Correlation between the Density of Acipenser sinensis and Its Environmental DNA
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. The Specific Primers and Probes for A. sinensis
2.2. Experiment Design and Sample Collection
2.3. eDNA Extraction and PCR Amplification
2.4. Preparation of the Recombinant Plasmid Standard
2.5. Amplification of Fluorescent Quantitative PCR
2.6. Data Analysis
3. Results
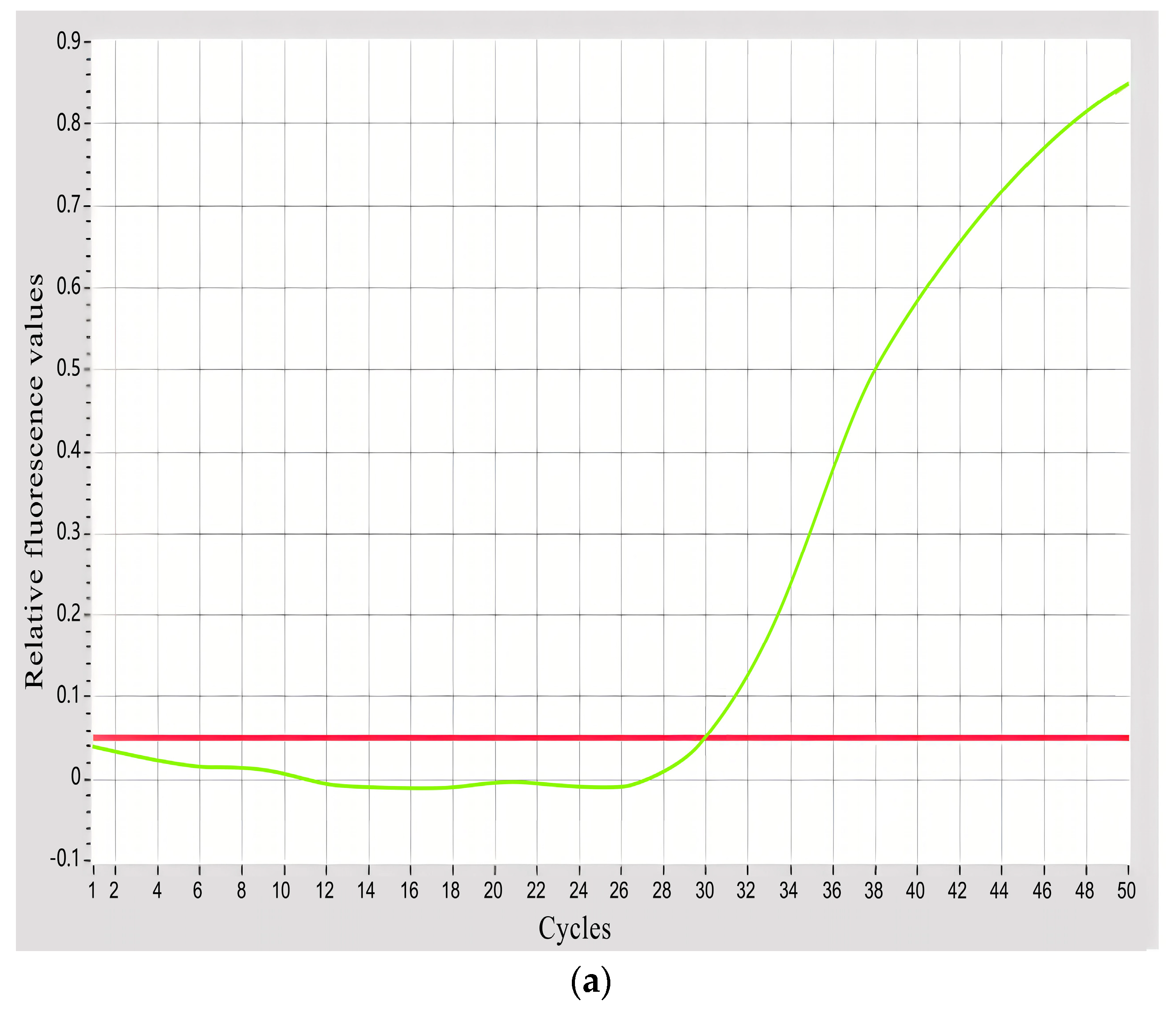
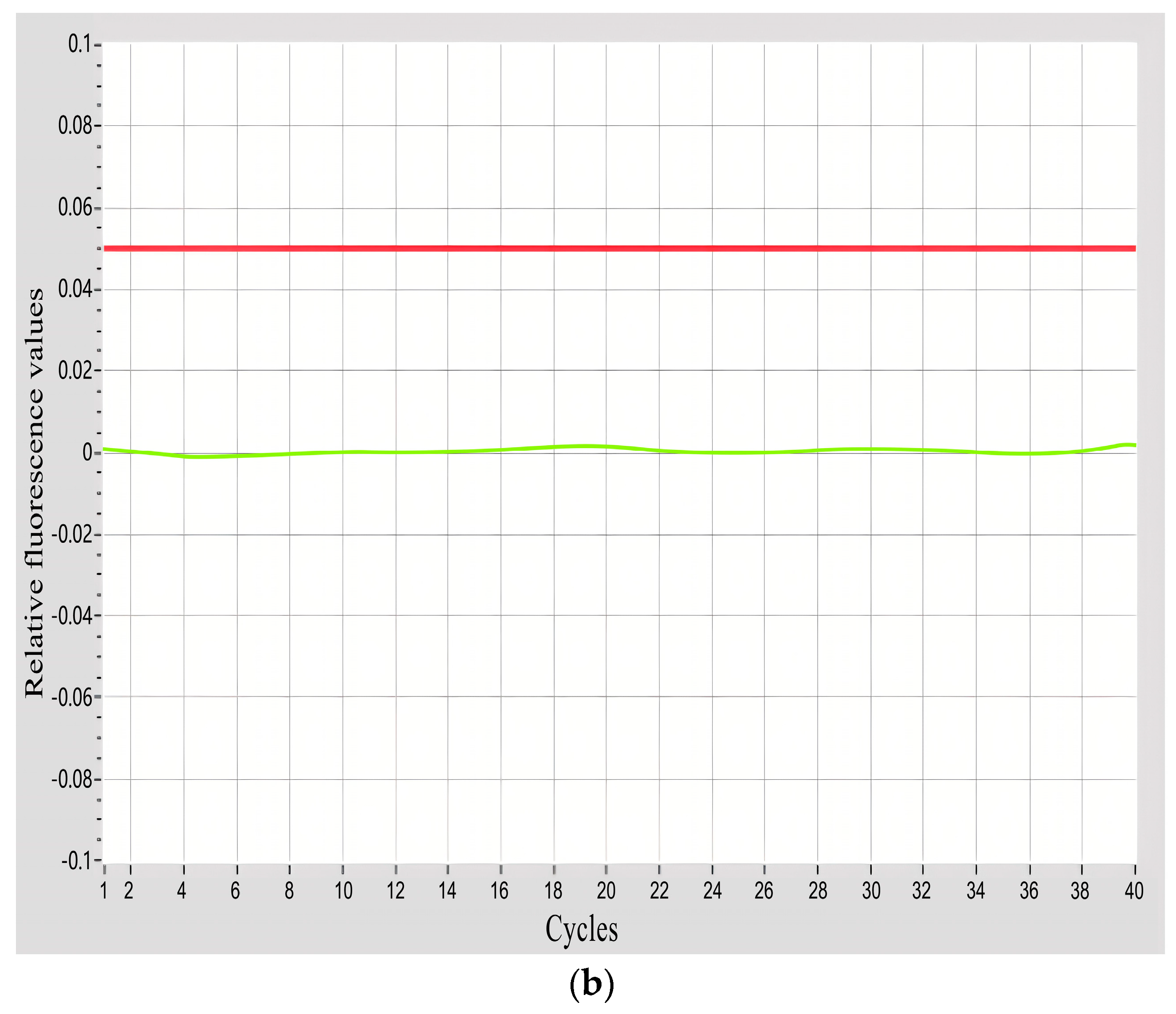
3.1. Specificity Verification of Primers and Probe
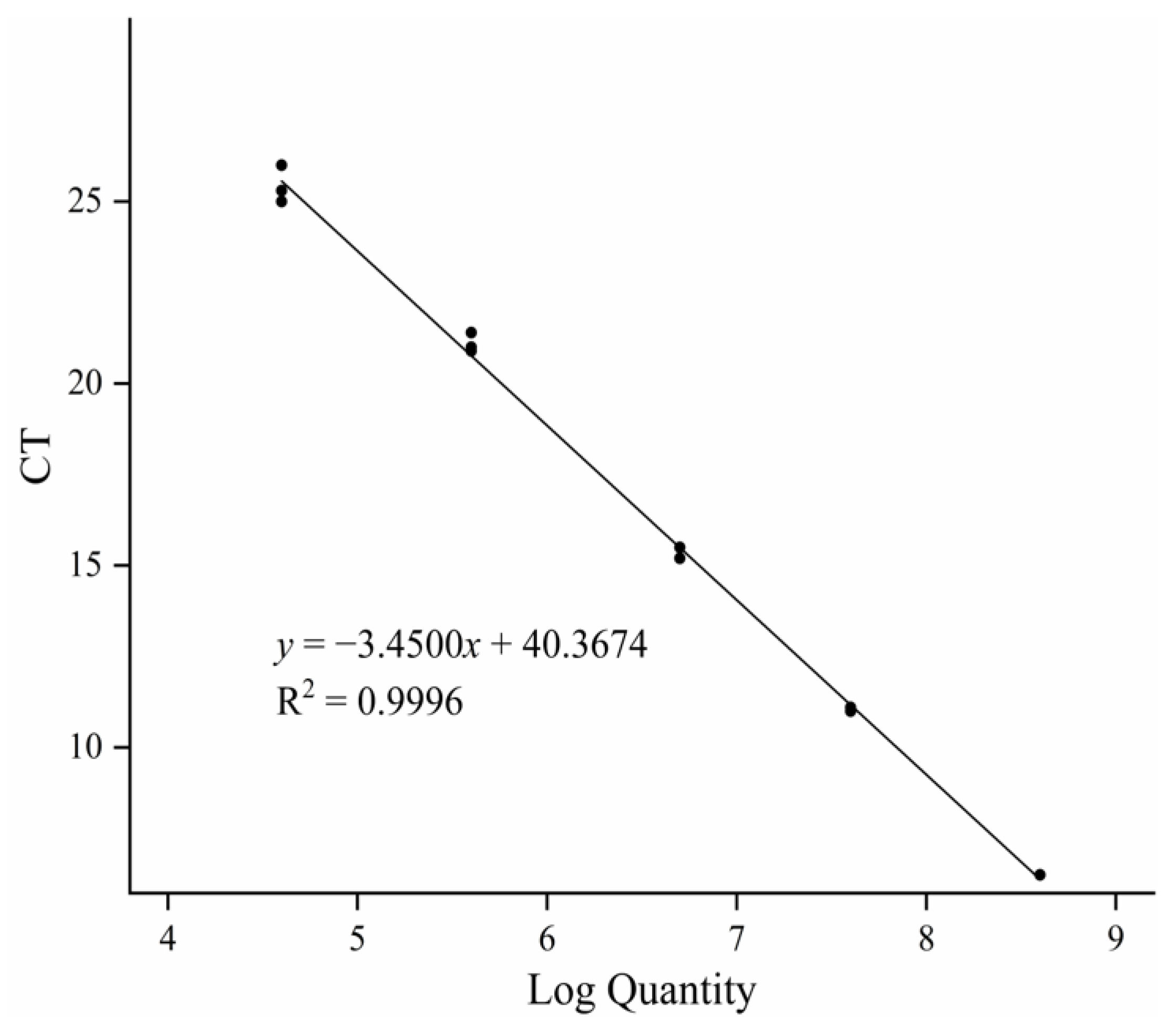
3.2. Establishment of Standard Curve and Regression Equation
3.3. Changing Trend of eDNA Content of A. sinensis
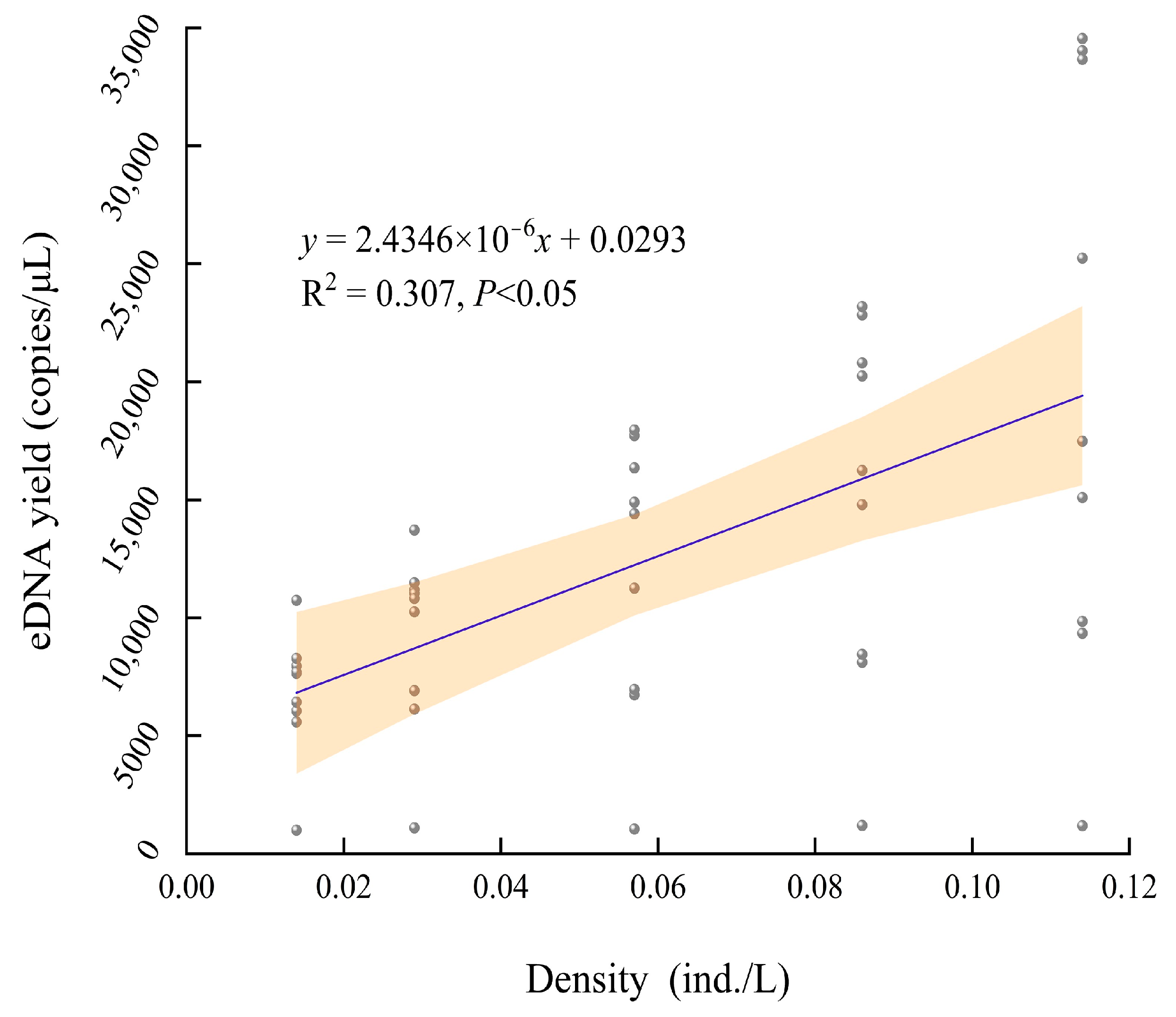
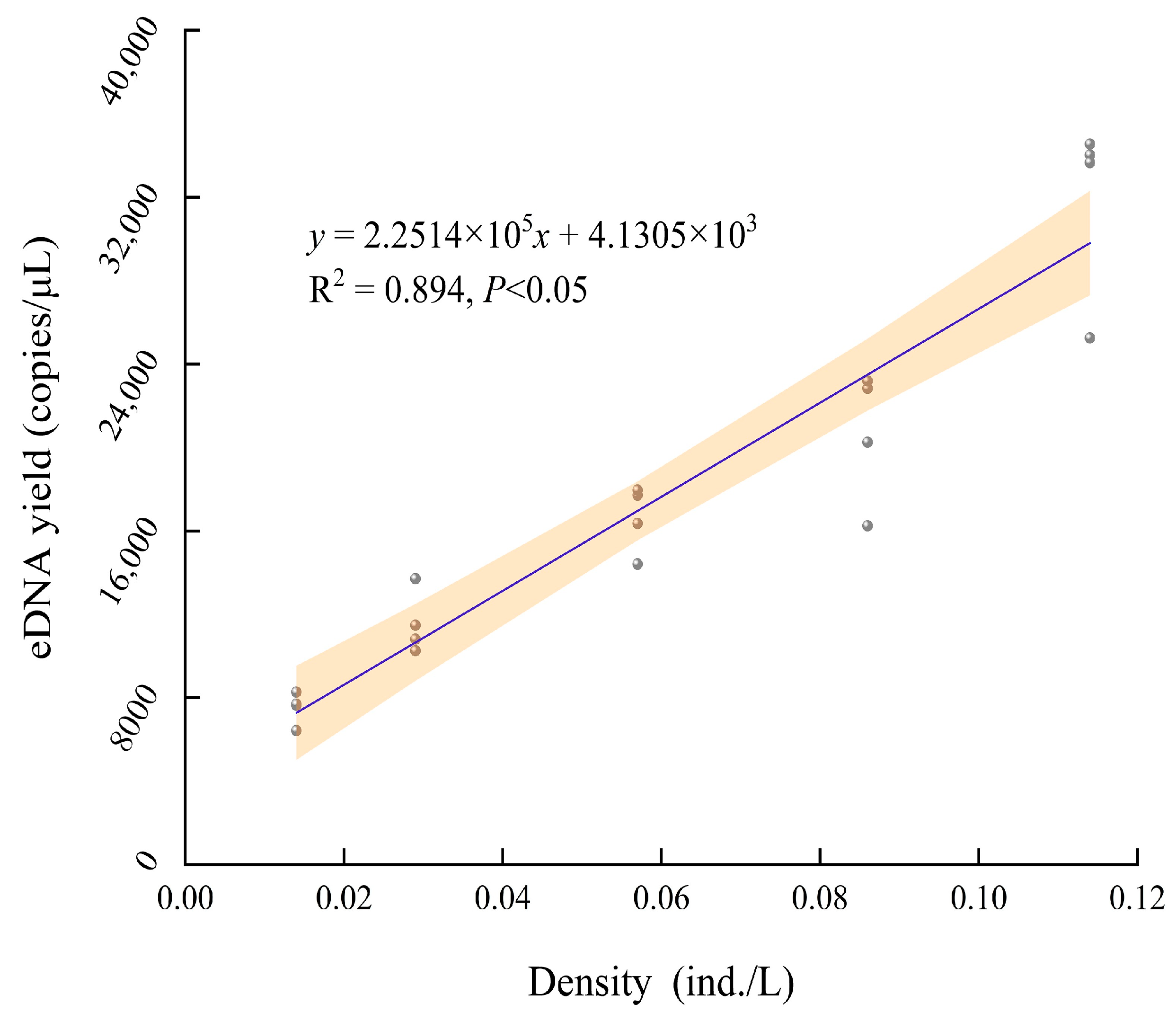
3.4. Correlation between eDNA Content and Density of A. sinensis
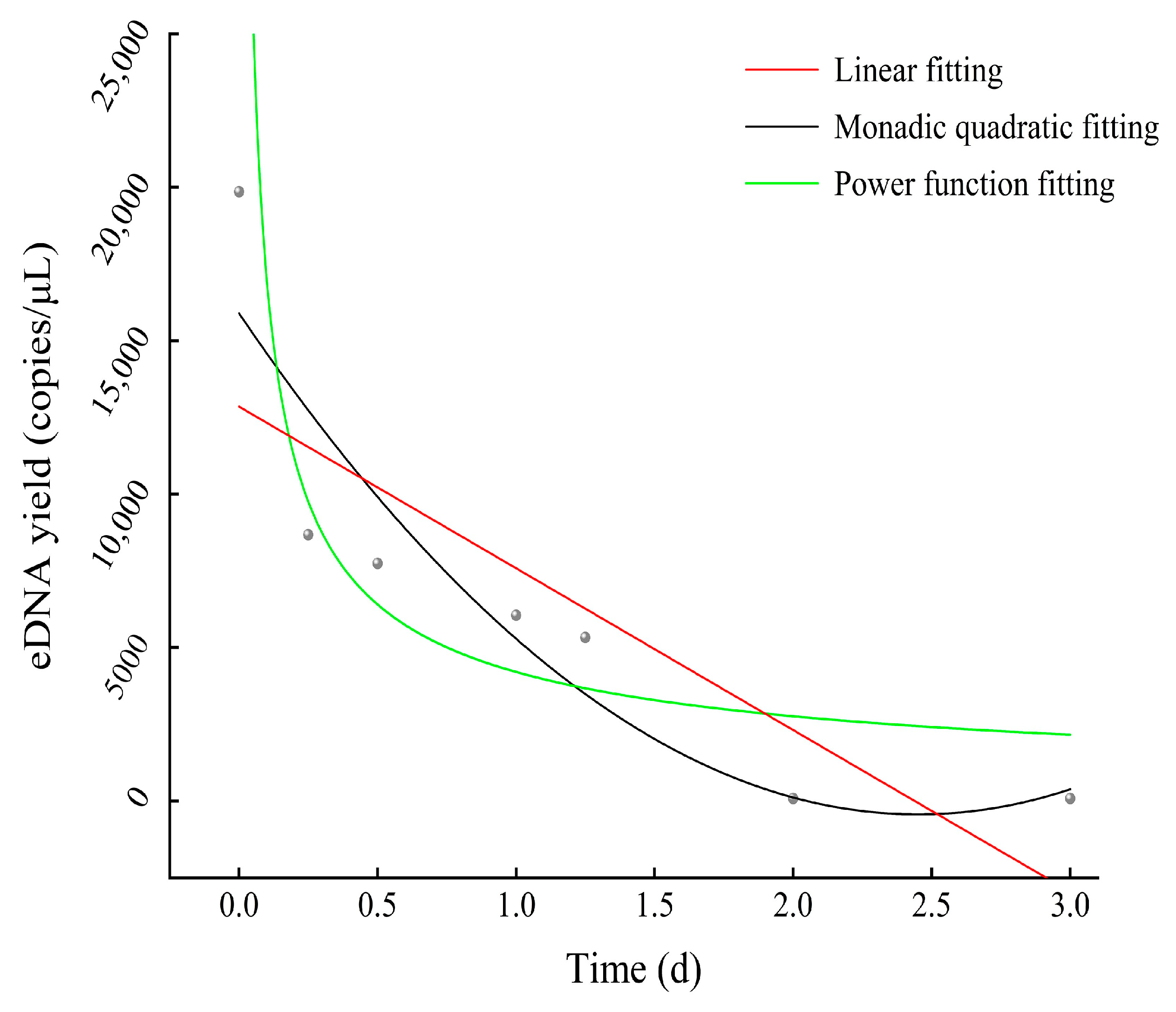
3.5. Degradation Results of eDNA of A. sinensis
4. Discussion
4.1. Changes in eDNA Content
4.2. Correlation between eDNA Content and Density
4.3. Degradation of eDNA in Water
4.4. Biomass Assessment of eDNA
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, C.S.; Xu, Y.G.; Deng, Z.L.; Yu, Z.T. Observation on the reproductive glands of adult Acipenser sinensis gray in Changjiang River below Gezhouba Dam. Acta Hydrobiol. Sin. 1985, 9, 164–170. (In Chinese) [Google Scholar]
- Gao, X.; Brosse, S.; Chen, Y.B.; Lek, S.; Chang, J.B. Effects of damming on population sustainability of Chinese sturgeon, Acipenser sinensis: Evaluation of optimal conservation measures. Environ. Biol. Fishes 2009, 86, 325–336. [Google Scholar] [CrossRef]
- Gao, X.; Lin, P.C.; Li, M.Z.; Duan, Z.H.; Liu, H.Z. Impact of the Three Gorges Dam on the spawning stock and natural reproduction of Chinese sturgeon in Changjiang River, China. Chin. J. Oceanol. Limnol. 2016, 34, 894–901. [Google Scholar] [CrossRef]
- Zhuang, P.; Zhao, F.; Zhang, T.; Chen, Y.; Liu, J.Y.; Zhang, L.Z.; Kynard, B. New evidence may support the persistence and adaptability of the near-extinct Chinese sturgeon. Biol. Conserv. 2016, 193, 66–69. [Google Scholar] [CrossRef]
- Yangtze River Basin Fishery Administration Office of the Ministry of Agriculture and Rural Affairs; the Changjiang Water Resources Commission of the Ministry of Water Resources; the Yangtze River Basin ecological Environment Supervision Administration of the Ministry of Ecology and Environment; The Changjiang River Adminstration of Navigational Affairs, Mot. The Bulletin of Aquatic Biological Resources and Habitat Status in the Yangtze River Basin; Ministry of Agriculture and Rural Affairs of the People’s Republic of China: Beijing, China, 2022. (In Chinese)
- Bohmann, K.; Evans, A.; Gilbert, M.T.P.; Carvalho, G.R.; Creer, S.; Knapp, M.; Yu, D.W.; Bruyn, M.D. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 2014, 29, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Anish, K.; Daniel, W.; Thomas, N.; Liza, B.; Claire, O.; Riley, P.; Ashley, C.; Katie, L.; Lauren, S. Quantification of Environmental DNA (eDNA) shedding and decay rates for three commercially harvested fish species and comparison between eDNA detection and trawl catches. Environ. DNA 2021, 3, 1142–1155. [Google Scholar]
- Mytilineou, C.; Herrmann, B.; Smith, C.J.; Mantopoulou, P.D.; Anastasopoulou, A.; Siapatis, A.; Sala, A.; Megalofonou, P.; Papadopoulou, N.; Vassilopoulou, V.; et al. Impacts on biodiversity from codend and fisher selection in bottom trawl fishing. Front. Mar. Sci. 2022, 9, 1021467. [Google Scholar] [CrossRef]
- Zhou, H.X.; Gan, W.X. Advances in fish labeling technology and its application in artificial proliferation and release. Hubei Agric. Sci. 2017, 56, 1206–1210. (In Chinese) [Google Scholar]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef]
- Rourke, M.L.; Fowler, A.M.; Hughes, J.M.; Broadhurst, M.K.; Dibattista, J.D.; Fielder, S.; Walburn, J.W.; Furlan, E.M. Environmental DNA (eDNA) as a tool for assessing fish biomass: A review of approaches and future considerations for resource surveys. Environ. DNA 2022, 4, 9–33. [Google Scholar] [CrossRef]
- Pfleger, M.O.; Rider, S.J.; Johnston, C.E.; Janosik, A.M. Saving the doomed: Using eDNA to aid in detection of rare sturgeon for conservation (Acipenseridae). Glob. Ecol. Conserv. 2016, 8, 99–107. [Google Scholar] [CrossRef]
- Mizumoto, H.; Mitsuzuka, T.; Araki, H. An environmental DNA survey on distribution of an endangered salmonid species, Parahucho perryi, in Hokkaido, Japan. Front. Ecol. Evol. 2020, 8, 569425. [Google Scholar] [CrossRef]
- Yu, D.; Shen, Z.Y.; Chang, T.; Li, S.; Liu, H.Z. Using environmental DNA methods to improve detectability in an endangered sturgeon (Acipenser sinensis) monitoring program. BMC Ecol. Evol. 2021, 21, 216. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Zhu, B.; Shi, F.; Shao, K.; Que, Y.F.; Li, W.T.; Li, W.; Jiao, W.J.; Tian, H.; Xu, D.M.; et al. Monitoring seasonal distribution of an endangered anadromous sturgeon in a large river using environmental DNA. Sci. Nat. 2018, 105, 62. [Google Scholar] [CrossRef] [PubMed]
- Spear, M.J.; Embke, H.S.; Krysan, P.J.; Zanden, M.J.V. Application of eDNA as a tool for assessing fish population abundance. Environ. DNA 2021, 3, 83–91. [Google Scholar] [CrossRef]
- Xin, Y.; Guo, Y.; Sun, M.J.; Yu, G.; Ma, Z.H.; Pei, K.; Qin, C.X. Optimal conditions to quantify the relationship between eDNA concentration and biomass in Acanthopagrus latus. Water 2022, 14, 3521. [Google Scholar] [CrossRef]
- Pont, D.; Rocle, M.; Valentini, A.; Civade, R.; Jean, P.; Maire, A.; Roset, N.; Schabuss, M.; Zornig, H.; Dejean, T. Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Sci. Rep. 2018, 8, 10361. [Google Scholar] [CrossRef]
- Zhang, J.M.; Ding, R.Y.; Wang, Y.R.; Wen, J.T. Experimental study on the response relationship between environmental DNA concentration and biomass of Schizothorax prenanti in still water. Front. Ecol. Evol. 2022, 10, 972680. [Google Scholar] [CrossRef]
- Takahara, T.; Minamoto, T.; Yamanaka, H.; Doi, H.; Kawabata, Z.I. Estimation of fish biomass using environmental DNA. PLoS ONE 2012, 7, e35868. [Google Scholar] [CrossRef]
- Itakura, H.; Wakiya, R.; Yamamoto, S.; Kaifu, K.; Sato, T.; Minamoto, T. Environmental DNA analysis reveals the spatial distribution, abundance, and biomass of Japanese eels at the river-basin scale. Aquat. Conserv. 2019, 29, 361–373. [Google Scholar] [CrossRef]
- Doi, H.; Katano, I.; Sakata, Y.; Rio, S.; Toshihiro, K.; Mariko, N.; Kousuke, I.; Koki, Y.; Koji, T. Detection of an endangered aquatic heteropteran using environmental DNA in a wetland ecosystem. R. Soc. Open Sci. 2017, 4, 170568. [Google Scholar] [CrossRef] [PubMed]
- Shogren, A.J.; Tank, J.L.; Egan, S.P.; Diogo, B.; Tenna, R. Riverine distribution of mussel environmental DNA reflects a balance among density, transport, and removal processes. Freshw. Biol. 2019, 64, 1467–1479. [Google Scholar] [CrossRef]
- Karlsson, E.; Ogonowski, M.; Sundblad, G.; Sundin, J.; Svensson, O.; Nousiainen, I.; Vasemägi, A. Strong positive relationships between eDNA concentrations and biomass in juvenile and adult pike (Esox lucius) under controlled conditions: Implications for monitoring. Environ. DNA 2022, 4, 881–893. [Google Scholar] [CrossRef]
- Sint, D.; Kolp, B.; Rennstam, R.O.; Füreder, L.; Traugott, M. The amount of environmental DNA increases with freshwater crayfish density and over time. Environ. DNA 2022, 4, 417–424. [Google Scholar] [CrossRef]
- Minamoto, T.; Fukuda, M.; Katsuhara, K.R.; Fujiwara, A.; Hidaka, S.; Yamamoto, S.; Takahashi, K.; Masuda, R. Environmental DNA reflects spatial and temporal jellyfish distribution. PLoS ONE 2017, 12, e0173073. [Google Scholar] [CrossRef] [PubMed]
- Jerde, C.L.; Chadderton, W.L.; Mahon, A.R.; Renshaw, M.A.; Corush, J.; Budny, M.L.; Mysorekar, S.; Lodge, D.M. Detection of Asian carp DNA as part of a great lakes basin-wide surveillance program. Can. J. Fish. Aquat. Sci. 2013, 70, 522–526. [Google Scholar] [CrossRef]
- Benoit, N.P.; Robinson, K.M.; Kellogg, C.T.E.; Lemay, M.A.; Hunt, B.P.V. Using qPCR of environmental DNA (eDNA) to estimate the biomass of juvenile Pacific salmon (Oncorhynchus spp.). Environ. DNA 2023, 5, 683–696. [Google Scholar] [CrossRef]
- Mauvisseau, Q.; Parrondo, M.; Fernández, M.P.; García, L.; Martínez, J.L.; García-Vázquez, E.; Borrell, Y.J. On the way for detecting and quantifying elusive species in the sea: The Octopus vulgaris case study. Fish. Res. 2017, 191, 41–48. [Google Scholar] [CrossRef]
- Gao, T.X.; Chen, Z.; Wang, X.Y.; Zhang, H.B.; Shi, H.L. Correlation between the density of cultured Sebastiscus marmoratus and its environmental DNA. Chin. J. Hydrobiol. 2022, 46, 1007–1015. (In Chinese) [Google Scholar]
- Lacoursière-Roussel, A.; Rosabal, M.; Bernatchez, L. Estimating fish abundance and biomass from eDNA concentrations: Variability among capture methods and environmental conditions. Mol. Ecol. Resour. 2016, 16, 1401–1414. [Google Scholar] [CrossRef]
- Stoeckle, M.Y.; Adolf, J.; Charlop-Powers, Z.; Dunton, K.J.; Hinks, G.; VanMorter, S.M. Trawl and eDNA assessment of marine fish diversity, seasonality, and relative abundance in coastal New Jersey, USA. ICES J. Mar. Sci. 2021, 78, 293–304. [Google Scholar] [CrossRef]
- Hansen, B.K.; Bekkevold, D.; Clausen, L.W.; Nielsen, E.E. The sceptical optimist: Challenges and perspectives for the application of environmental DNA in marine fisheries. Fish Fish. 2018, 19, 751–768. [Google Scholar] [CrossRef]
- Stewart, K.A. Understanding the effects of biotic and abiotic factors on sources of aquatic environmental DNA. Biodivers. Conserv. 2019, 28, 983–1001. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Strickler, K.M.; Pilliod, D.S. Moving environmental DNA methods from concept to practice for monitoring aquatic macroorganisms. Biol. Conserv. 2015, 183, 1–3. [Google Scholar] [CrossRef]
- Pilliod, D.S.; Goldberg, C.S.; Arkle, R.S.; Waits, L.P. Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Can. J. Fish. Aquat. Sci. 2013, 70, 1123–1130. [Google Scholar] [CrossRef]
- Baldigo, B.P.; Sporn, L.A.; George, S.D.; Ball, J.A. Efficacy of environmental DNA to detect and quantify brook trout populations in headwater streams of the Adirondack Mountains, New York. Trans. Am. Fish. Soc. 2017, 146, 99–111. [Google Scholar] [CrossRef]
- Yamamoto, S.; Minami, K.; Fukaya, K.; Takahashi, K.; Sawada, H.; Murakami, H.; Tsuji, S.; Hashizume, H.; Kubonaga, S.; Horiuchi, T.; et al. Environmental DNA as a ‘snapshot’of fish distribution: A case study of Japanese jack mackerel in Maizuru Bay, Sea of Japan. PLoS ONE 2016, 11, e0149786. [Google Scholar]
- Peltier, H.; Dabin, W.; Daniel, P.; Canneyt, O.V.; Dorémus, G.; Huon, M.; Ridoux, V. The significance of stranding data as indicators of cetacean populations at sea: Modelling the drift of cetacean carcasses. Ecol. Indic. 2012, 18, 278–290. [Google Scholar] [CrossRef]
- Li, M.; Shan, X.J.; Wang, W.J.; Ding, X.S.; Dai, F.G.; Lü, D.; Wu, H.H. Studying the retention time of Fenneropenaeus chinensis eDNA in water. Prog. Fish. Sci. 2020, 41, 51–57. (In Chinese) [Google Scholar]
- Rees, H.C.; Maddison, B.C.; Middleditch, D.J.; Patmore, J.R.M.; Gough, K.C. The detection of aquatic animal species using environmental DNA–a review of eDNA as a survey tool in ecology. J. Appl. Ecol. 2014, 51, 1450–1459. [Google Scholar] [CrossRef]
- Roussel, J.M.; Paillisson, J.M.; Treguier, A.; Petit, E. The downside of eDNA as a survey tool in water bodies. J. Appl. Ecol. 2015, 52, 823–826. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 2016, 17, 1–17. [Google Scholar] [CrossRef]
- Geerts, A.N.; Boets, P.; Van den Heede, S.; Goethals, P.; Van der Heyden, C. A search for standardized protocols to detect alien invasive crayfish based on environmental DNA (eDNA): A lab and field evaluation. Ecol. Indic. 2018, 84, 564–572. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 2014, 48, 1819–1827. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Sepulveda, A.; Ray, A.; Baumgardt, J.; Waits, L.P. Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrgus antipodarum). Freshw. Sci. 2013, 32, 792–800. [Google Scholar] [CrossRef]
- Cerco, C.F.; Schultz, M.T.; Noel, M.R.; Skahill, B.; Kim, S.C. A fate and transport model for Asian carp environmental DNA in the Chicago area waterways system. J. Great Lakes Res. 2018, 44, 813–823. [Google Scholar] [CrossRef]
- Strickler, K.M.; Fremier, A.K.; Goldberg, C.S. Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol. Conserv. 2015, 183, 85–92. [Google Scholar] [CrossRef]
- Yan, H.G.; Dong, Z.L.; Ma, T.T.; Zhang, L.B.; Wang, X.Y.; Ye, H.; Yao, W.Z.; He, W.P. Detection and biomass assessment of Procypris rabaudi based on environmental DNA. J. Fish. China 2020, 46, 1018–1026. (In Chinese) [Google Scholar]
- Wood, S.A.; Biessy, L.; Latchford, J.L.; Zaiko, A.; von Ammon, U.; Audrezet, F.; Cristescu, M.E.; Pochon, X. Release and degradation of environmental DNA and RNA in a marine system. Sci. Total Environ. 2020, 704, 135314. [Google Scholar] [CrossRef]
- Rice, C.J.; Larson, E.R.; Taylor, C.A. Environmental DNA detects a rare large river crayfish but with little relation to local abundance. Freshw. Biol. 2018, 63, 443–455. [Google Scholar] [CrossRef]

| Time | Linear Fitting | R2 (p < 0.05) 1 |
|---|---|---|
| 6 h | y = 4955.51x + 529.96 | 0.983 |
| 12 h | y = 5511.27x + 508.16 | 0.936 |
| 1 d | y = 7953.28x + 1550.44 | 0.768 |
| 2 d | y = 9626.96x + 704.68 | 0.867 |
| 3 d | y = 4966.78x + 2583.90 | 0.986 |
| 4 d | y = 2455.37x + 3372.55 | 0.834 |
| 5 d | y = 3774.58x + 3566.41 | 0.979 |
| 7 d | y = 5354.20x + 3335.19 | 0.971 |
| Items | Linear Function | Monadic Quadratic Function | Power Function |
|---|---|---|---|
| AIC | 127.8203 | 137.0612 | 108.2885 |
| BIC | 119.6581 | 116.8449 | 95.6638 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Feng, G.; Zhu, J.; Jiang, W. Correlation between the Density of Acipenser sinensis and Its Environmental DNA. Biology 2024, 13, 19. https://doi.org/10.3390/biology13010019
Wang X, Feng G, Zhu J, Jiang W. Correlation between the Density of Acipenser sinensis and Its Environmental DNA. Biology. 2024; 13(1):19. https://doi.org/10.3390/biology13010019
Chicago/Turabian StyleWang, Xiaojing, Guangpeng Feng, Jiazhi Zhu, and Wei Jiang. 2024. "Correlation between the Density of Acipenser sinensis and Its Environmental DNA" Biology 13, no. 1: 19. https://doi.org/10.3390/biology13010019
APA StyleWang, X., Feng, G., Zhu, J., & Jiang, W. (2024). Correlation between the Density of Acipenser sinensis and Its Environmental DNA. Biology, 13(1), 19. https://doi.org/10.3390/biology13010019






