J. Plant Pathol. 2015, 143, 85–98. [Google Scholar] [CrossRef]
- Freitas, J.C.O.; Viana, A.P.; Santos, E.A.; Paiva, C.L.; Silva, F.H.L.; Souza, M.M. Sour passion fruit breeding: Strategy applied to individual selection in segregating population of Passiflora resistant to Cowpea aphid-borne mosaic virus (CABMV). Sci. Hortic. 2016, 211, 241–247. [Google Scholar] [CrossRef]
- Zhou, H.L.; Zheng, Y.Y.; Zheng, J.Z.; Zheng, K.B. High-Quality Varieties of Passion Fruit and Supporting Cultivation Techniques. South China Fruits 2015, 44, 121–124. [Google Scholar] [CrossRef]
- Trevisan, F.; Mendes, B.M.J.; Maciel, S.C.; Vieira, M.L.C.; Meletti, L.M.M.; Rezende, J.A.M. Resistance to Passion fruit woodiness virus in transgenic passionflower expressing the virus coat protein gene. Plant Dis. 2006, 90, 1026–1030. [Google Scholar] [CrossRef]
- Alfenas, P.F.; Braz, A.S.; Torres, L.B.; Santana, E.N.; Verônica, A.; Nascimento, S.D.; Carvalho, M.G.; Otoni, W.C.; Zerbini, F. M Transgenic passionfruit expressing RNA derived from Cowpea aphid-borne mosaic virus is resistant to passionfruit woodiness disease. Fitopatol. Bras. 2005, 30, 33–38. [Google Scholar] [CrossRef]
- Correa, M.F.; Pinto, A.P.C.; Rezende, J.A.M. Genetic transformation of sweet passion fruit (Passiflora alata) and reactions of the transgenic plants to Cowpea aphid borne mosaic virus. Eur. J. Plant Pathol. 2015, 143, 813–821. [Google Scholar] [CrossRef]
- Da Silva, M.L.; Pinto, D.L.P.; Passos, A.B.; Marcelino-Guimarães, F.C.; Rossi, A.A.B.; Krause, W.; de Carvalho, I.F.; Batista, D.S.; Rocha, D.I.; Otoni, W.C. Novel and efficient transformation of wild passion fruit (Passiflora cincinnata Mast.) using sonication-assisted Agrobacterium-mediated transformation. In Vitro Cell Dev. Biol. Plant 2021, 57, 380–386. [Google Scholar] [CrossRef]
- Rizwan, H.M.; Yang, Q.; Yousef, A.F.; Zhang, X.; Sharif, Y.; Kaijie, J.; Shi, M.; Li, H.; Munir, N.; Yang, X.; et al. Establishment of a Novel and Efficient Agrobacterium-Mediated in Planta Transformation System for Passion Fruit (Passiflora edulis). Plants 2021, 10, 2459. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.K.; Upadhyay, S.K.; Mishra, M.; Saurabh, S.; Singh, R.; Singh, H.; Thakur, N.; Rai, P.; Pandey, P.; Hans, A.L.; et al. Expression of an insecticidal fern protein in cotton protects against whitefly. Nat. Biotechnol. 2016, 34, 1046–1051. [Google Scholar] [CrossRef]
- Javaid, S.; Amin, I.; Jander, G.; Mukhtar, Z.; Saeed, N.A.; Mansoor, S. A transgenic approach to control hemipteran insects by expressing insecticidal genes under phloem-specific promoters. Sci. Rep. 2016, 6, 34706. [Google Scholar] [CrossRef]
- Twayana, M.; Girija, A.M.; Mohan, V.; Shah, J. Phloem: At the center of action in plant defense against aphids. J. Plant Physiol. 2022, 273, 153695. [Google Scholar] [CrossRef]
- Siqueira-Júnior, C.L.; Jardim, B.C.; Urményi, T.P.; Vicente, A.C.; Hansen, E.; Otsuki, K.; da Cunha, M.; Madureira, H.C.; de Carvalho, D.R.; Jacinto, T. Wound response in passion fruit (Passiflora f. edulis flavicarpa) plants: Gene characterization of a novel chloroplast-targeted allene oxide synthase up-regulated by mechanical injury and methyl jasmonate. Plant Cell Rep. 2008, 27, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Tahir, M.N.; Asad, S.; Bilal, R.; Van Eck, J.; Jander, G.; Mansoor, S. RNAi-Mediated Simultaneous Resistance Against Three RNA Viruses in Potato. Mol. Biotechnol. 2017, 59, 73–83. [Google Scholar] [CrossRef]
- Paul, S.; de la Fuente-Jiménez, J.L.; Manriquez, C.G.; Sharma, A. Identification, characterization and expression analysis of passion fruit (Passiflora edulis) microRNAs. 3 Biotech 2020, 10, 25. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.; Huang, D.; Xing, W.; Wu, B.; Wei, Q.; Xu, Y.Y.; Zhan, R.L.; Ma, F.N.; Song, S. Research progress on the MYB transcription factors in tropical fruit. Trop. Plants 2022, 1, 5. [Google Scholar] [CrossRef]
- Ghanim, M.; Kontsedalov, S. Gene expression in pyriproxyfen-resistant Bemisia tabaci Q biotype. Pest Manag. Sci. 2007, 63, 776–783. [Google Scholar] [CrossRef]
- Kaleem Ullah, R.M.; Gao, F.; Sikandar, A.; Wu, H. Insights into the Effects of Insecticides on Aphids (Hemiptera: Aphididae): Resistance Mechanisms and Molecular Basis. Int. J. Mol. Sci. 2023, 24, 6750. [Google Scholar] [CrossRef]
- Wang, Z.; Yan, H.; Yang, Y.; Wu, Y. Biotype and insecticide resistance status of the whitefly Bemisia tabaci from China. Pest Manag. Sci. 2010, 66, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, J.D.; Che, W.N.; Luo, C. First report of field resistance to cyantraniliprole, a new anthranilic diamide insecticide, on Bemisia tabaci MED in China. J. Integr. Agric. 2018, 17, 158–163. [Google Scholar] [CrossRef]
- Gressent, F.; Da Silva, P.; Eyraud, V.; Karaki, L.; Royer, C. Pea albumin 1 subunit b (PA1b), a promising bioinsecticide of plant origin. Toxins 2011, 3, 1502–1517. [Google Scholar] [CrossRef]
- Li, Q.Z. Efficacy Analysis of Several Chemical Agents for the Control of Passion Fruit Leaf Spot Virus Disease. South China Agric. 2017, 11, 9–10. [Google Scholar] [CrossRef]
- Rendina, N.; Nuzzaci, M.; Scopa, A.; Cuypers, A.; Sofo, A. Chitosan-elicited defense responses in Cucumber mosaic virus (CMV)-infected tomato plants. J. Plant Physiol. 2019, 234–235, 9–17. [Google Scholar] [CrossRef]
- Gangireddygari, V.S.R.; Chung, B.N.; Cho, I.S.; Yoon, J.Y. Inhibitory effect of chitosan and phosphate cross-linked chitosan against cucumber mosaic virus and pepper mild mottle virus. Plant Pathol. J. 2021, 37, 632–640. [Google Scholar] [CrossRef]
- Wang, J.; Wang, H.Y.; Xia, X.M.; Li, P.P.; Wang, K.Y. Inhibitory effect of sulfated lentinan and lentinan against tobacco mosaic virus (TMV) in tobacco seedlings. Int. J. Biol. Macromol. 2013, 61, 264–269. [Google Scholar] [CrossRef]
- Fan, H.; Yan, X.; Fu, M.; Liu, D.; Awan, A.W.; Chen, P.; Rasheed, S.M.; Gao, L.; Zhang, R. Interactive Effect of Biological Agents Chitosan, Lentinan and Ningnanmycin on Papaya Ringspot Virus Resistance in Papaya (Carica papaya L.). Molecules 2022, 27, 7474. [Google Scholar] [CrossRef]
- Huang, M.; Wu, Z.; Li, J.; Ding, Y.; Chen, S.; Li, X. Plant Protection against Viruses: An Integrated Review of Plant Immunity Agents. Int. J. Mol. Sci. 2023, 24, 4453. [Google Scholar] [CrossRef]
- Chen, J.X.; Luo, X.; Chen, Y.F.; Wang, Y.; Peng, J.; Xing, Z.F. Recent Research Progress: Discovery of Anti-Plant Virus Agents Based on Natural Scaffold. Front. Chem. 2022, 26, 926202. [Google Scholar] [CrossRef] [PubMed]
- Chong, Y.H.; Do, D.H.; Cheng, H.W.; Raja, J.A.J.; Ngo, X.T.; Hwang, S.G.; Yeh, S.D. Generation of attenuated mutants of East Asian Passiflora virus for disease management by cross protection. Mol. Plant Microbe Interact. 2023, 36, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Do, D.H.; Nguyen, T.B.; Ha, V.C.; Raja, J.A.J.; Yeh, S.D. Generation of attenuated Passiflora mottle virus through modification of the helper component-protease for cross protection. Phytopathology 2023, 113, 1605–1614. [Google Scholar] [CrossRef] [PubMed]
- Ziebell, H.; Payne, T.; Berry, J.O.; Walsh, J.A.; Carr, J.P. A cucumber mosaic virus mutant lacking the 2b counter-defence protein gene provides protection against wild-type strains. J. Gen. Virol. 2007, 88, 2862–2871. [Google Scholar] [PubMed]
- Maneechoat, P.; Takeshita, M.; Uenoyama, M.; Nakatsukasa, M.; Kuroda, A.; Furuya, N.; Tsuchiya, K. A single amino acid at N-terminal region of the 2b protein of cucumber mosaic virus strain m1 has a pivotal role in virus attenuation. Virus Res. 2015, 197, 67–74. [Google Scholar] [CrossRef]
- Wang, L.; Shi, W.; Aziz, A.; Wang, X.; Liu, H.; Shen, W.; Cui, H.; Dai, Z. Mutating the arginine residue within the FRNK motif of telosma mosaic virus (TelMV) HC-Pro protein attenuates viral infection and confers effective protection against TelMV in passion fruit (Passiflora edulis). Pest Manag. Sci. 2024, 80, 5256–5265. [Google Scholar] [CrossRef]

| Genus | Name | Abbreviation | GenBank | Symptoms of Virus-Infected Passion Fruit | References |
|---|---|---|---|---|---|
| Potyviridae, Potyvirus (PVY) | Passion fruit woodiness virus | PWV | JF427619.1 | Leaf twisting, lignification of fruits, and smaller fruit. | [13] |
| Cowpea aphid-borne mosaic virus | CABMV | MH151199 | Leaf wrinkled and twisted, with greenish, ringed spots; fruit lignified and malformed; slow plant development. | [14] | |
| Passion fruit ringspot virus | PFRSV | -- | Mottled and ringed spots; severe leaf deformation; fruit asymptomatic. | [15,16] | |
| Passion fruit mottle virus | PaMV | MG087836.1 | Leaves lightly mottled; fruit epidermis mottled. | [17] | |
| Soybean mosaic virus | SMV | KY249377.1 | Leaves mottled, wrinkled, and deformed. | [18] | |
| Passiflora virus Y | PaVY | AY461661.1 | Yellow-green mottling, ring spots, faded green spots, and curled leaves. | [19] | |
| East Asian Passiflora virus | EAPV | MG650164.1 | Vein necrosis and rugosity of the upper trifoliate leaves; misshapen, woody and pitted fruit; stunted vegetative growth. | [20] | |
| Ugandan Passiflora virus | UPV | NC_076338.1 | Foliar mosaics, vein clearing, fruit hardening, and malformation | [21] | |
| Malaysian Passiflora virus | MPV | EU035271.1 | - | [12] | |
| Bean yellow mosaic virus | BYMV | FM180011 | Deformed leaves with mosaic. | [22] | |
| Telosma mosaic virus | TeMV | MW297551.1 | Deformation of flowers, leaves, and leaflets; fading green leaves with mosaic; small fruits; uneven colouring of fruits. | [23] | |
| Watermelon mosaic virus | WMV | KX512320.1 | Mosaic and severe leaf crumpling. | [24] | |
| Passiflora chlorosis virus | PaCV | NC_043156.1 | Chlorotic. | [24] | |
| Passion fruit severe mottle-associated virus | PFMoAV or PaMoV | MK449340 | Severe foliar mosaic; stunted growth; mottling, yellowing, and distortion of leaves; small, woody, and twisted fruits. | [25,26] | |
| East Asian Passiflora distortion virus | EAPDV | LC379162 | Mosaic and curled leaves; fruit deformed. | [27] | |
| Geminiviridae, Begomovirus | Passion fruit little leaf mosaic virus | PLLMV | AY167566 | Severely yellowed and greatly reduced foliage; small, mostly misshapen fruits; fewer fruits set on a single plant; drastic reduction in foliage layer and plant growth. | [28] |
| Passion fruit leaf distortion virus | PLDV | KT899302 | Yellow foliage and leaf deformation. | [29] | |
| Euphorbia mosaic virus | EuMV | KJ647290.1 | Mottled yellowing, distortion, and apical necrosis of leaves; bright foliar mosaic begins with light mottling, followed by necrotic spots, leaf distortion, and flower abortion. | [30] | |
| Euphorbia leaf curl virus | EuLCV | KC161185 | Systematically mottled and malformed leaves; yellowing, twisting, and necrosis at the top of the leaves; striped concave surfaces on the surface of immature fruits. | [31] | |
| Papaya leaf curl Guangdong virus | PaLCuGdV | KY884675 | Mosaic patterning, mottling, yellowing, crumpling, and twisting of leaves. | [31] | |
| Passion fruit leaf mottle virus | PLMV | -- | Severe curling, twisting, and mottling of leaves and fruits. | [15] | |
| Ramie mosaic virus | RamMV | KC171652.1 | Stunting, mosaic, and yellow or necrotic spots. | [32] | |
| Tomato yellow leaf curl virus | TYLCV | MW814910 | [15] | ||
| Papaya leaf curl China virus | PaLCuCNV | KX273343. | [15,33] | ||
| Passion fruit severe leaf distortion virus | PSLDV | FJ972767 | Dwarfing, leaf twisting, and greenish coloration. | [34] | |
| Passion fruit chlorotic mottle virus | PCMoV | NC_040706.1 | Chlorosis, wrinkling, and leaf distortion. | [35] | |
| Melochia yellow mosaic virus | MelYMV | MG461177.1 | Mosaic, yellow spots, and leaf curling and deformities. | [36] | |
| Cotton leaf curl Multan virus | CLCuMuV | KX656801.1 | Leaf curling and vein swelling. | [37] | |
| Sida mottle Alagoas virus | SiMAV | KX896427.1 | Severe mosaic with yellow spots, leaf deformities, and blisters. | [38] | |
| Geminiviridae, other genus | Giant granadilla malformation virus | GGMV | -- | [33] | |
| Cucumovirus | Cucumber mosaic virus | CMV | LC654689.1 | Mosaic and yellow spots on leaves; severely curled, raised, and whitened epidermis of fruit. | [39] |
| Tobamovirus | Passion fruit mosaic virus | PafMV | NC_015552.1 | [40] | |
| Maracuja mosaic virus | MarMV | NC_008716.1 | Mosaics or mottling; necrotic spots. | [41] | |
| Cilevirus | Passion fruit green spot virus | PfGSV | NC_055653 | Leaves mottled, faded green spots; yellow spots on senescent leaves with green bands of veins; green spots on fruit and older leaves; and, in severe cases, deadly necrotic lesions around the stems. | [42] |
| Hibiscus strain of Citrus Leprosis Virus C2 | CiLV-C2H | KC626783 | Green spots on young leaves. | [43] | |
| Carlavirus | Passiflora latent virus | PLV | OK274270.1 | Inconspicuous systemic mosaic; senescent leaves mottled; faded green spots; systemic faded green necrosis of leaves and mottling of upper leaves; and black annular blotches on the surface of ripe fruit. | [44] |
| Crinivirus | Lettuce chlorosis virus | LCV | FJ380119.1 | Slight yellowing, mosaic, leaf distortion, and yellow spots. | [45] |
| Tymovirus | Passion fruit yellow mosaic virus | PFYMV | MW393830.1 | Mosaic, vein mottling, wilting, and leaflet deformation. | [46] |
| Purple passion fruit leaf deformation virus | PpLDV | ON542230.1 | Leaf curling, leaf distortion, and ruffling. | [47] | |
| Nepovirus | Tomato ringspot virus | ToRSV | FJ577800.1 | [40] | |
| Rhabdoviridae | Passion fruit vein clearing virus | PaVCV | -- | Reduction in leaf area and fruit size in addition to bright veins on the leaves. | [48] |
| Purple granadilla mosaic virus | PGMV | -- | Mildly linear leaves; small, deformed, and woody fruits. | [11] | |
| Citrus-associated rhabdovirus | CiaRV | -- | Yellow and green spots. | [49] | |
| Polerovirus | Cucurbit aphid-borne yellows virus | CABYV | OP909796.1 | Wrinkling, mosaic, leaf and fruit deformation, blistering, yellow spots, vein whitening, purple leaves, yellowing and thickening of old leaves, and reduction in fruit number. | [50] |
| Roymovirus | Passiflora edulis symptomless virus | PeSV | MT271639.1 | [51] |
| Name | Protein Description | Action Mechanism | Related Research |
|---|---|---|---|
| P1-Protease | Serine protease | The first (N-terminal) mature protein of all monopartite viruses, which is highly polymorphic and the most variable and least conserved region in the genome, plays a role in influencing intercellular virus spread and determining host range. | P1 can enhance the activity of HC-Pro in the genus Potyvirus, interfering with host defence mechanisms and inducing the production of HSP70 heat shock proteins [57]. |
| Helper component-proteinase, HC-Pro | Rich in cysteine protease motifs, it is essential for aphid transmission | KLSC and PTK MOBS in HC-Pro and DAG MOBS in CP can promote aphid transmission of SMV [58]. | HC-Pro can inhibit host plant gene silencing by binding to double-stranded RNA (dsRNAs) and suppressing Dicer processing and the accumulation of 21-nucleotide short interfering RNA (siRNA) [59]. HC-Pro also plays a significant role in reducing photosynthetic rates after PVY infection in plants [60]. |
| Cylindrical inclusion, CI | CI protein possesses ATP binding and RNA helicase activities, as well as NTPase activity, and is also a component of the viral replication complex | CI protein is involved in viral replication through its helicase domain and C-terminal region. The N-terminal sequence is associated with intercellular movement. It may assist in virus genome replication by binding to RNA through its helicase domain and C-terminal region, thereby unwinding RNA double strands. | Likely involved in intercellular movement through the formation of cone structures on plasmodesmata (PD) and interaction with the capsid protein (CP) [61]. |
| P3-Protease | Transcriptional slippage at a single-nucleotide insertion site within the P3 cistron generates an additional peptide, P3N-PIPO | Regulates viral replication, movement, and pathogenesis. | P3N-PIPO localises to plasmodesmata (PD), interacts with the CI protein, and facilitates intercellular movement of the virus in susceptible hosts [62]. P3 is a determinant of virulence in soybean mosaic virus (SMV) [63]. |
| 6K1 | Located at the periphery of infected cells, rich in hydrophobic amino acids, and associated with membrane binding | 6K1 may be involved in intercellular movement [58,64]. | 6K1 inhibits JA-dependent defence and suppresses aphid reproduction [65]. |
| VPg | It plays a crucial role in the translation or replication of positive-strand RNA viruses, serving as an intrinsically disordered protein, a characteristic that endows it with the ability to bind to multiple proteins | VPg exists in various precursor forms, such as 6K2-VPg-NIa-Pro, which is recruited into the viral replication complex; VPg-Pro-Pol serves as a primer for replication; VPg covalently binds to the 5′ end of RNA, serving as a determinant of virulence [66,67]. | VPg interacts with the eukaryotic translation initiation factor 4E (eIF4E), playing a crucial role in virus RNA replication [66,68]. |
| Nuclear inclusion b-protease, NIb | An RNA-dependent RNA polymerase responsible for replicating the viral genome | Recruited into the Viral Replication Complex (VRC) through interaction with the VPg domain of 6K2-VPg-Pro. | NIb is also crucial for the formation of the viral replication complexes (VRCs) and is involved in multiple virus-host interactions. For example, NIb acts as an inhibitor of host defence responses [69] and engages in an arms-race-like antagonism with NPR1 (Nonexpresser of Pathogenesis-Related Genes 1, which is a major regulatory factor in salicylic acid-mediated plant local and systemic acquired resistance) [70]. |
| Nuclear Inclusion a-protease, NIa-Pro | A cysteine protease with trypsin-like activity associated with the small ribonucleic acid virus 3C proteinase | The small ribonucleic acid virus 3C protease can cleave hundreds of host proteins to facilitate viral infection [71], and NIb is released by the NIa protease [69]. | NIa often exists in stable intermediate forms, such as the previously mentioned 6K2-VPg-NIa-Pro [67,71]. NIa also participates in RNA replication, interacting with viral RNA-dependent RNA polymerase (RdRp) and viral RNA to stimulate viral RNA replication [71]. |
| Coat protein, CP | The main structural protein of the virion | CP (Coat Protein) has a conserved DAG motif near the N-terminus of the protein, which is involved in the interaction between CP and HCPro, associated with aphid transmission [72]. The coat protein is also involved in virus replication, movement, symptom expression, RNA encapsidation, and other processes [23,72]. | Usually plays a role in the production and spread of symptoms [54]. |
| Name | Protein Description | Action Mechanism and Related Research | |
|---|---|---|---|
| DNA-A | AV1/CP/V1 | Coat protein | As the CP protein, it is also involved in the intracellular transport of viral DNA and transmission by insects. |
| AV2/MP/V2 | Movement proteins, not present in the bipartite Begomoviruses [77] | AV2 is a potent suppressor of Post-Transcriptional Gene Silencing (PTGS) and Transcriptional Gene Silencing (TGS), mediating the nuclear export of CP [78,79]. | |
| AC1/Rep/C1 | Replication initiator protein | AC1 is crucial for replication and may play a key role in the recruitment and assembly of the viral replication mechanisms. | |
| AC2/TrAP/C2 | Transcription activator protein | AC2 is a pathogenic factor that suppresses host defences and is also a gene silencing suppressor, interfering with the ubiquitination pathway and jasmonic acid signalling [80]. | |
| AC3/Ren/C3 | Replication enhancer protein | C3 can enhance viral replication, increasing the amount of virus accumulated in the host [79]. | |
| AC4/C4 | Multifunctional protein, an inhibitor of RNA silencing. | C4 is a determinant of symptoms and one of the main means of plant defence, with the amino acid sequence of this protein showing the greatest variability. It is involved in the suppression of RNA silencing (PTGS and TGS) and has the ability to disrupt JA (jasmonic acid) signalling [74]. Recent research has found that the C4 protein also participates in regulating the severity of leaf curling during symptom development [81]. | |
| AC5/C5 | Present in some monopartite viruses, it is a determinant of virulence | It can suppress transcriptional gene silencing induced by single-stranded RNA, aiding in viral infection [82,83,84]. | |
| DNA-B | BV1/NSP | Nuclear shuttle protein | Involved in the development of symptoms [76]. |
| BC1/MP | Movement protein | Involved in viral movement [76]. | |
| Other small ORFs, of which the largest is named V3. | Located in the Golgi apparatus, it acts as an RNA silencing suppressor | It is essential for complete viral infection [78]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, W.; Ma, F.; Zhang, X.; Tan, Y.; Han, T.; Ding, J.; Wu, J.; Xing, W.; Wu, B.; Huang, D.; et al. Research Progress on Viruses of Passiflora edulis. Biology 2024, 13, 839. https://doi.org/10.3390/biology13100839
Wu W, Ma F, Zhang X, Tan Y, Han T, Ding J, Wu J, Xing W, Wu B, Huang D, et al. Research Progress on Viruses of Passiflora edulis. Biology. 2024; 13(10):839. https://doi.org/10.3390/biology13100839
Chicago/Turabian StyleWu, Wenhua, Funing Ma, Xiaoyan Zhang, Yuxin Tan, Te Han, Jing Ding, Juyou Wu, Wenting Xing, Bin Wu, Dongmei Huang, and et al. 2024. "Research Progress on Viruses of Passiflora edulis" Biology 13, no. 10: 839. https://doi.org/10.3390/biology13100839
APA StyleWu, W., Ma, F., Zhang, X., Tan, Y., Han, T., Ding, J., Wu, J., Xing, W., Wu, B., Huang, D., Zhang, S., Xu, Y., & Song, S. (2024). Research Progress on Viruses of Passiflora edulis. Biology, 13(10), 839. https://doi.org/10.3390/biology13100839






