Integrative Analysis of ATAC-Seq and RNA-Seq through Machine Learning Identifies 10 Signature Genes for Breast Cancer Intrinsic Subtypes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Preparation and Integration
2.2. Integrative Analysis between RNA-Seq and ATAC-Seq in GDC TCGA-BRCA
2.3. Data Preprocessing and Feature Selection
2.4. Construction and Evaluation of Machine Learning Algorithms
2.5. Validation with Independent Gene Expression Datasets
2.6. GO and KEGG Enrichment Analysis
2.7. Motif Discovery Analysis
3. Results
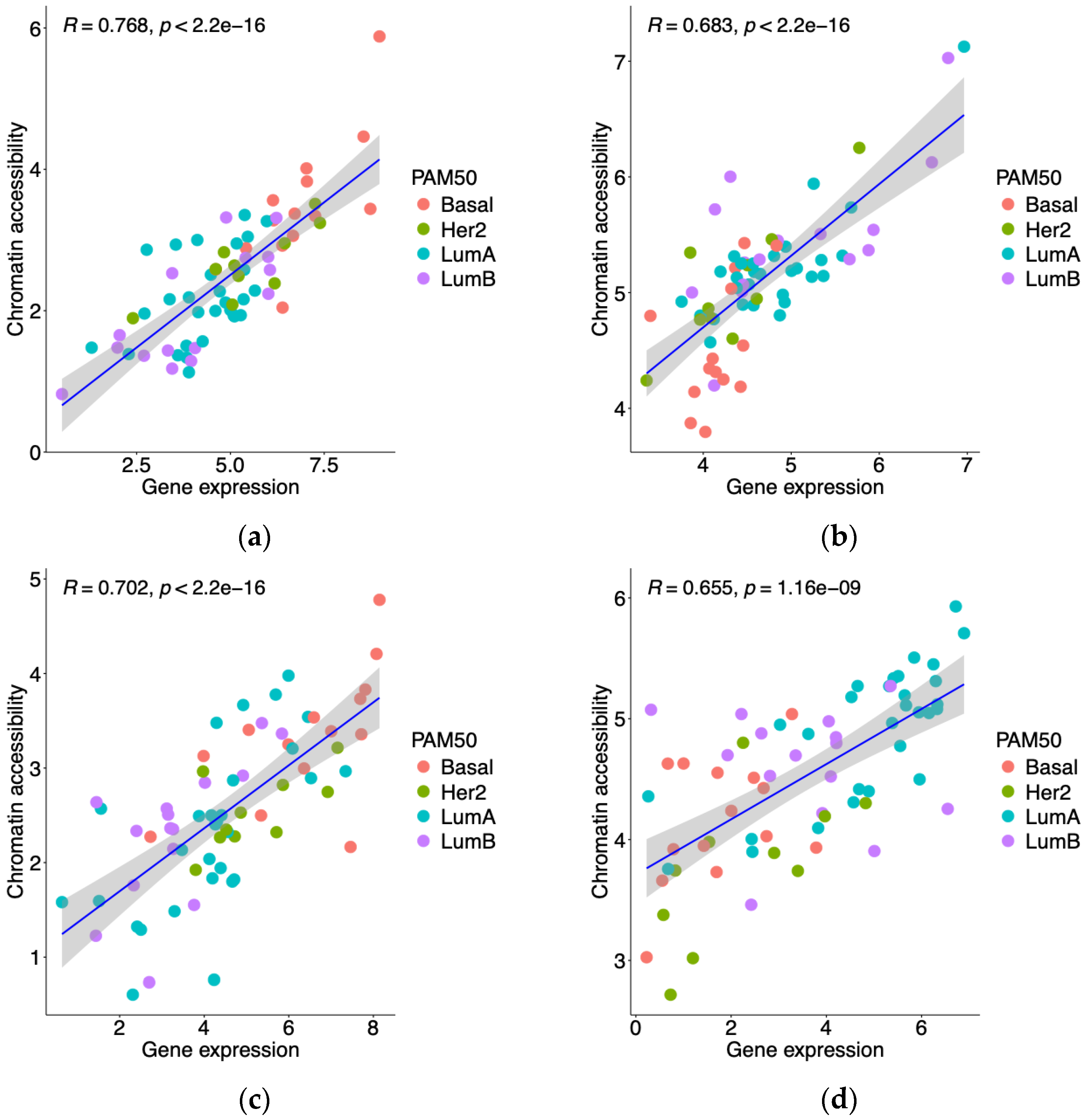
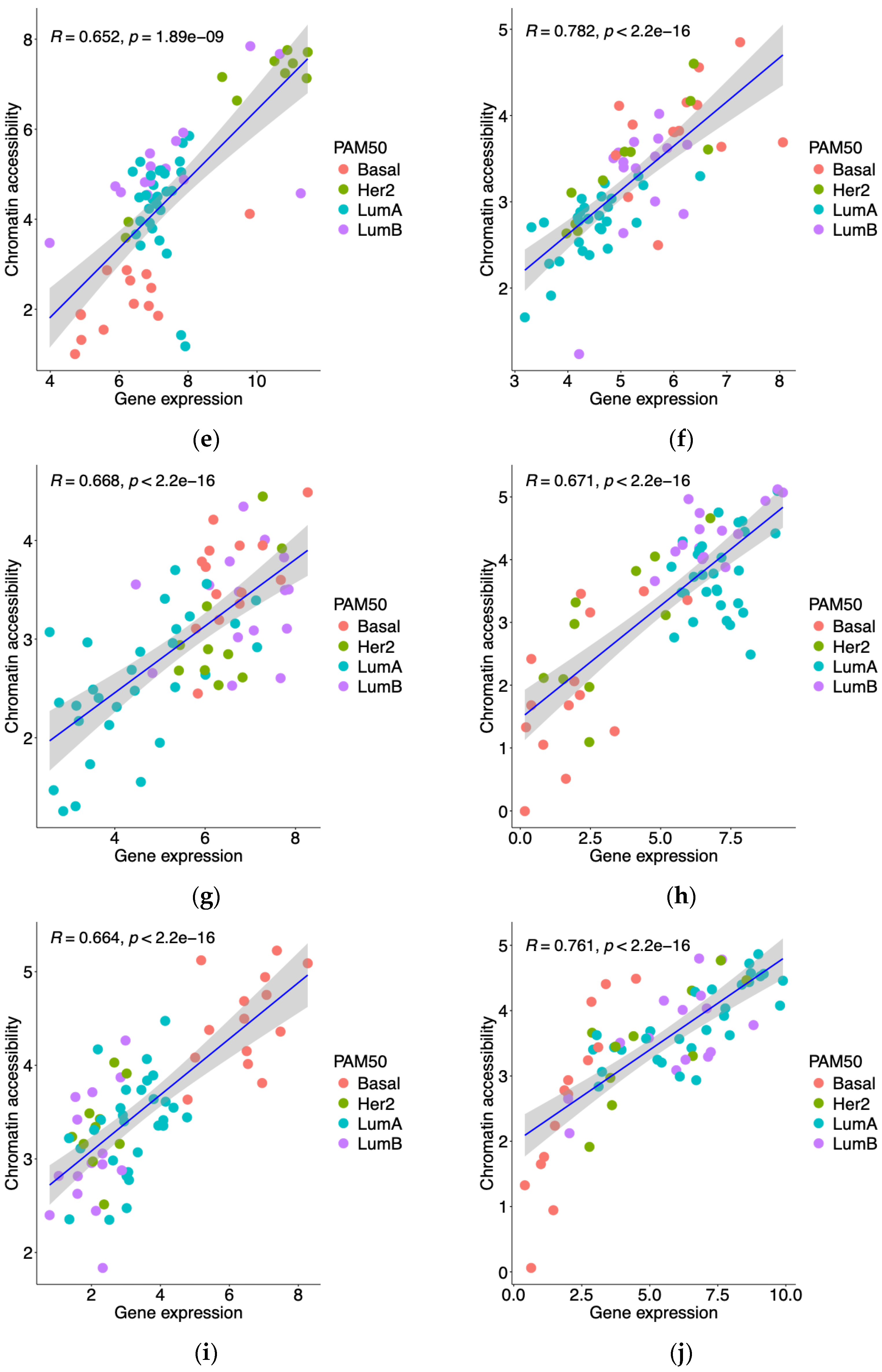
3.1. Integrative Analysis of Gene Expression and Promoter Accessibility in Breast Cancer Patients
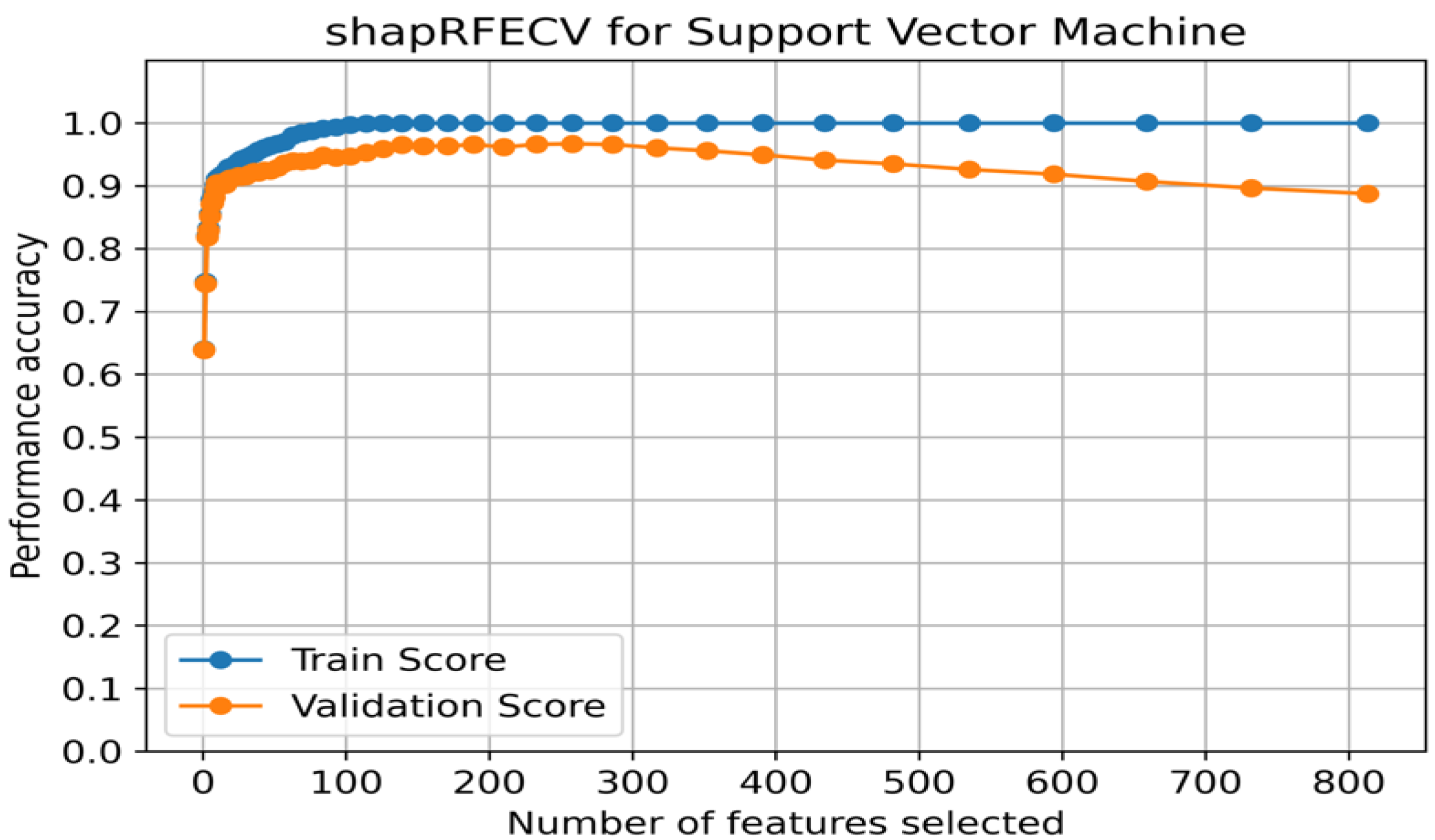
3.2. Recursive Feature Selection for Breast Cancer Subtype Prediction
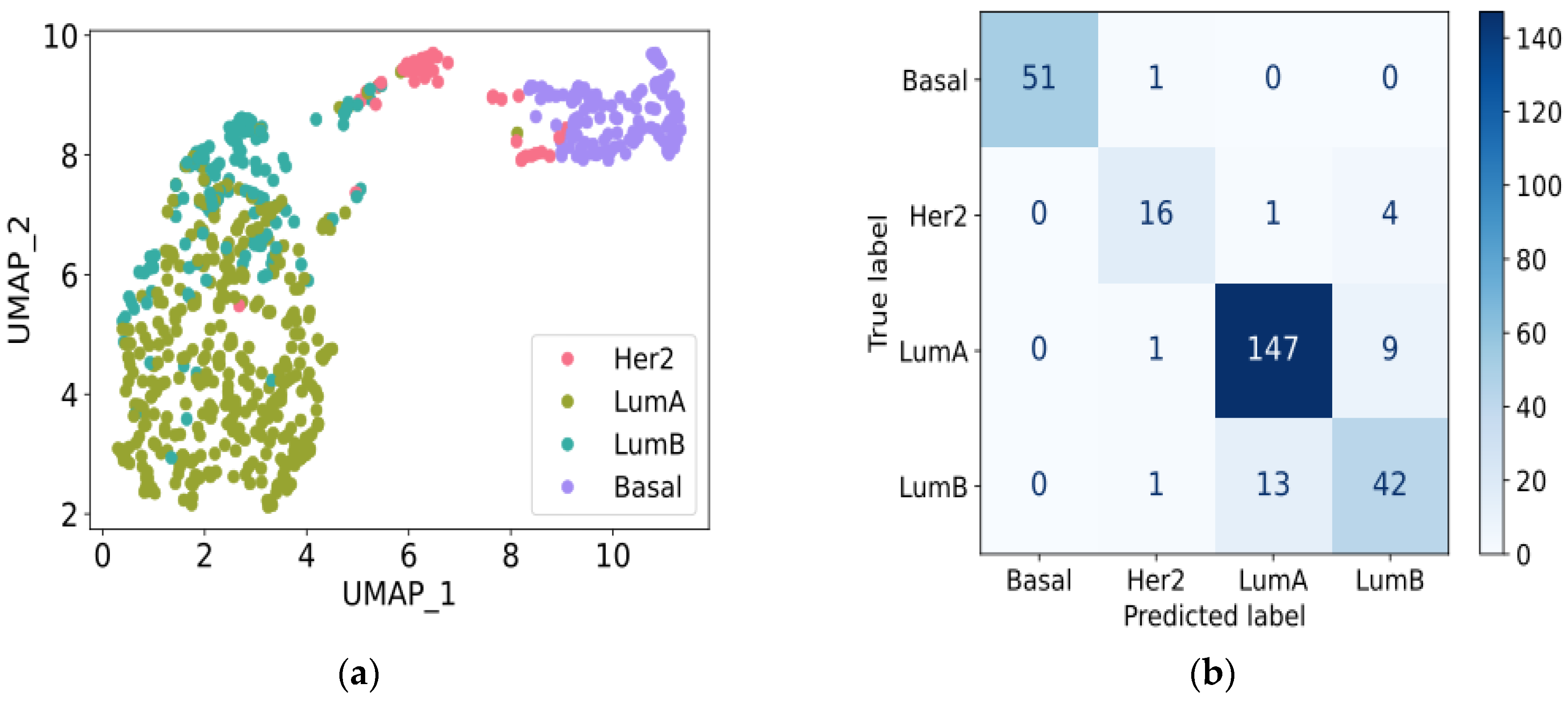
3.3. Prediction Performance with the 10 Selected Genes
3.4. External Validation Using Other Breast Cancer Gene Expression Data
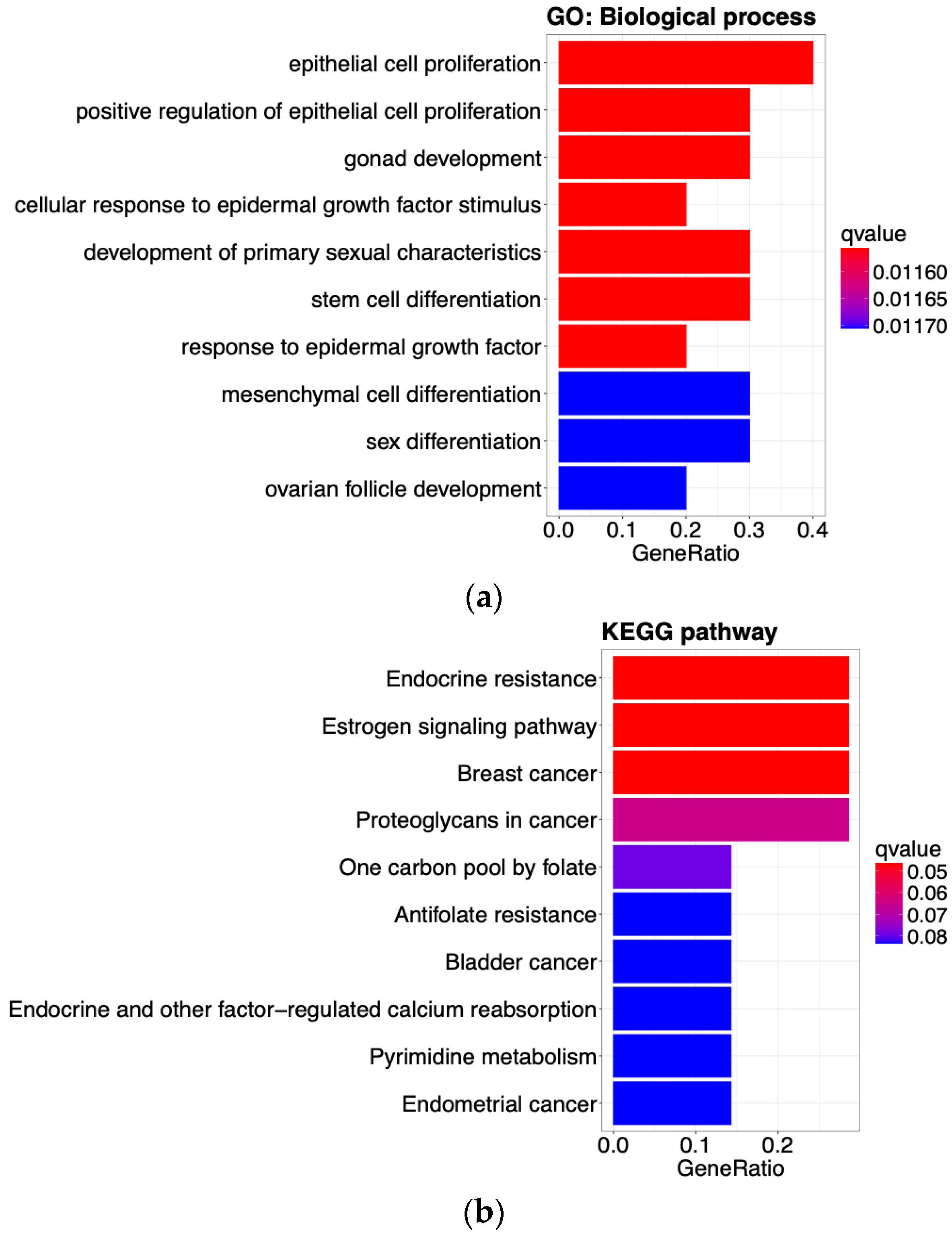
3.5. Identification of Enriched Gene Sets and Motif Discovery Using the 10 Selected Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lukasiewicz, S.; Czeczelewski, M.; Forma, A.; Baj, J.; Sitarz, R.; Stanislawek, A. Breast Cancer-Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies—An Updated Review. Cancers 2021, 13, 4287. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, C.L.T.; Larsson, A.-M.; Forsare, C.; Aaltonen, K.; Jansson, S.; Bradshaw, R.; Bendahl, P.-O.; Rydén, L. PAM50 intrinsic subtype profiles in primary and metastatic breast cancer show a significant shift toward more aggressive subtypes with prognostic implications. Cancers 2021, 13, 1592. [Google Scholar] [CrossRef]
- Okimoto, L.Y.S.; Mendonca-Neto, R.; Nakamura, F.G.; Nakamura, E.F.; Fenyo, D.; Silva, C.T. Few-shot genes selection: Subset of PAM50 genes for breast cancer subtypes classification. BMC Bioinform. 2024, 25, 92. [Google Scholar] [CrossRef] [PubMed]
- Easton, D.F.; Pooley, K.A.; Dunning, A.M.; Pharoah, P.D.; Thompson, D.; Ballinger, D.G.; Struewing, J.P.; Morrison, J.; Field, H.; Luben, R.; et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007, 447, 1087–1093. [Google Scholar] [CrossRef]
- Zhang, H.; Ahearn, T.U.; Lecarpentier, J.; Barnes, D.; Beesley, J.; Qi, G.; Jiang, X.; O’Mara, T.A.; Zhao, N.; Bolla, M.K.; et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat. Genet. 2020, 52, 572–581. [Google Scholar] [CrossRef]
- Behravan, H.; Hartikainen, J.M.; Tengstrom, M.; Kosma, V.M.; Mannermaa, A. Predicting breast cancer risk using interacting genetic and demographic factors and machine learning. Sci. Rep. 2020, 10, 11044. [Google Scholar] [CrossRef] [PubMed]
- Obeagu, E.I.; Obeagu, G.U. Breast cancer: A review of risk factors and diagnosis. Medicine 2024, 103, e36905. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Choi, J.-Y.; Choi, J.; Chung, S.; Song, N.; Park, S.K.; Han, W.; Noh, D.-Y.; Ahn, S.-H.; Lee, J.W. Gene-Environment interactions relevant to estrogen and risk of breast cancer: Can gene-environment interactions be detected only among candidate SNPs from genome-wide association studies? Cancers 2021, 13, 2370. [Google Scholar] [CrossRef] [PubMed]
- Franco, H.L.; Nagari, A.; Malladi, V.S.; Li, W.; Xi, Y.; Richardson, D.; Allton, K.L.; Tanaka, K.; Li, J.; Murakami, S.; et al. Enhancer transcription reveals subtype-specific gene expression programs controlling breast cancer pathogenesis. Genome Res. 2018, 28, 159–170. [Google Scholar] [CrossRef]
- Ochoa, S.; de Anda-Jauregui, G.; Hernandez-Lemus, E. Multi-Omic Regulation of the PAM50 Gene Signature in Breast Cancer Molecular Subtypes. Front. Oncol. 2020, 10, 845. [Google Scholar] [CrossRef]
- Minnoye, L.; Marinov, G.K.; Krausgruber, T.; Pan, L.; Marand, A.P.; Secchia, S.; Greenleaf, W.J.; Furlong, E.E.M.; Zhao, K.; Schmitz, R.J.; et al. Chromatin accessibility profiling methods. Nat. Rev. Methods Primers 2021, 1, 10. [Google Scholar] [CrossRef] [PubMed]
- Grandi, F.C.; Modi, H.; Kampman, L.; Corces, M.R. Chromatin accessibility profiling by ATAC-seq. Nat. Protoc. 2022, 17, 1518–1552. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. Clinical implications of chromatin accessibility in human cancers. Oncotarget 2020, 11, 1666–1678. [Google Scholar] [CrossRef] [PubMed]
- Holder, L.B.; Haque, M.M.; Skinner, M.K. Machine learning for epigenetics and future medical applications. Epigenetics 2017, 12, 505–514. [Google Scholar] [CrossRef]
- Rauschert, S.; Raubenheimer, K.; Melton, P.E.; Huang, R.C. Machine learning and clinical epigenetics: A review of challenges for diagnosis and classification. Clin. Epigenet. 2020, 12, 51. [Google Scholar] [CrossRef]
- Luo, L.; Gribskov, M.; Wang, S. Bibliometric review of ATAC-Seq and its application in gene expression. Brief. Bioinform. 2022, 23, bbac061. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Brueffer, C.; Vallon-Christersson, J.; Grabau, D.; Ehinger, A.; Hakkinen, J.; Hegardt, C.; Malina, J.; Chen, Y.; Bendahl, P.O.; Manjer, J.; et al. Clinical Value of RNA Sequencing-Based Classifiers for Prediction of the Five Conventional Breast Cancer Biomarkers: A Report from the Population-Based Multicenter Sweden Cancerome Analysis Network-Breast Initiative. JCO Precis. Oncol. 2018, 2, 1–18. [Google Scholar] [CrossRef]
- Tekpli, X.; Lien, T.; Rossevold, A.H.; Nebdal, D.; Borgen, E.; Ohnstad, H.O.; Kyte, J.A.; Vallon-Christersson, J.; Fongaard, M.; Due, E.U.; et al. An independent poor-prognosis subtype of breast cancer defined by a distinct tumor immune microenvironment. Nat. Commun. 2019, 10, 5499. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef] [PubMed]
- Corces, M.R.; Granja, J.M.; Shams, S.; Louie, B.H.; Seoane, J.A.; Zhou, W.; Silva, T.C.; Groeneveld, C.; Wong, C.K.; Cho, S.W.; et al. The chromatin accessibility landscape of primary human cancers. Science 2018, 362, eaav1898. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, M.C.; Konate, M.M.; Chen, L.; Das, B.; Karlovich, C.; Williams, P.M.; Evrard, Y.A.; Doroshow, J.H.; McShane, L.M. TPM, FPKM, or Normalized Counts? A Comparative Study of Quantification Measures for the Analysis of RNA-seq Data from the NCI Patient-Derived Models Repository. J. Transl. Med. 2021, 19, 269. [Google Scholar] [CrossRef]
- Sundararajan, M.; Najmi, A. The many Shapley values for model explanation. In Proceeding of the 37th International Conference on Machine Learning (ICML2020), Vienna, Austria, 12–18 July 2020; PMLR 119. pp. 9269–9278. Available online: https://proceedings.mlr.press/v119/sundararajan20b.html (accessed on 15 August 2024).
- Lin, W.-C.; Tsai, C.-F.; Hu, Y.-H.; Jhang, J.-S. Clustering-based undersampling in class-imbalanced data. Inf. Sci. 2017, 409, 17–26. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Lai, Y. A statistical method for the conservative adjustment of false discovery rate (q-value). BMC Bioinform. 2017, 18, 69. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Machanick, P.; Bailey, T.L. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 2011, 27, 1696–1697. [Google Scholar] [CrossRef] [PubMed]
- Rauluseviciute, I.; Riudavets-Puig, R.; Blanc-Mathieu, R.; Castro-Mondragon, J.A.; Ferenc, K.; Kumar, V.; Lemma, R.B.; Lucas, J.; Cheneby, J.; Baranasic, D.; et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res 2024, 52, D174–D182. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Hwang, K.B. An empirical evaluation of sampling methods for the classification of imbalanced data. PLoS ONE 2022, 17, e0271260. [Google Scholar] [CrossRef]
- Mohammed, H.; D’Santos, C.; Serandour, A.A.; Ali, H.R.; Brown, G.D.; Atkins, A.; Rueda, O.M.; Holmes, K.A.; Theodorou, V.; Robinson, J.L. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013, 3, 342–349. [Google Scholar] [CrossRef]
- Zhang, Y.; Chan, H.L.; Garcia-Martinez, L.; Karl, D.L.; Weich, N.; Slingerland, J.M.; Verdun, R.E.; Morey, L. Estrogen induces dynamic ERalpha and RING1B recruitment to control gene and enhancer activities in luminal breast cancer. Sci. Adv. 2020, 6, eaaz7249. [Google Scholar] [CrossRef]
- Metovic, J.; Borella, F.; D’Alonzo, M.; Biglia, N.; Mangherini, L.; Tampieri, C.; Bertero, L.; Cassoni, P.; Castellano, I. FOXA1 in Breast Cancer: A Luminal Marker with Promising Prognostic and Predictive Impact. Cancers 2022, 14, 4699. [Google Scholar] [CrossRef]
- Mehrgou, A.; Ebadollahi, S.; Jameie, B.; Teimourian, S. Analysis of subtype-specific and common Gene/MiRNA expression profiles of four main breast cancer subtypes using bioinformatic approach; Characterization of four genes, and two MicroRNAs with possible diagnostic and prognostic values. Inform. Med. Unlocked 2020, 20, 100425. [Google Scholar] [CrossRef]
- Liu, M.; Mo, F.; Song, X.; He, Y.; Yuan, Y.; Yan, J.; Yang, Y.; Huang, J.; Zhang, S. Exosomal hsa-miR-21-5p is a biomarker for breast cancer diagnosis. PeerJ 2021, 9, e12147. [Google Scholar] [CrossRef]
- Walian, P.J.; Hang, B.; Mao, J.H. Prognostic significance of FAM83D gene expression across human cancer types. Oncotarget 2016, 7, 3332–3340. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, Y.; Guo, L.; Wang, Y.; Miao, T.; Ma, L.; Wei, Q.; Lin, X.; Mao, J.H.; Zhang, P. The FBXW7-binding sites on FAM83D are potential targets for cancer therapy. Breast Cancer Res. 2024, 26, 37. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Huang, Z.; Qian, X.; Wang, Y.; Fang, C.; Chen, R.; Zhang, X.; Xiao, Z.; Wang, Q.; Yu, B.; et al. Pan-cancer analysis of the FAM83 family and its association with prognosis and tumor microenvironment. Front. Genet. 2022, 13, 919559. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Jimenez, N.; Sklias, A.; Ecsedi, S.; Cahais, V.; Degli-Esposti, D.; Jay, A.; Ancey, P.B.; Woo, H.D.; Hernandez-Vargas, H.; Herceg, Z. Lowly methylated region analysis identifies EBF1 as a potential epigenetic modifier in breast cancer. Epigenetics 2017, 12, 964–972. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Guo, W.; Dong, B.; Wang, Y.; Deng, P.; Wang, C.; Liu, J.; Zhang, Q.; Grosschedl, R.; Yu, Z.; et al. EBF1 promotes triple-negative breast cancer progression by surveillance of the HIF1alpha pathway. Proc. Natl. Acad. Sci. USA 2022, 119, e2119518119. [Google Scholar] [CrossRef] [PubMed]
- Normann, L.S.; Aure, M.R.; Leivonen, S.K.; Haugen, M.H.; Hongisto, V.; Kristensen, V.N.; Maelandsmo, G.M.; Sahlberg, K.K. MicroRNA in combination with HER2-targeting drugs reduces breast cancer cell viability in vitro. Sci. Rep. 2021, 11, 10893. [Google Scholar] [CrossRef]
- Zheng, F.; Du, F.; Qian, H.; Zhao, J.; Wang, X.; Yue, J.; Hu, N.; Si, Y.; Xu, B.; Yuan, P. Expression and clinical prognostic value of m6A RNA methylation modification in breast cancer. Biomark. Res. 2021, 9, 28. [Google Scholar] [CrossRef]
- Vitiello, M.; Valentino, T.; De Menna, M.; Crescenzi, E.; Francesca, P.; Rea, D.; Arra, C.; Fusco, A.; De Vita, G.; Cerchia, L. PATZ1 is a target of miR-29b that is induced by Ha-Ras oncogene in rat thyroid cells. Sci. Rep. 2016, 6, 25268. [Google Scholar] [CrossRef]
- Grassilli, S.; Bertagnolo, V.; Brugnoli, F. Mir-29b in Breast Cancer: A Promising Target for Therapeutic Approaches. Diagnostics 2022, 12, 2139. [Google Scholar] [CrossRef]
- Rhee, J.K.; Kim, K.; Chae, H.; Evans, J.; Yan, P.; Zhang, B.T.; Gray, J.; Spellman, P.; Huang, T.H.; Nephew, K.P.; et al. Integrated analysis of genome-wide DNA methylation and gene expression profiles in molecular subtypes of breast cancer. Nucleic Acids Res. 2013, 41, 8464–8474. [Google Scholar] [CrossRef]
- List, M.; Hauschild, A.C.; Tan, Q.; Kruse, T.A.; Mollenhauer, J.; Baumbach, J.; Batra, R. Classification of breast cancer subtypes by combining gene expression and DNA methylation data. J. Integr. Bioinform. 2014, 11, 236. [Google Scholar] [CrossRef]
- Bichindaritz, I.; Liu, G.; Bartlett, C. Integrative survival analysis of breast cancer with gene expression and DNA methylation data. Bioinformatics 2021, 37, 2601–2608. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.M.; Selvarajoo, K. Advancing drug-response prediction using multi-modal and -omics machine learning integration (MOMLIN): A case study on breast cancer clinical data. Brief. Bioinform. 2024, 25, bbae300. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zhang, W.; Cao, H.; Li, G.; Du, W. Classifying Breast Cancer Subtypes Using Deep Neural Networks Based on Multi-Omics Data. Genes 2020, 11, 888. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Chae, H. moBRCA-net: A breast cancer subtype classification framework based on multi-omics attention neural networks. BMC Bioinform. 2023, 24, 169. [Google Scholar] [CrossRef]
- Huang, Y.; Zeng, P.; Zhong, C. Classifying breast cancer subtypes on multi-omics data via sparse canonical correlation analysis and deep learning. BMC Bioinform. 2024, 25, 132. [Google Scholar] [CrossRef]

| Models | Accuracy | Specificity | Sensitivity | F1 Score | AUROC |
|---|---|---|---|---|---|
| SVM | 0.904 | 0.960 | 0.888 | 0.894 | 0.983 |
| LR | 0.905 | 0.961 | 0.889 | 0.895 | 0.984 |
| RF | 0.862 | 0.939 | 0.824 | 0.846 | 0.970 |
| NB | 0.858 | 0.941 | 0.833 | 0.842 | 0.969 |
| AdaBoost | 0.843 | 0.935 | 0.819 | 0.831 | 0.937 |
| MLP | 0.896 | 0.957 | 0.874 | 0.882 | 0.981 |
| Models | Accuracy | Specificity | Sensitivity | F1 Score | AUROC |
|---|---|---|---|---|---|
| GSE96058 (Illumina Nextseq 500) | 0.901 | 0.957 | 0.856 | 0.872 | 0.983 |
| GSE81538 | 0.888 | 0.960 | 0.904 | 0.896 | 0.985 |
| GSE135298 | 0.878 | 0.955 | 0.845 | 0.824 | 0.991 |
| Average | 0.889 | 0.957 | 0.868 | 0.864 | 0.986 |
| Motifs | E-Value | Known or Similar Motifs |
|---|---|---|
 | 6.0 × 10−05 | EBF1 |
 | 1.5 × 10−03 | - |
 | 5.4 × 10−03 | ZNF770/PATZ1 |
 | 9.9 × 10−03 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.-W.; Rhee, J.-K. Integrative Analysis of ATAC-Seq and RNA-Seq through Machine Learning Identifies 10 Signature Genes for Breast Cancer Intrinsic Subtypes. Biology 2024, 13, 799. https://doi.org/10.3390/biology13100799
Park J-W, Rhee J-K. Integrative Analysis of ATAC-Seq and RNA-Seq through Machine Learning Identifies 10 Signature Genes for Breast Cancer Intrinsic Subtypes. Biology. 2024; 13(10):799. https://doi.org/10.3390/biology13100799
Chicago/Turabian StylePark, Jeong-Woon, and Je-Keun Rhee. 2024. "Integrative Analysis of ATAC-Seq and RNA-Seq through Machine Learning Identifies 10 Signature Genes for Breast Cancer Intrinsic Subtypes" Biology 13, no. 10: 799. https://doi.org/10.3390/biology13100799
APA StylePark, J.-W., & Rhee, J.-K. (2024). Integrative Analysis of ATAC-Seq and RNA-Seq through Machine Learning Identifies 10 Signature Genes for Breast Cancer Intrinsic Subtypes. Biology, 13(10), 799. https://doi.org/10.3390/biology13100799








