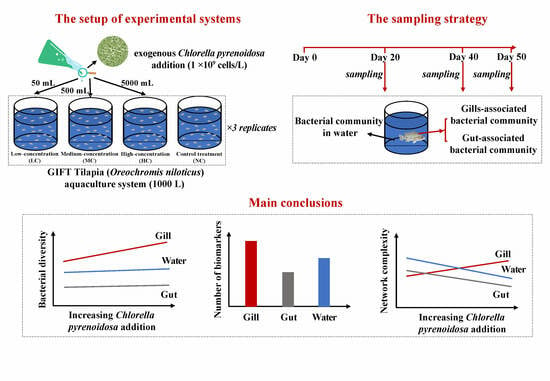
The Gill-Associated Bacterial Community Is More Affected by Exogenous Chlorella pyrenoidosa Addition than the Bacterial Communities of Water and Fish Gut in GIFT Tilapia (Oreochromis niloticus) Aquaculture System
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Sampling Work and Measurements of Water Quality
2.3. DNA Extraction, PCR, and Sequencing
2.4. Statistical Analysis
3. Results
3.1. Effects of the Chlorella pyrenoidosa Addition on Bacterial Community Diversity in Different Microhabitats
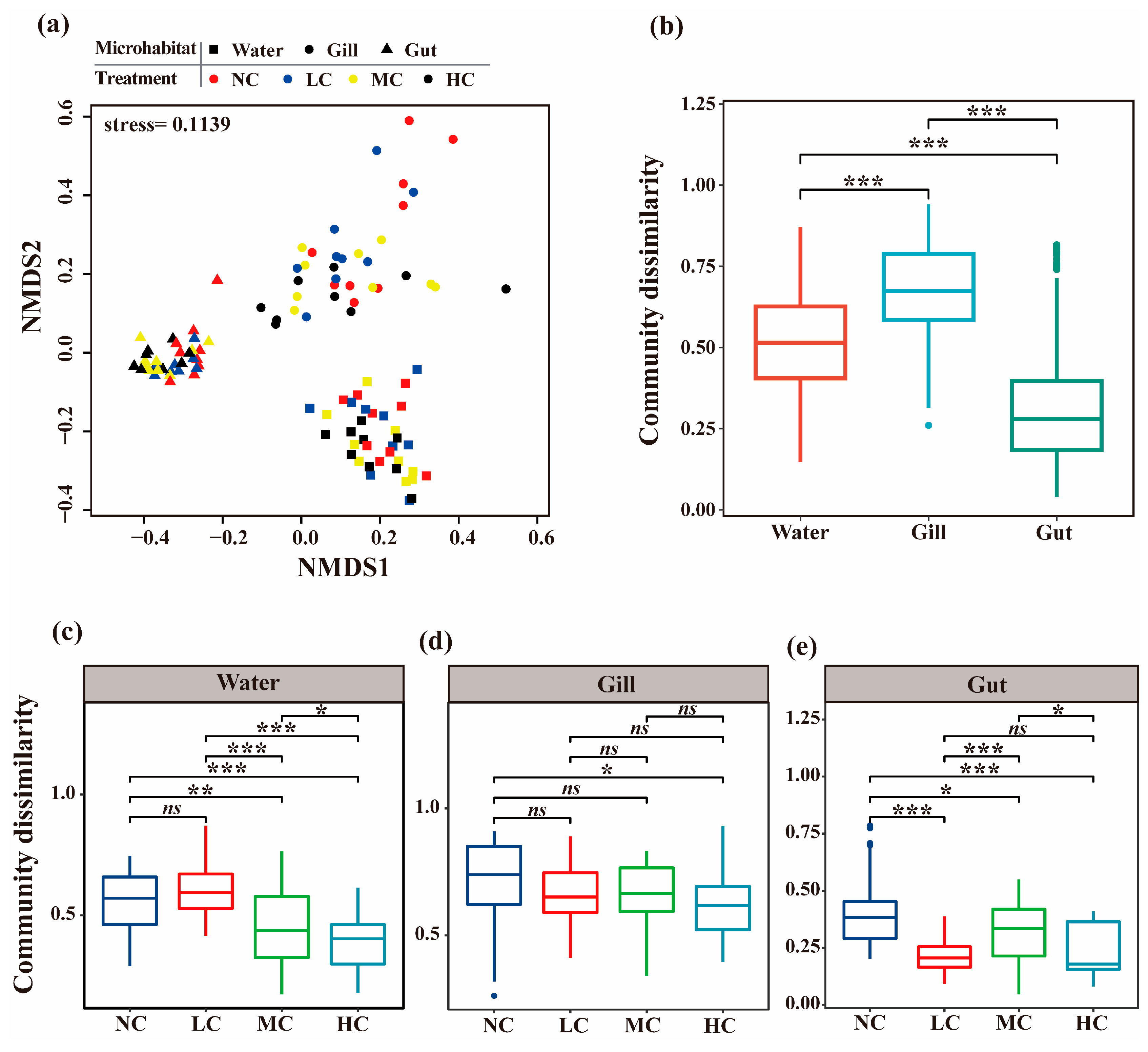
3.2. The Bacterial Community Structure of Different Microhabitats Affected by the Chlorella pyrenoidosa Addition
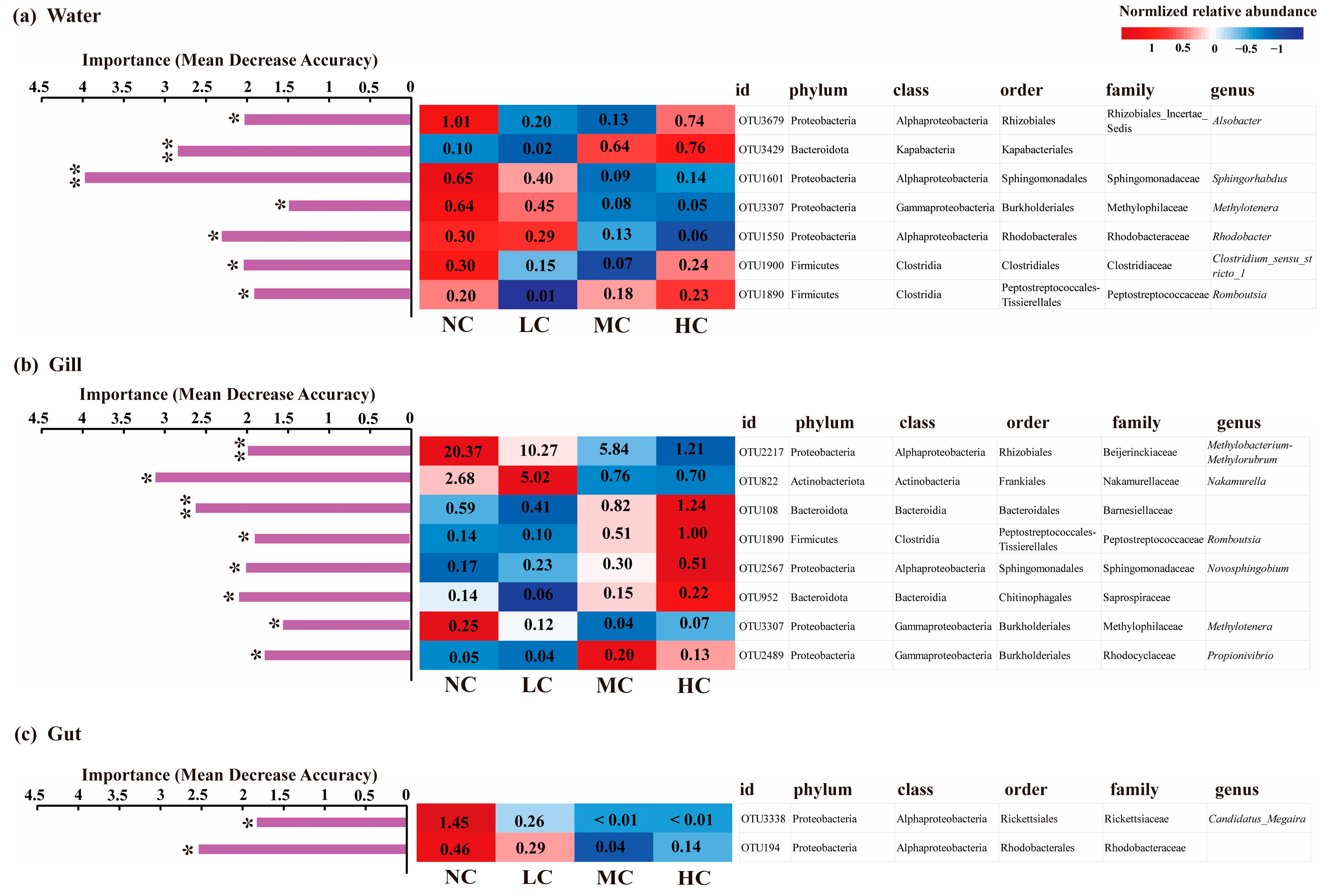
3.3. The Taxonomic Compositions of Bacterial Communities in Different Microhabitats Affected by the Chlorella pyrenoidosa Addition
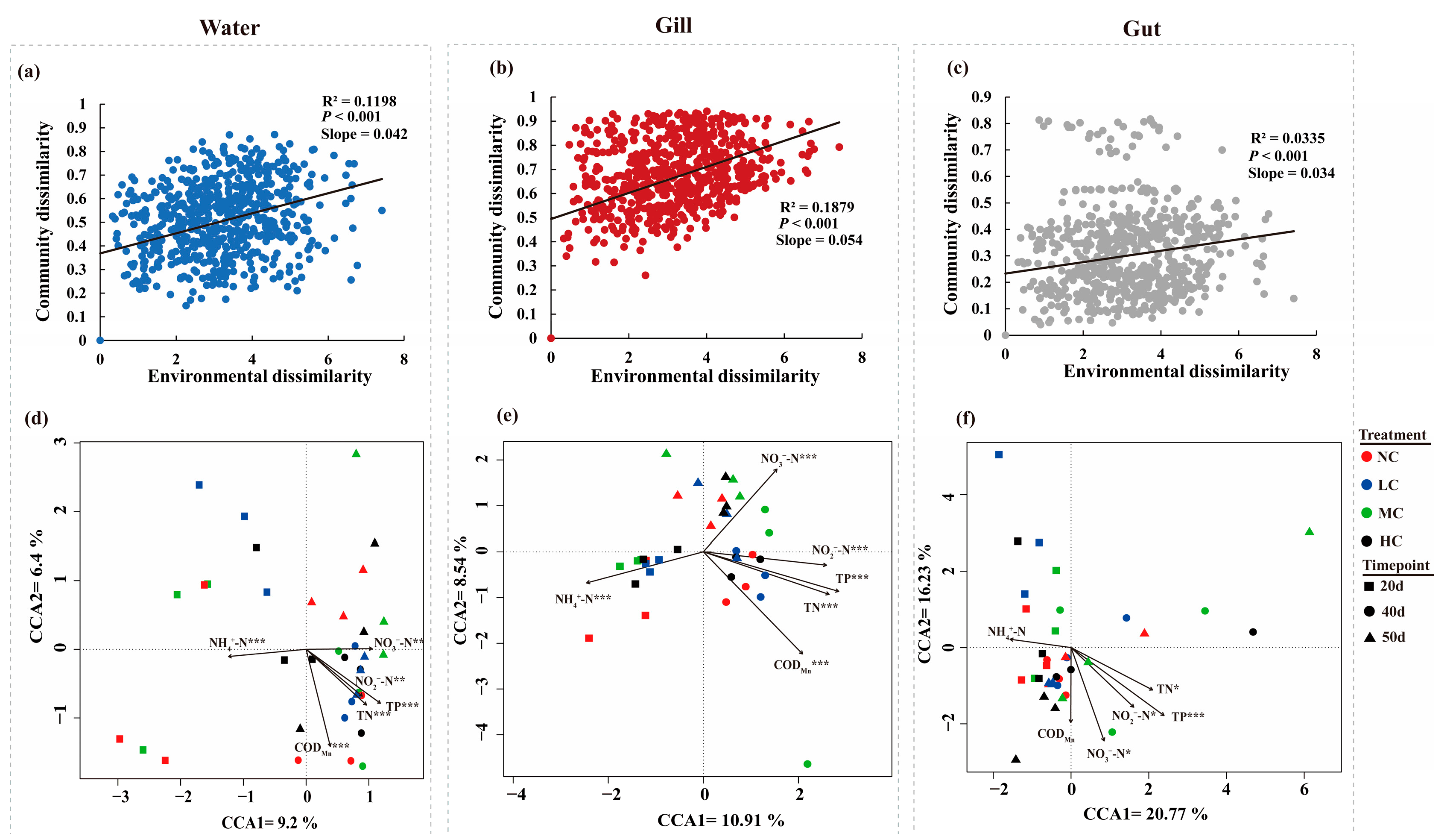
3.4. The Relationships between the Water Quality and Bacterial Communities in Different Microhabitats
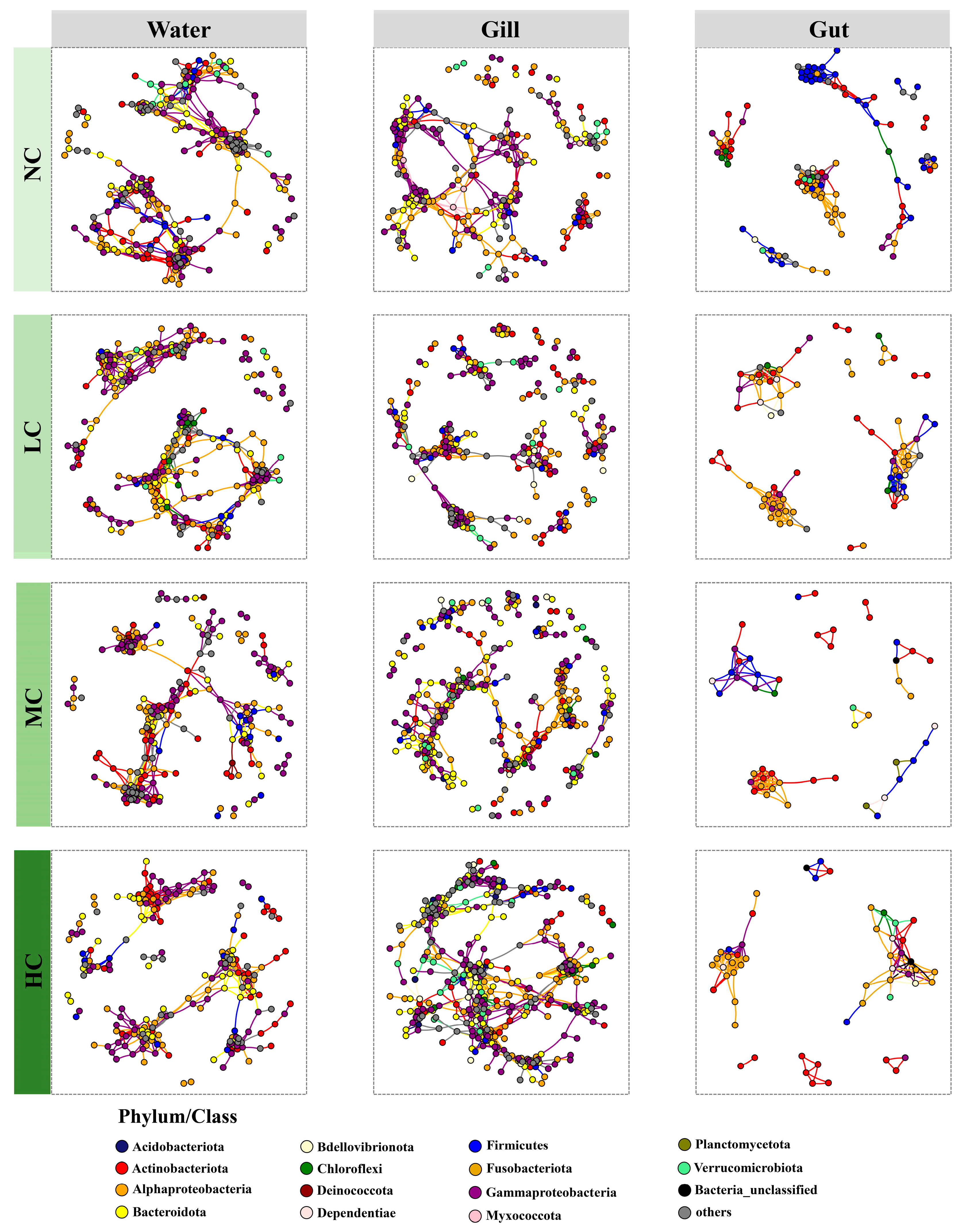
3.5. The Co-Occurrence Patterns of Bacterial Communities in Different Microhabitats Influenced by the Chlorella pyrenoidosa Addition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, L.M.; Li, H.K.; Lu, Q.; Zhou, W.G. Emerging trends of culturing microalgae for fish-rearing environment protection. J. Chem. Technol. Biotechnol. 2021, 96, 31–37. [Google Scholar] [CrossRef]
- Han, P.; Lu, Q.; Fan, L.L.; Zhou, W.G. A review on the use of microalgae for sustainable aquaculture. Appl. Sci. 2019, 9, 2377. [Google Scholar] [CrossRef]
- Nagappan, S.; Das, P.; AbdulQuadir, M.; Thaher, M.; Khan, S.; Mahata, C.; Al-Jabri, H.; Vatland, A.K.; Kumar, G. Potential of microalgae as a sustainable feed ingredient for aquaculture. J. Biotechnol. 2021, 341, 1–20. [Google Scholar] [CrossRef]
- Silfronio Melo, R.C.; de Souza Santos, L.P.; Morais Brito, A.P.; Gouveia, A.d.A.; Marcal, C.; Cavalli, R.O. Use of the microalga Nannochloropsis occulata in the rearing of newborn longsnout seahorse Hippocampus reidi (Syngnathidae) juveniles. Aquac. Res. 2016, 47, 3934–3941. [Google Scholar] [CrossRef]
- de Souza, F.P.; Suzuki de Lima, E.C.; Urrea-Rojas, A.M.; Suphoronski, S.A.; Facimoto, C.T.; Bezerra Junior, J.d.S.; Silva de Oliveira, T.E.; Pereira, U.d.P.; Di Santis, G.W.; Lopes de Oliveira, C.A.; et al. Effects of dietary supplementation with a microalga (Schizochytrium sp.) on the hemato-immunological, and intestinal histological parameters and gut microbiota of Nile tilapia in net cages. PLoS ONE 2020, 15, e0226977. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.F.; Qian, J.; He, Y.; Leng, Y.Y.; Zhou, W.G. Could Chlorella pyrenoidosa be exploited as an alternative nutrition source in aquaculture feed? A study on the nutritional values and anti-nutritional factors. Front. Nutr. 2022, 9, 1069760. [Google Scholar] [CrossRef]
- Xu, W.; Gao, Z.; Qi, Z.T.; Qiu, M.; Peng, J.Q.; Shao, S. Effect of Dietary Chlorella on the Growth Performance and Physiological Parameters of Gibel carp, Carassius auratus gibelio. Turk. J. Fish. Aauatic Sci. 2014, 14, 53–57. [Google Scholar] [CrossRef]
- Que, J.; Rao, Y.; Xu, X.; Ding, L.; Zhang, H.; Zhang, A.; Li, Y. Effects of Chlorella pyrenoidosa on growth performance, immune function, digestive enzyme activities and muscle quality of Ctenopharyngodon idellus. Fish. Sci. 2022. in press (In Chinese) [Google Scholar] [CrossRef]
- Yuan, D.N.; Wang, L.; Wang, H.X.; Miao, R.L.; Wang, Y.L.; Jin, H.; Tan, L.; Wei, C.J.; Hu, Q.; Gong, Y.C. Application of microalgae Scenedesmus acuminatus enhances water quality in rice-crayfish culture. Front. Bioeng. Biotechnol. 2023, 11, 1143622. [Google Scholar] [CrossRef]
- Zhang, Z.M.; Li, D.P.; Xu, W.T.; Tang, R.; Li, L. Microbiome of co-cultured fish exhibits host selection and niche differentiation at the organ scale. Front. Microbiol. 2019, 10, 02576. [Google Scholar] [CrossRef]
- Seymour, J.R.; Amin, S.A.; Raina, J.B.; Stocker, R. Zooming in on the phycosphere: The ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2017, 2, 17065. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.J. Interactions between bacteria and algae in aquatic ecosystems. Annu. Rev. Ecol. Syst. 1982, 13, 291–314. [Google Scholar] [CrossRef]
- Camarena-Gomez, M.T.; Lipsewers, T.; Piiparinen, J.; Eronen-Rasimus, E.; Perez-Quemalinos, D.; Hoikkala, L.; Sobrino, C.; Spilling, K. Shifts in phytoplankton community structure modify bacterial production, abundance and community composition. Aquat. Microb. Ecol. 2018, 81, 149–170. [Google Scholar] [CrossRef]
- de Bruijn, I.; Liu, Y.Y.; Wiegertjes, G.F.; Raaijmakers, J.M. Exploring fish microbial communities to mitigate emerging diseases in aquaculture. FEMS Microbiol. Ecol. 2018, 94, fix161. [Google Scholar] [CrossRef]
- Gomez, D.; Sunyer, J.O.; Salinas, I. The mucosal immune system of fish: The evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739. [Google Scholar] [CrossRef]
- Cui, X.Y.; Zhang, Q.R.; Zhang, Q.D.; Zhang, Y.Y.; Chen, H.; Liu, G.Q.; Zhu, L.F. Research progress of the gut microbiome in hybrid fish. Microorganisms 2022, 10, 891. [Google Scholar] [CrossRef]
- Sylvain, F.E.; Leroux, N.; Normandeau, E.; Holland, A.; Bouslama, S.; Mercier, P.L.; Val, A.L.; Derome, N. Genomic and environmental factors shape the active gill bacterial community of an Amazonian Teleost Holobiont. Microbiol. Spectr. 2022, 10, e02064-22. [Google Scholar] [CrossRef]
- Sylvain, F.-E.; Cheaib, B.; Llewellyn, M.; Correia, T.G.; Fagundes, D.B.; Val, A.L.; Derome, N. pH drop impacts differentially skin and gut microbiota of the Amazonian fish tambaqui (Colossoma macropomum). Sci. Rep. 2016, 6, 32032. [Google Scholar] [CrossRef]
- Fan, L.M.; Hu, G.D.; Qiu, L.P.; Meng, S.L.; Wu, W.; Zheng, Y.; Song, C.; Li, D.D.; Chen, J.Z. Variations in bacterioplankton communities in aquaculture ponds and the influencing factors during the peak period of culture. Environ. Pollut. 2020, 258, 113656. [Google Scholar] [CrossRef]
- Pratte, Z.A.; Besson, M.; Hollman, R.D.; Stewart, F.J. The Gills of reef fish support a distinct microbiome influenced by host-specific factors. Appl. Environ. Microbiol. 2018, 84, e00063-18. [Google Scholar] [CrossRef]
- Bereded, N.K.; Abebe, G.B.; Fanta, S.W.; Curto, M.; Waidbacher, H.; Meimberg, H.; Domig, K.J. The gut bacterial microbiome of Nile tilapia (Oreochromis niloticus) from lakes across an altitudinal gradient. BMC Microbiol. 2022, 22, 87. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Song, X.X.; Cao, X.H.; He, L.Y.; Liu, S.S.; Yu, Z.M. Healthier communities of phytoplankton and bacteria Achieved via the application of modified clay in shrimp aquaculture ponds. Int. J. Environ. Res. Public Health 2021, 18, 11569. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Xu, R.Z.; Shen, X.X.; Gao, P.; Xue, Z.X.; Huang, D.C.; Jin, G.Q.; Li, C.; Cao, J.S. The response of sediment microbial communities to temporal and site-specific variations of pollution in interconnected aquaculture pond and ditch systems. Sci. Total Environ. 2022, 806, 150498. [Google Scholar] [CrossRef] [PubMed]
- Barberan, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; Dan McGlinn, P.R.; Minchin, R.B.; O’Hara, G.L.; Simpson, P.; Solymos, M.; et al. R Package Version 2.0-10. Community Ecology Package, Vegan. 2013. Available online: http://CRAN.R-project.org/package=vegan) (accessed on 18 May 2017).
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef]
- Tackmann, J.; Arora, N.; Schmidt, T.S.B.; Rodrigues, J.F.M.; von Mering, C. Ecologically informed microbial biomarkers and accurate classification of mixed and unmixed samples in an extensive cross-study of human body sites. Microbiome 2018, 6, 192. [Google Scholar] [CrossRef]
- Clauset, A.; Newman, M.E.J.; Moore, C. Finding community structure in very large networks. Phys. Rev. E 2004, 70, 066111. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJournal Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Maina, J.N. Structure, function and evolution of the gas exchangers: Comparative perspectives. J. Anat. 2002, 201, 281–304. [Google Scholar] [CrossRef] [PubMed]
- Wilkie, M.P. Ammonia excretion and urea handling by fish gills: Present understanding and future research challenges. J. Exp. Zool. 2002, 293, 284–301. [Google Scholar] [CrossRef] [PubMed]
- Kuang, T.; He, A.; Lin, Y.; Huang, X.; Liu, L.; Zhou, L. Comparative analysis of microbial communities associated with the gill, gut, and habitat of two filter-feeding fish. Aquac. Rep. 2020, 18, 100501. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, Q.; Lin, Y.; Hao, J.; Wang, S.; Zhang, J.; Li, A. Taxonomic and functional characteristics of the gill and gastrointestinal microbiota and its correlation with Intestinal metabolites in NEW GIFT strain of farmed adult Nile Tilapia (Oreochromis niloticus). Microorganisms 2021, 9, 617. [Google Scholar] [CrossRef] [PubMed]
- Tarnecki, A.M.; Brennan, N.P.; Schloesser, R.W.; Rhody, N.R. Shifts in the skin-associated microbiota of hatchery-reared Common Snook Centropomus undecimalis during acclimation to the wild. Microb. Ecol. 2019, 77, 770–781. [Google Scholar] [CrossRef]
- Vestrum, R.I.; Attramadal, K.J.K.; Vadstein, O.; Gundersen, M.S.; Bakke, I. Bacterial community assembly in Atlantic cod larvae (Gadus morhua): Contributions of ecological processes and metacommunity structure. FEMS Microbiol. Ecol. 2020, 96, fiaa163. [Google Scholar] [CrossRef]
- Zhou, D.; Zhang, T.; Ren, L.; Fang, D.-A.; Xu, D.-P. Differential study of microbiota in the gill and intestine of silver carp (Hypophthalmichthys molitrix) from the algae-dominated and hydrophyte-dominated areas of Taihu Lake, China. Fishes 2022, 7, 304. [Google Scholar] [CrossRef]
- Xu, H.; Zhao, D.; Zeng, J.; Mao, Z.; Gu, X.; Wu, Q.L. Evaluating the effects of aquaculture on the freshwater lake from the perspective of plankton communities: The diversity, co-occurrence patterns and their underlying mechanisms. Environ. Pollut. 2022, 309, 119741. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, Z.; He, S.; Liu, Y.; Cao, Y.; Shi, P.; Yao, B.; Ringo, E. Identification of the adherent microbiota on the gills and skin of poly-cultured gibel carp (Carassius auratus gibelio) and bluntnose black bream (Megalobrama amblycephala Yih). Aquac. Res. 2010, 41, e72–e83. [Google Scholar] [CrossRef]
- Minich, J.J.; Poore, G.D.; Jantawongsri, K.; Johnston, C.; Bowie, K.; Bowman, J.; Knight, R.; Nowak, B.; Allen, E.E. Microbial ecology of Atlantic salmon (Salmo salar) hatcheries: Impacts of the built environment on fish mucosal microbiota. Appl. Environ. Microbiol. 2020, 86, e00411-20. [Google Scholar] [CrossRef]
- Serra, V.; Gammuto, L.; Petroni, G.; Ciurli, A.; Chiellini, C. Identification of the bacteria associated to the phycosphere of the Chlorella-like strain SEC_LI_ChL_1. Algal Res.-Biomass Biofuels Bioprod. 2022, 67, 102869. [Google Scholar] [CrossRef]
- Kayani, M.U.R.; Zaidi, S.S.A.; Feng, R.; Yu, K.; Qiu, Y.; Yu, X.; Chen, L.; Huang, L. Genome-resolved characterization of structure and potential functions of the zebrafish stool microbiome. Front. Cell. Infect. Microbiol. 2022, 12, 910766. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Zhang, R.; Luo, L.; Wang, S.; Xu, W.; Zhao, Z. Effects of thermal stress on the antioxidant capacity, blood biochemistry, intestinal microbiota and metabolomic responses of Luciobarbus capito. Antioxidants 2023, 12, 198. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lin, Y.; Wei, Z.; Wu, Z.; Zhang, Q.; Hao, J.; Wang, S.; Li, A. Edwardsiella ictaluri almost completely occupies the gut microbiota of fish suffering from enteric septicemia of catfish (Esc). Fishes 2023, 8, 30. [Google Scholar] [CrossRef]
- Huang, J.N.; Zhang, Y.; Xu, L.; He, K.X.; Wen, B.; Yang, P.W.; Ding, J.Y.; Li, J.Z.; Ma, H.C.; Gao, J.Z.; et al. Microplastics: A tissue-specific threat to microbial community and biomarkers of discus fish (Symphysodon aequifasciatus). J. Hazard. Mater. 2022, 424, 127751. [Google Scholar] [CrossRef]
- Green, P.N.; Ardley, J.K. Review of the genus Methylobacterium and closely related organisms: A proposal that some Methylobacterium species be reclassified into a new genus, Methylorubrum gen. nov. ISME J. 2018, 68, 2727–2748. [Google Scholar] [CrossRef]
- Tsagkari, E.; Keating, C.; Couto, J.M.; Sloan, W.T. A keystone Methylobacterium strain in biofilm formation in drinking water. Water 2017, 9, 778. [Google Scholar] [CrossRef]
- Boutin, S.; Sauvage, C.; Bernatchez, L.; Audet, C.; Derome, N. Inter individual variations of the fish skin microbiota: Host genetics basis of mutualism? PLoS ONE 2014, 9, e102649. [Google Scholar] [CrossRef]
- Korotkova, N.; Chistoserdova, L.; Lidstrom, M.E. Poly-beta-hydroxybutyrate biosynthesis in the facultative methylotroph Methylobacterium extorquens AM1: Identification and mutation of gap11, gap20, and phaR. J. Bacteriol. 2002, 184, 6174–6181. [Google Scholar] [CrossRef]
- Halet, D.; Defoirdt, T.; Van Damme, P.; Vervaeren, H.; Forrez, I.; Van de Wiele, T.; Boon, N.; Sorgeloos, P.; Bossier, P.; Verstraete, W. Poly-beta-hydroxybutyrate-accumulating bacteria protect gnotobiotic Artemia franciscana from pathogenic Vibrio campbellii. FEMS Microbiol. Ecol. 2007, 60, 363–369. [Google Scholar] [CrossRef]
- Zheng, T.; Tao, Y.; Lu, S.; Qiang, J.; Xu, P. Integrated transcriptome and 16S rDNA analyses reveal that transport stress induces oxidative stress and immune and metabolic disorders in the intestine of hybrid yellow catfish (Tachysurus fulvidraco female × Pseudobagrus vachellii male). Antioxidants 2022, 11, 1737. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.Y.; Xu, H.M.; Zhao, D.Y.; Zeng, J.; Wu, Q.L.L. Aquaculture drives distinct patterns of planktonic and sedimentary bacterial communities: Insights into co-occurrence pattern and assembly processes. Environ. Microbiol. 2022, 24, 4079–4093. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.Y.; Zhao, D.Y.; Xu, H.M.; Huang, R.; Zeng, J.; Yu, Z.B. Heterogeneity of interactions of microbial communities in regions of Taihu Lake with different nutrient loadings: A network analysis. Sci. Rep. 2018, 8, 8890. [Google Scholar] [CrossRef] [PubMed]
- Riera, J.L.; Baldo, L. Microbial co-occurrence networks of gut microbiota reveal community conservation and diet-associated shifts in cichlid fishes. Anim. Microbiome 2020, 2, 36. [Google Scholar] [CrossRef]
- Philippot, L.; Griffiths, B.S.; Langenheder, S. Microbial community resilience across ecosystems and multiple disturbances. Microbiol. Mol. Biol. Rev. 2021, 85, 10–1128. [Google Scholar] [CrossRef]
- Shade, A.; Read, J.S.; Welkie, D.G.; Kratz, T.K.; Wu, C.H.; McMahon, K.D. Resistance, resilience and recovery: Aquatic bacterial dynamics after water column disturbance. Environ. Microbiol. 2011, 13, 2752–2767. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berglyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature Methods 2013, 10, 57-U11. [Google Scholar] [CrossRef] [PubMed]
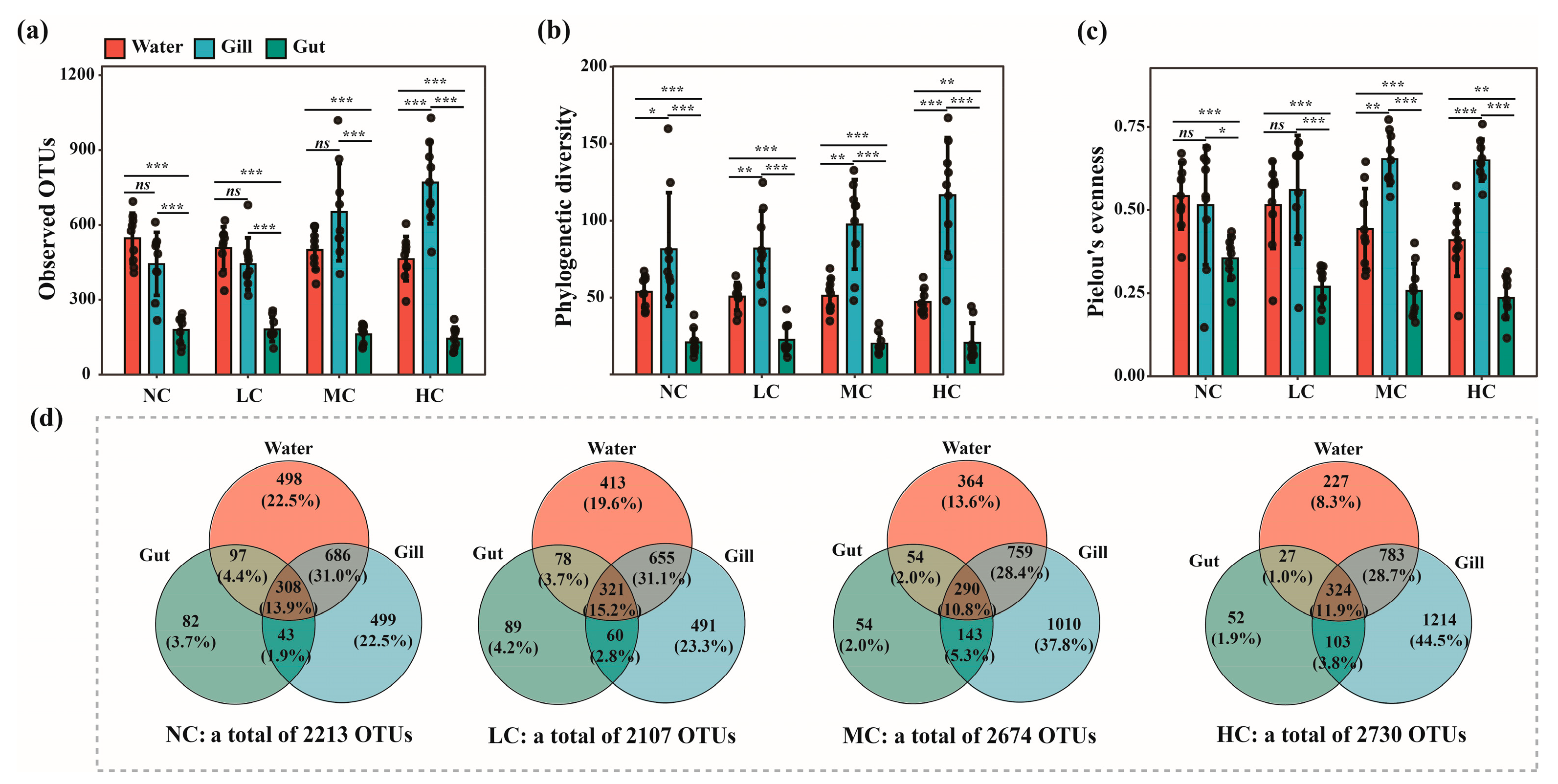
| Observed OTUs | Phylogenetic Diversity | Pielou’s Evenness | ||||
|---|---|---|---|---|---|---|
| F | p | F | p | F | p | |
| Microhabitat | 160.5 | <0.001 | 224.9 | <0.001 | 86.6 | <0.001 |
| Treatment | 3.8 | 0.014 | 2.6 | >0.05 | 0.7 | >0.05 |
| Time | 0.7 | >0.05 | 15.1 | <0.001 | 0.5 | >0.05 |
| Microhabitat × Treatment | 9.6 | <0.001 | 4.7 | <0.001 | 4.5 | <0.001 |
| Microhabitat × Time | 3.2 | 0.018 | 11.8 | <0.001 | 6.3 | <0.001 |
| Treatment × Time | 0.6 | >0.05 | 2.5 | 0.033 | 0.9 | >0.05 |
| Microhabitat × Treatment × Time | 0.7 | >0.05 | 1.6 | >0.05 | 0.3 | >0.05 |
| Df | Sum of Sqs | R2 | F | p (>F) | |
|---|---|---|---|---|---|
| Microhabitat | 2 | 17.4 | 0.53 | 81.5 | 0.001 |
| Treatment | 3 | 0.9 | 0.03 | 2.7 | 0.002 |
| Time | 2 | 1.5 | 0.05 | 7.1 | 0.001 |
| Microhabitat × Treatment | 6 | 1.3 | 0.04 | 2.0 | 0.004 |
| Microhabitat × Time | 4 | 2.1 | 0.06 | 5.0 | 0.001 |
| Treatment × Time | 6 | 0.9 | 0.03 | 1.3 | 0.118 |
| Microhabitat × Treatment × Time | 12 | 1.3 | 0.04 | 1.0 | 0.445 |
| Residual | 72 | 7.7 | 0.23 | ||
| Total | 107 | 33.0 | 1.00 |
| Water | ||||
|---|---|---|---|---|
| NC | LC | MC | HC | |
| No. of nodes | 277 | 256 | 223 | 214 |
| No. of edges | 2182 | 1309 | 1216 | 870 |
| Average degree | 15.8 | 10.2 | 10.9 | 8.1 |
| Graph density | 0.06 | 0.04 | 0.05 | 0.04 |
| Transitivity | 0.81 *** | 0.70 *** | 0.78 *** | 0.70 *** |
| Average path length | 7.23 *** | 4.61 *** | 4.16 *** | 5.13 *** |
| Network diameter | 20.3 *** | 10.0 *** | 10.8 *** | 14.6 *** |
| Modularity | 0.67 *** | 0.71 *** | 0.69 *** | 0.77 *** |
| Assortativity | 0.81 *** | 0.62 *** | 0.76 *** | 0.59 *** |
| Gill | ||||
| NC | LC | MC | HC | |
| No. of nodes | 217 | 233 | 286 | 413 |
| No. of edges | 762 | 981 | 982 | 2777 |
| Average degree | 7.0 | 8.4 | 6.9 | 13.4 |
| Graph density | 0.03 | 0.04 | 0.02 | 0.03 |
| Transitivity | 0.66 *** | 0.74 *** | 0.64 *** | 0.73 *** |
| Average path length | 4.95 *** | 3.72 *** | 8.22 *** | 7.27 *** |
| Network diameter | 11.9 *** | 11.1 *** | 21.1 *** | 18.0 *** |
| Modularity | 0.76 *** | 0.77 *** | 0.80 *** | 0.74 *** |
| Assortativity | 0.66 *** | 0.72 *** | 0.58 *** | 0.73 *** |
| Gut | ||||
| NC | LC | MC | HC | |
| No. of nodes | 107 | 75 | 49 | 53 |
| No. of edges | 480 | 240 | 101 | 164 |
| Average degree | 9.0 | 6.4 | 4.1 | 6.2 |
| Graph density | 0.08 | 0.09 | 0.09 | 0.12 |
| Transitivity | 0.79 *** | 0.71 *** | 0.83 *** | 0.78 *** |
| Average path length | 2.92 *** | 2.01 *** | 1.73 *** | 1.90 *** |
| Network diameter | 9.7 *** | 4.6 | 4.6 | 4.5 |
| Modularity | 0.65 *** | 0.68 *** | 0.66 *** | 0.59 *** |
| Assortativity | 0.71 *** | 0.44 *** | 0.74 *** | 0.64 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, S.; Xu, H.; Qin, L.; Chen, X.; Qiu, L.; Li, D.; Song, C.; Fan, L.; Hu, G.; Xu, P. The Gill-Associated Bacterial Community Is More Affected by Exogenous Chlorella pyrenoidosa Addition than the Bacterial Communities of Water and Fish Gut in GIFT Tilapia (Oreochromis niloticus) Aquaculture System. Biology 2023, 12, 1209. https://doi.org/10.3390/biology12091209
Meng S, Xu H, Qin L, Chen X, Qiu L, Li D, Song C, Fan L, Hu G, Xu P. The Gill-Associated Bacterial Community Is More Affected by Exogenous Chlorella pyrenoidosa Addition than the Bacterial Communities of Water and Fish Gut in GIFT Tilapia (Oreochromis niloticus) Aquaculture System. Biology. 2023; 12(9):1209. https://doi.org/10.3390/biology12091209
Chicago/Turabian StyleMeng, Shunlong, Huimin Xu, Lu Qin, Xi Chen, Liping Qiu, Dandan Li, Chao Song, Limin Fan, Gengdong Hu, and Pao Xu. 2023. "The Gill-Associated Bacterial Community Is More Affected by Exogenous Chlorella pyrenoidosa Addition than the Bacterial Communities of Water and Fish Gut in GIFT Tilapia (Oreochromis niloticus) Aquaculture System" Biology 12, no. 9: 1209. https://doi.org/10.3390/biology12091209
APA StyleMeng, S., Xu, H., Qin, L., Chen, X., Qiu, L., Li, D., Song, C., Fan, L., Hu, G., & Xu, P. (2023). The Gill-Associated Bacterial Community Is More Affected by Exogenous Chlorella pyrenoidosa Addition than the Bacterial Communities of Water and Fish Gut in GIFT Tilapia (Oreochromis niloticus) Aquaculture System. Biology, 12(9), 1209. https://doi.org/10.3390/biology12091209