A Bacillus subtilis Strain ZJ20 with AFB1 Detoxification Ability: A Comprehensive Analysis
Abstract
Simple Summary
Abstract
1. Introduction
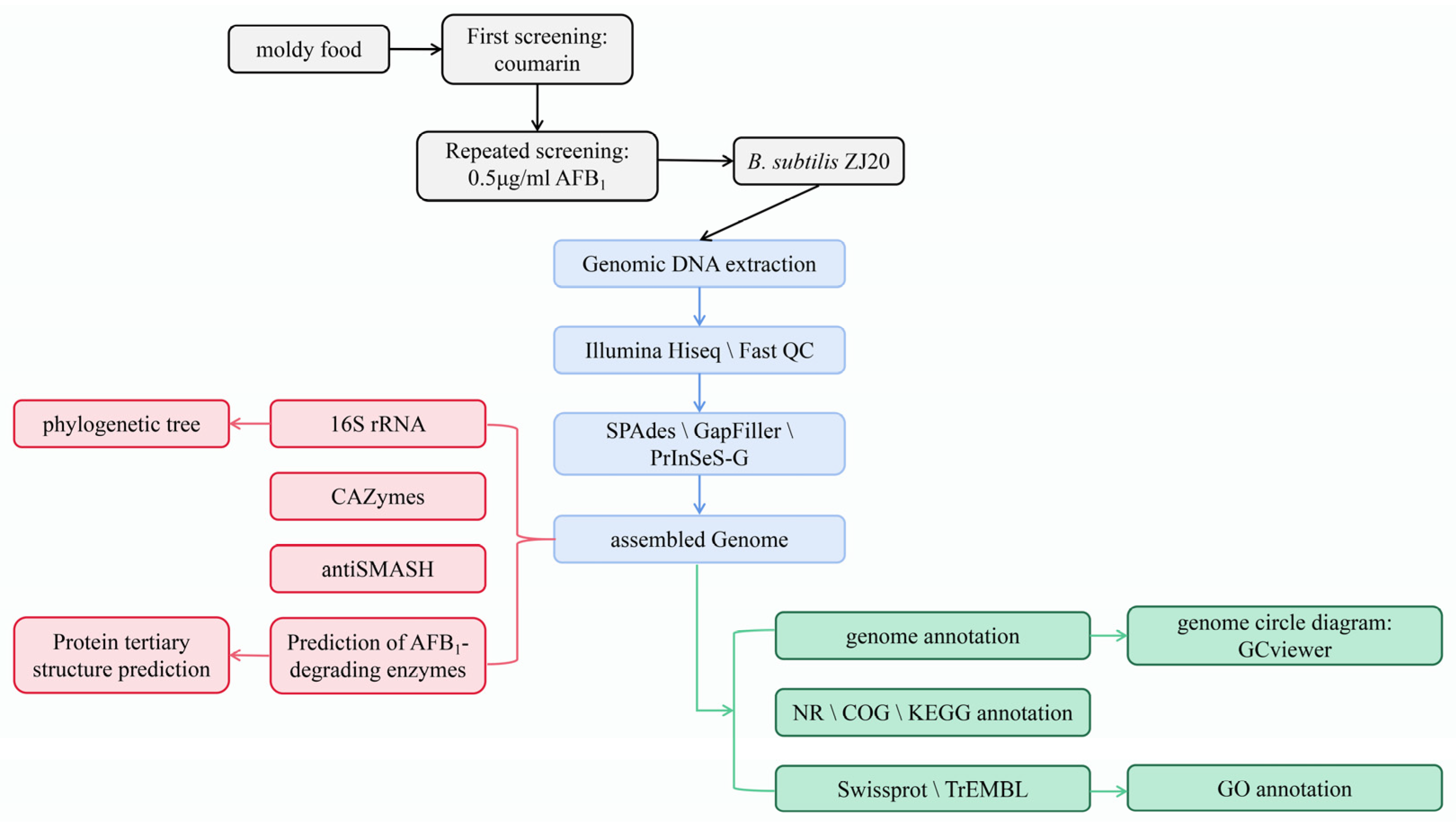
2. Materials and Methods
2.1. Screening of AFB1-Degrading Strains
2.2. Detection of AFB1 Degradation Ability
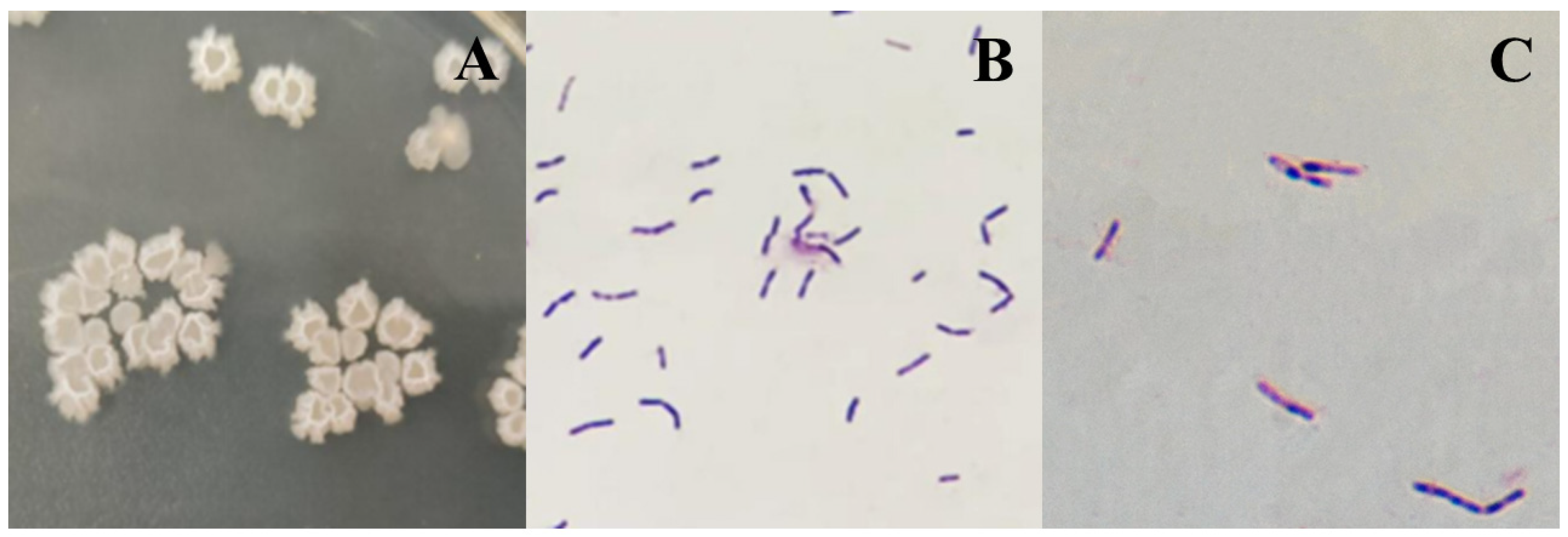
2.3. General Characteristics of ZJ20
2.4. Genome Sequence Determination and Assembly of Strain ZJ20
2.5. Molecular Confirmation of B. subtilis ZJ20
2.6. Functional Annotation of the ZJ20 Genome
2.7. Prediction of Genes Encoding for CAZymes and Secondary Metabolites in B. subtilis ZJ20
2.8. Mining of Potential Aflatoxin AFB1-Degrading Enzymes of B. subtilis ZJ20
3. Results
3.1. Screening of AFB1-Degrading Strains
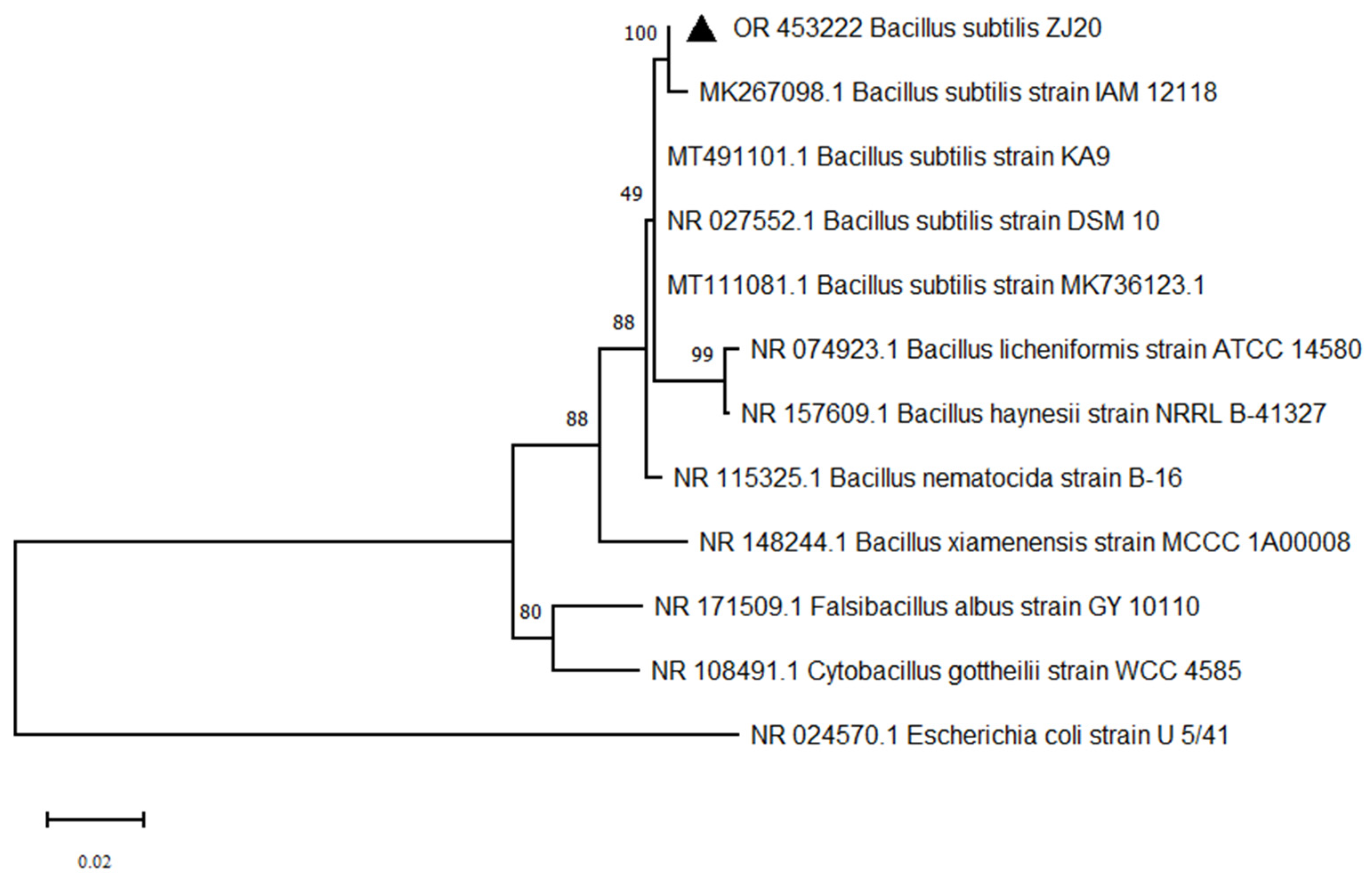
3.2. Bacillus Identification
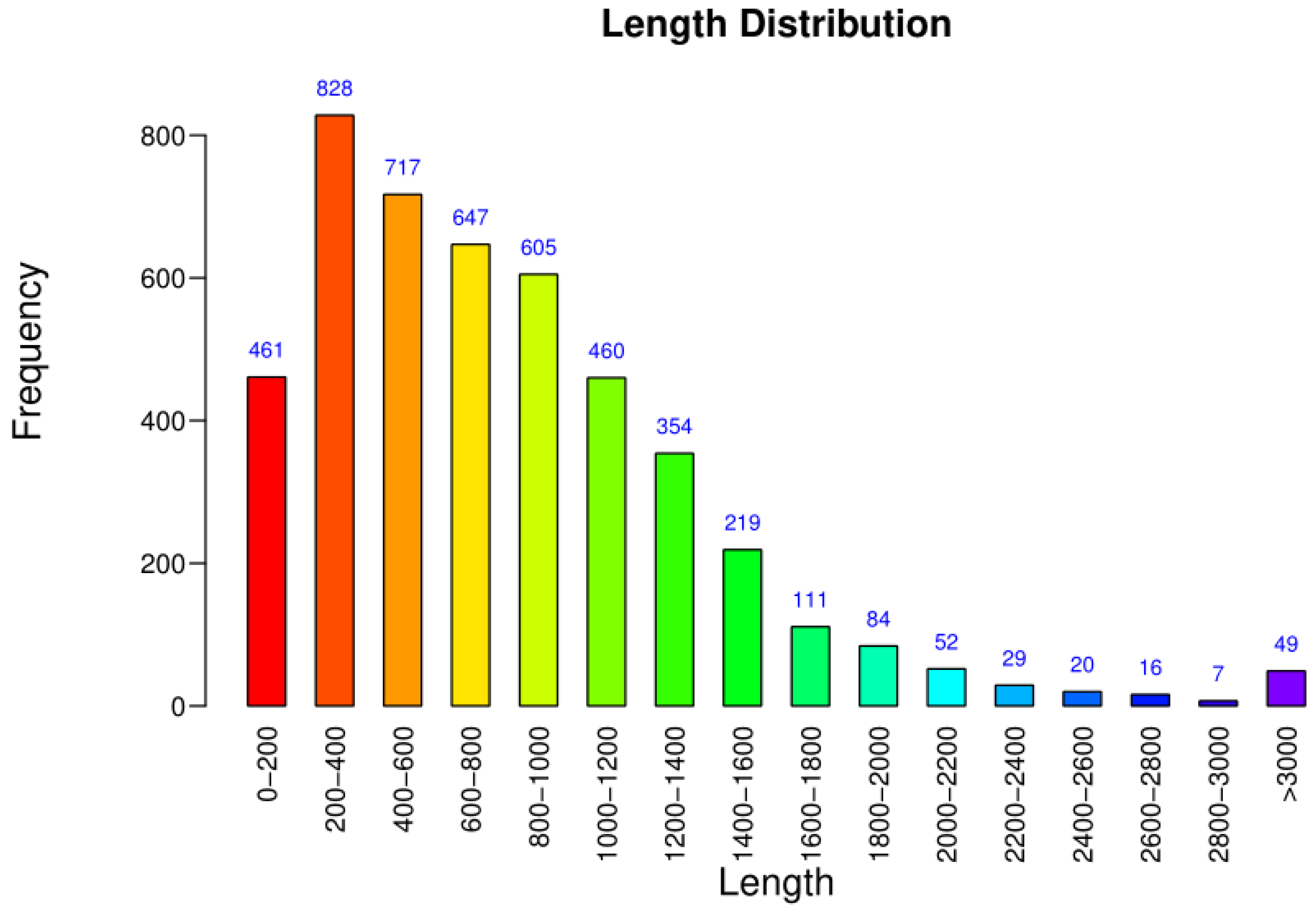
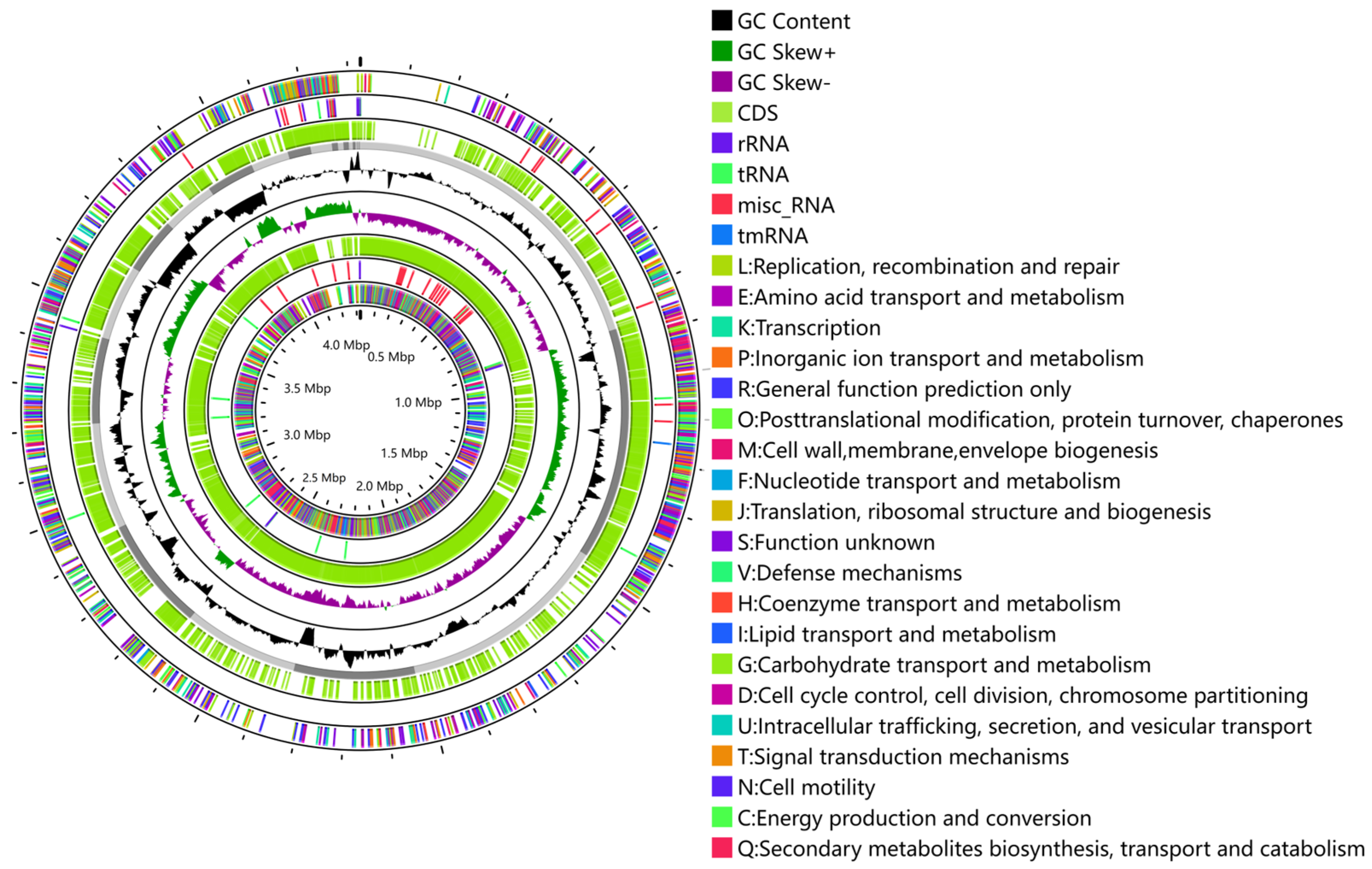
3.3. Basic Characteristics of the Genome
3.4. COG Annotation Results
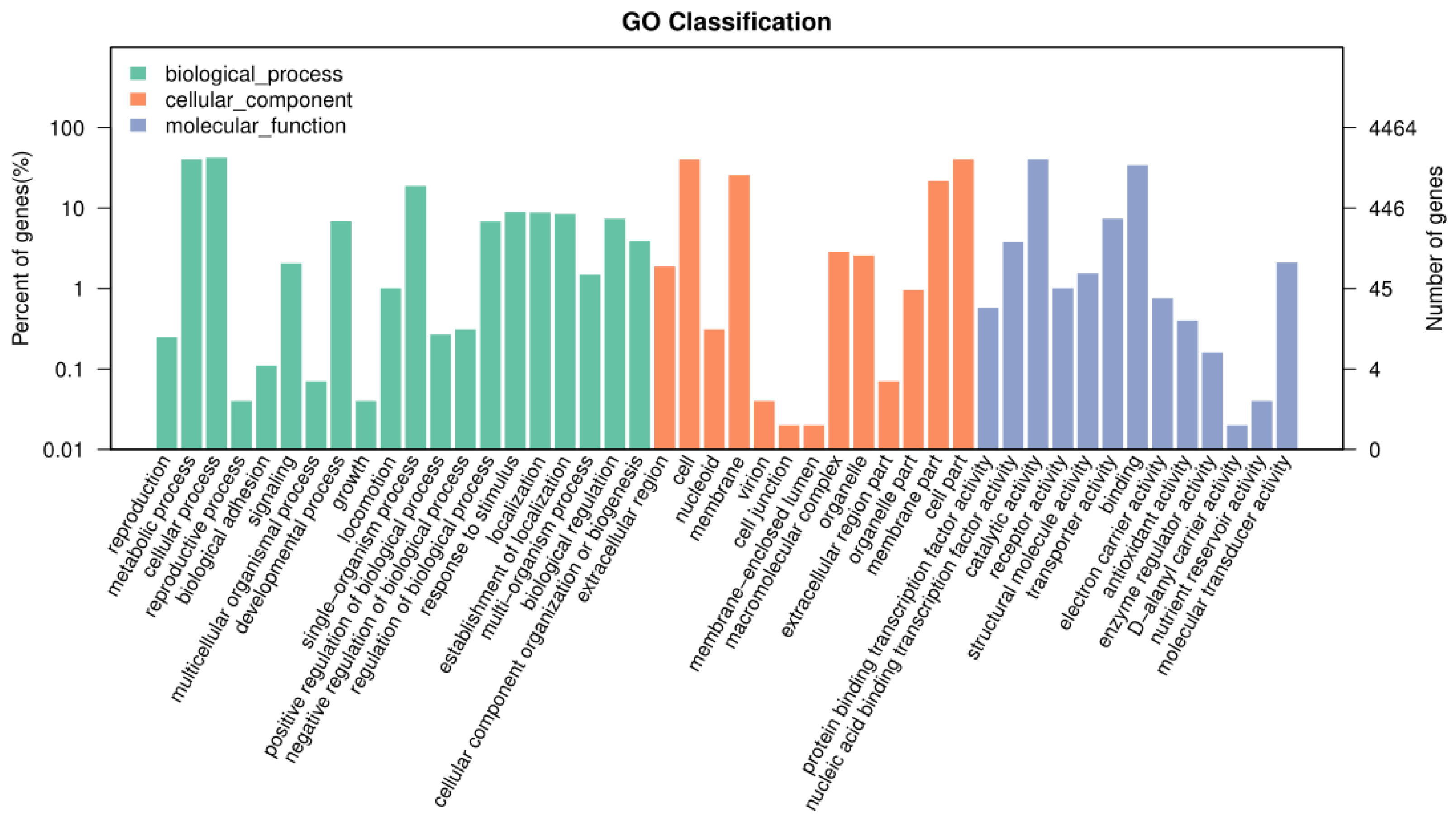
3.5. GO Annotation Results
3.6. KEGG Annotation Results
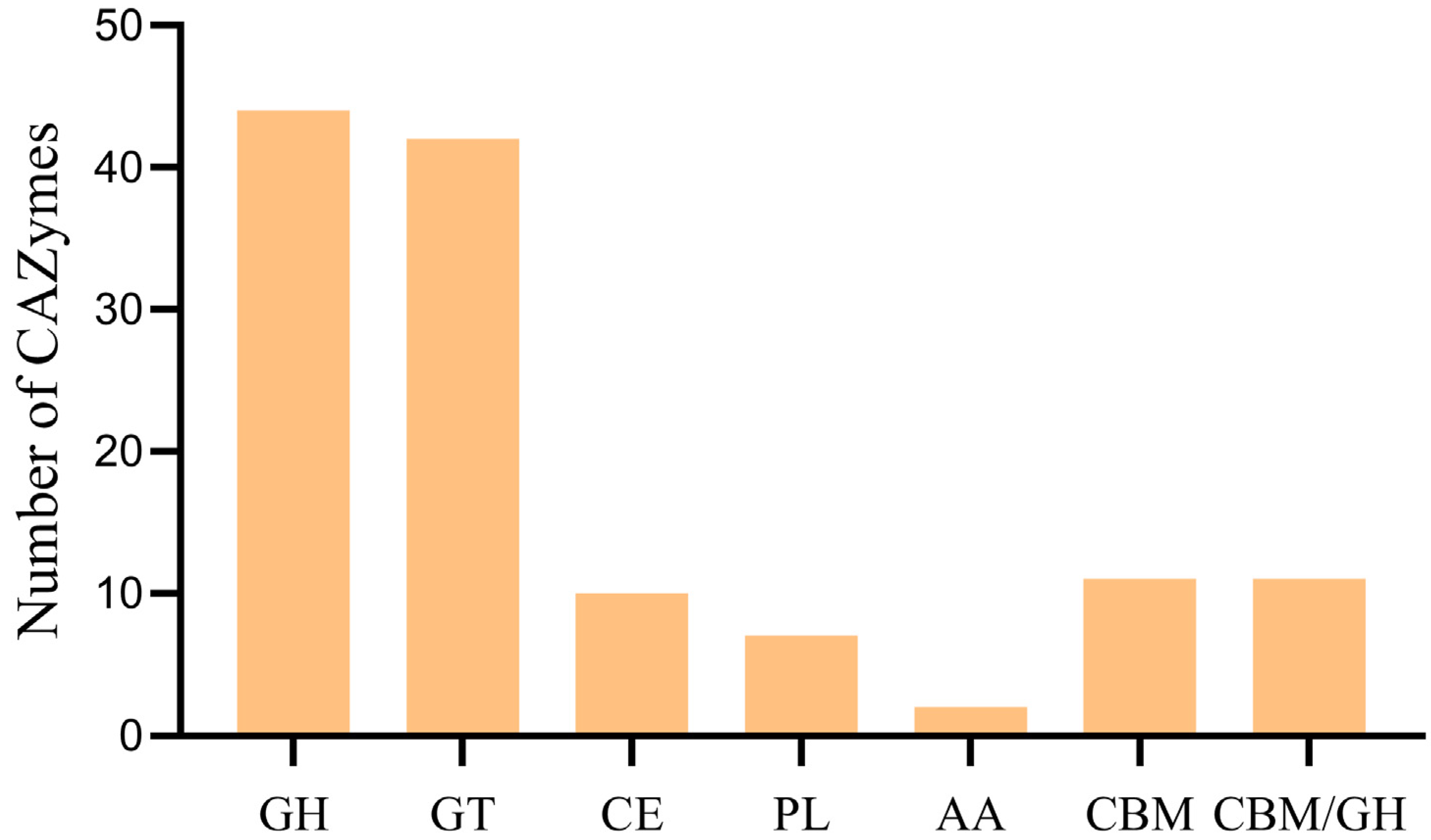
3.7. CAZy Database Annotation Result
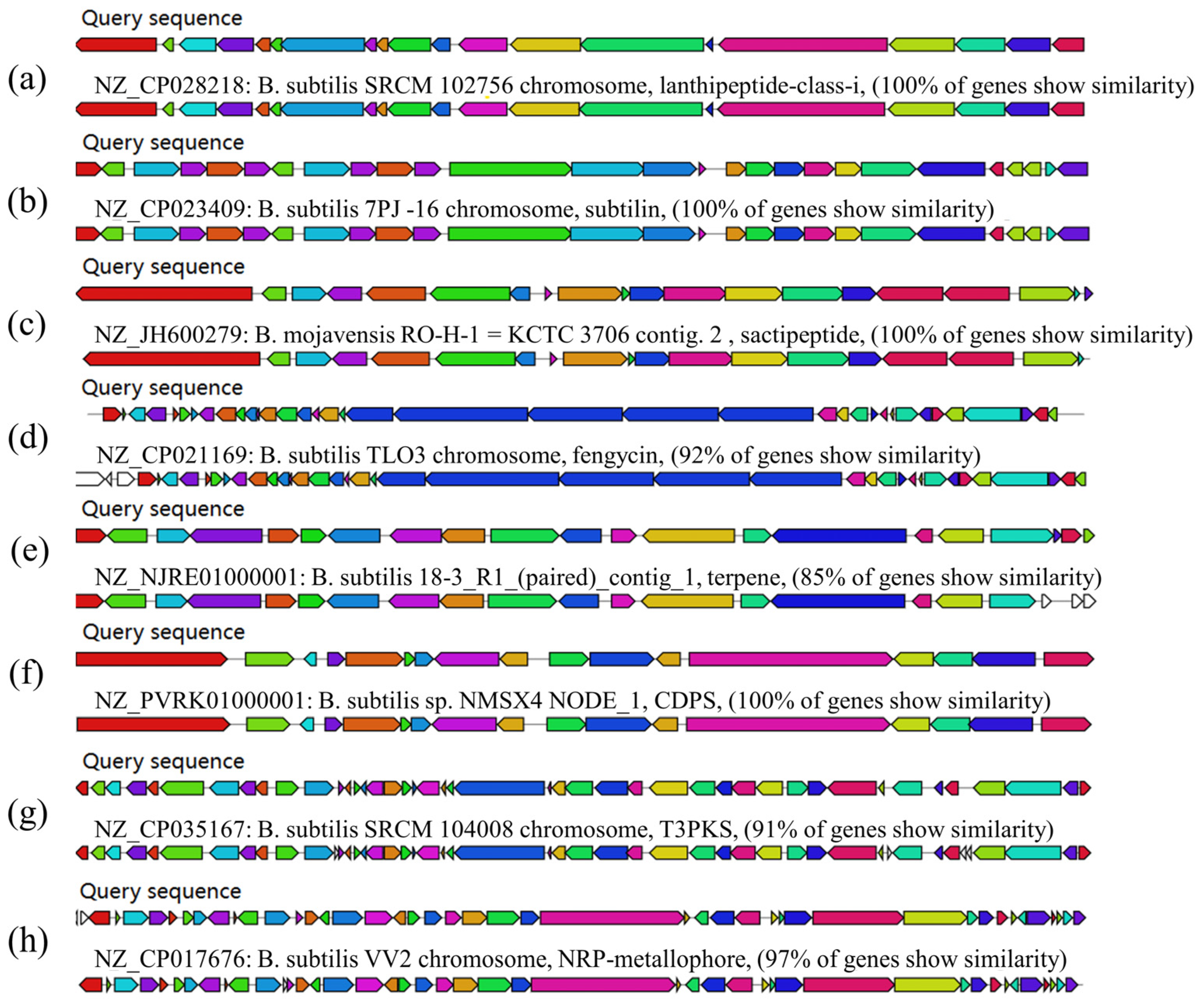
3.8. Prediction of Secondary Metabolites of B. subtilis ZJ20
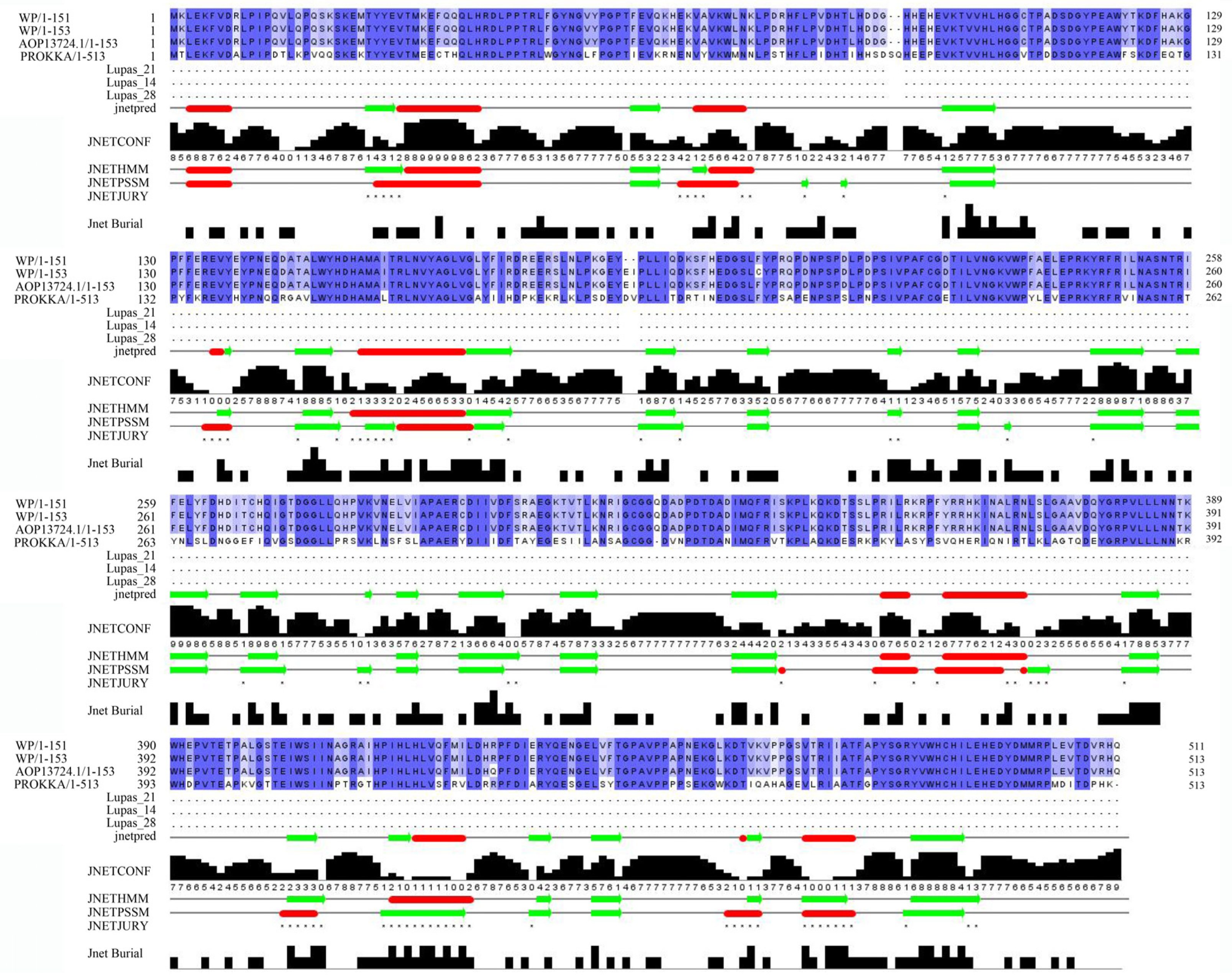
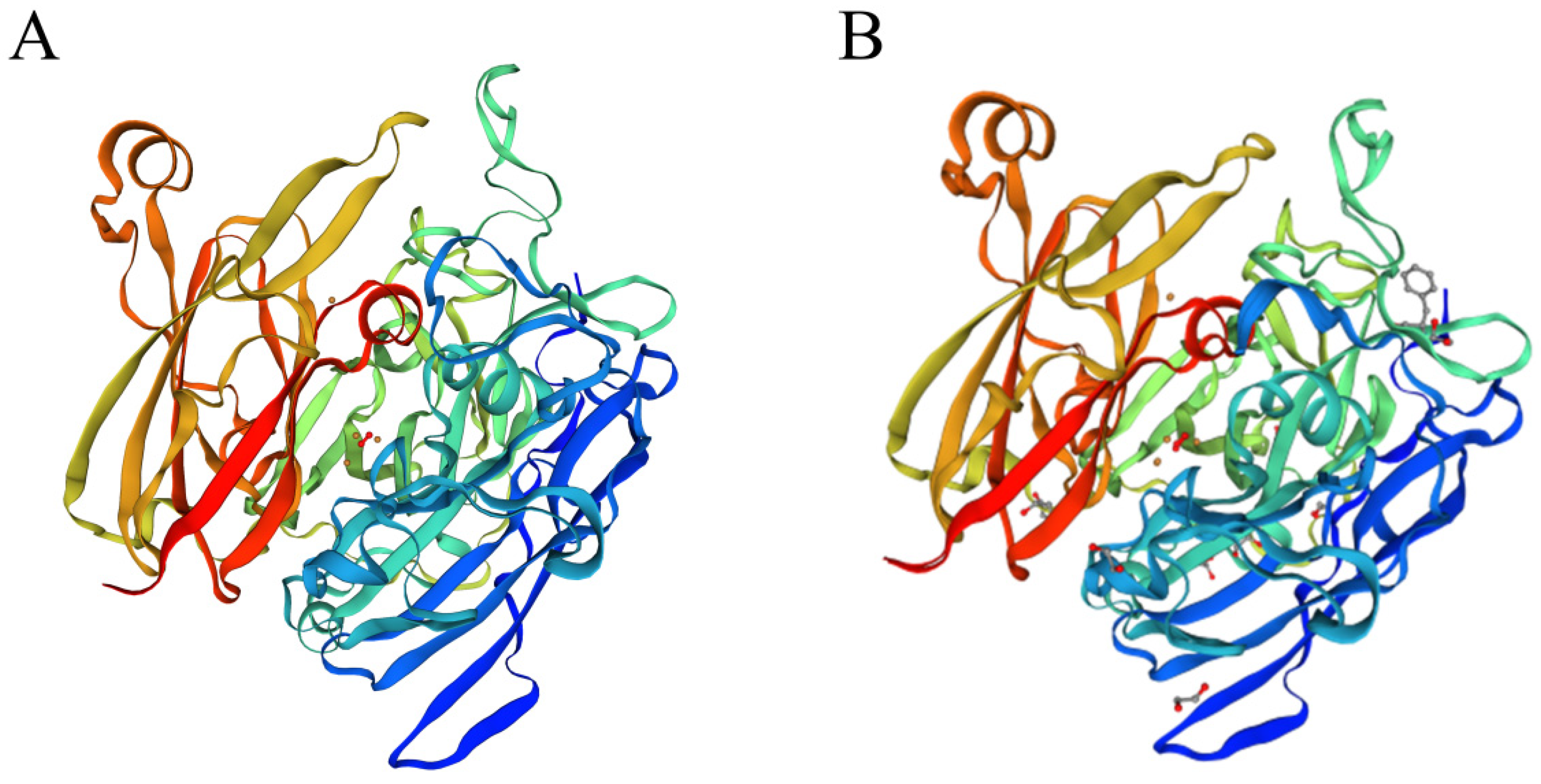
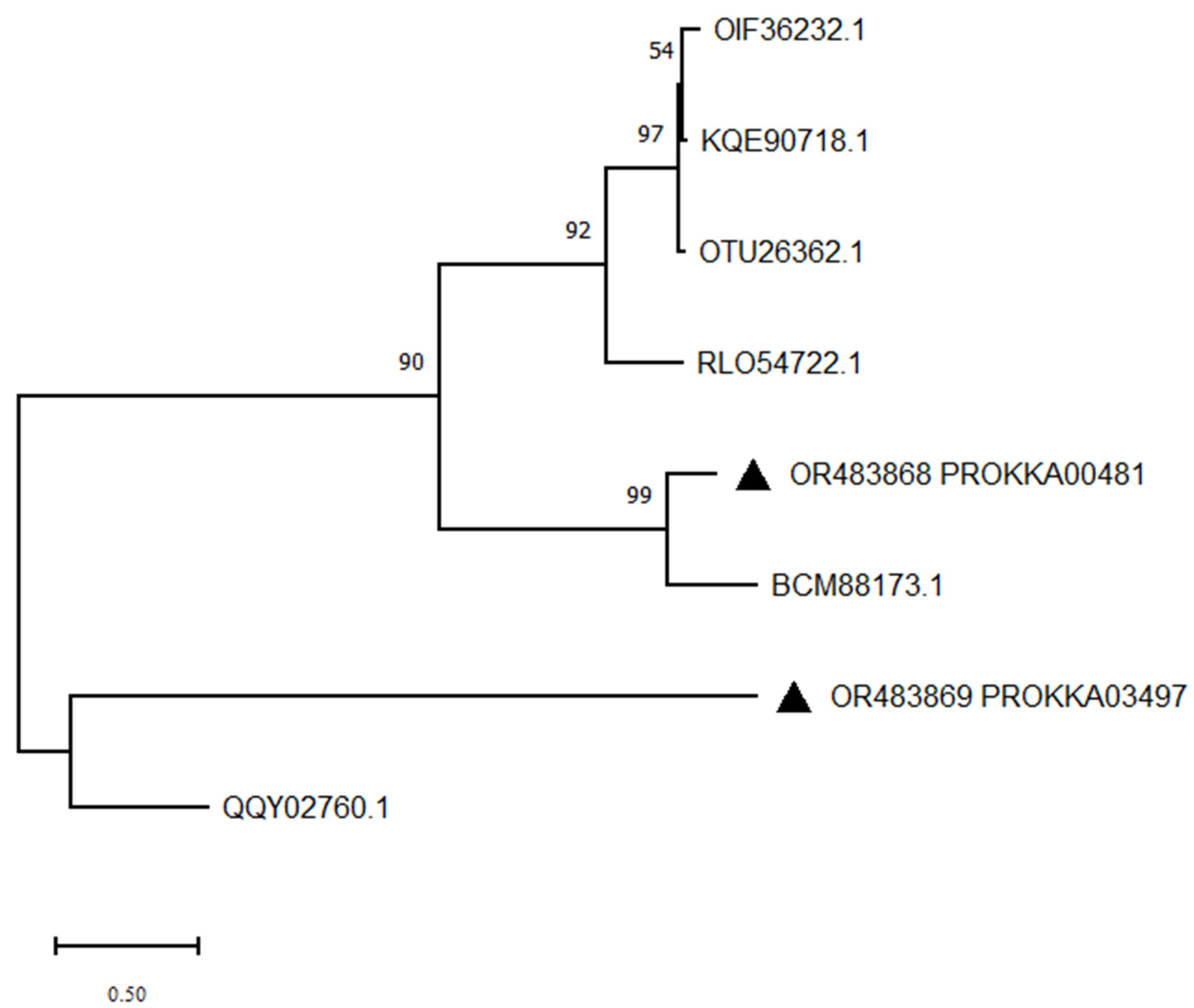
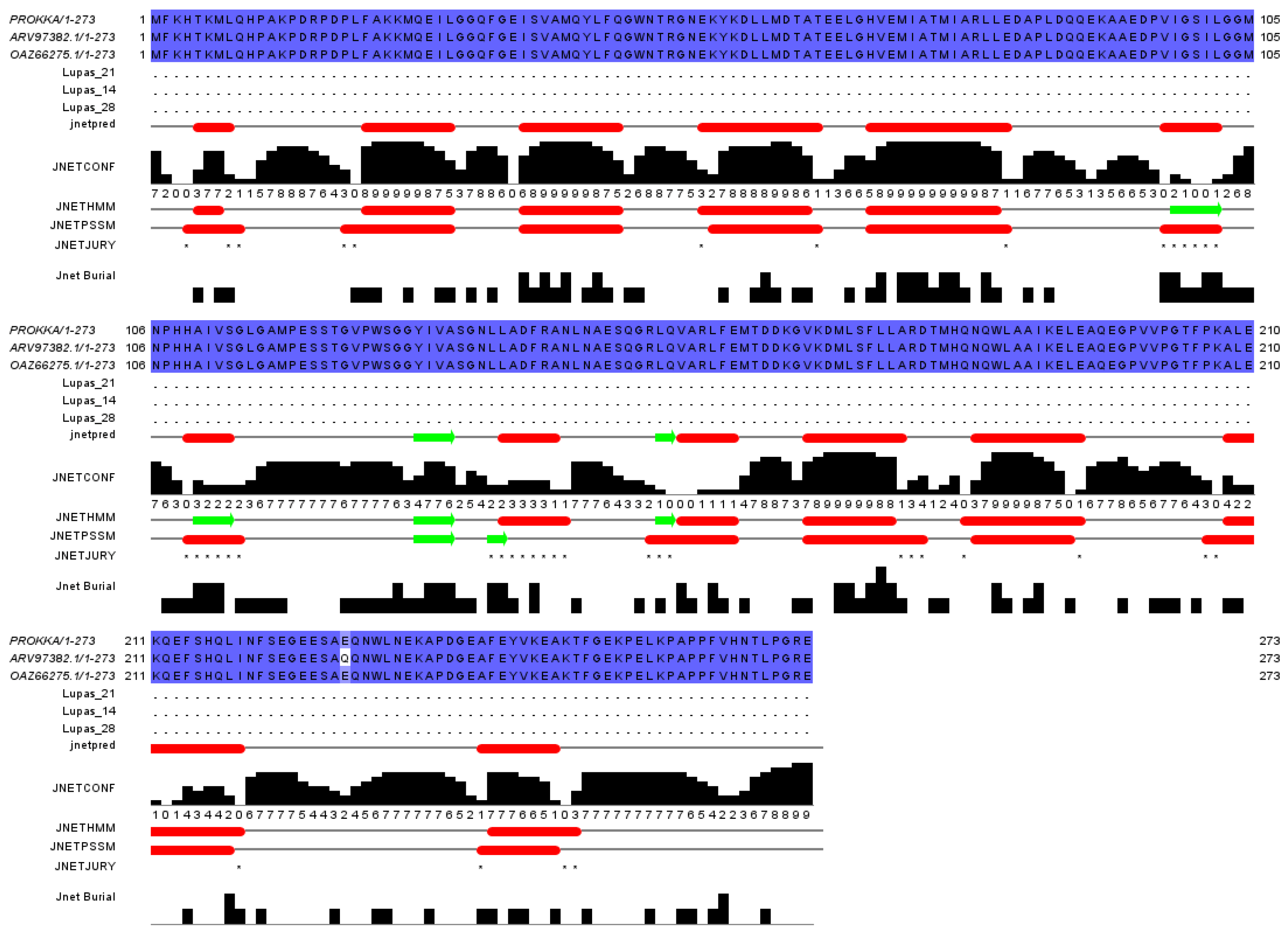
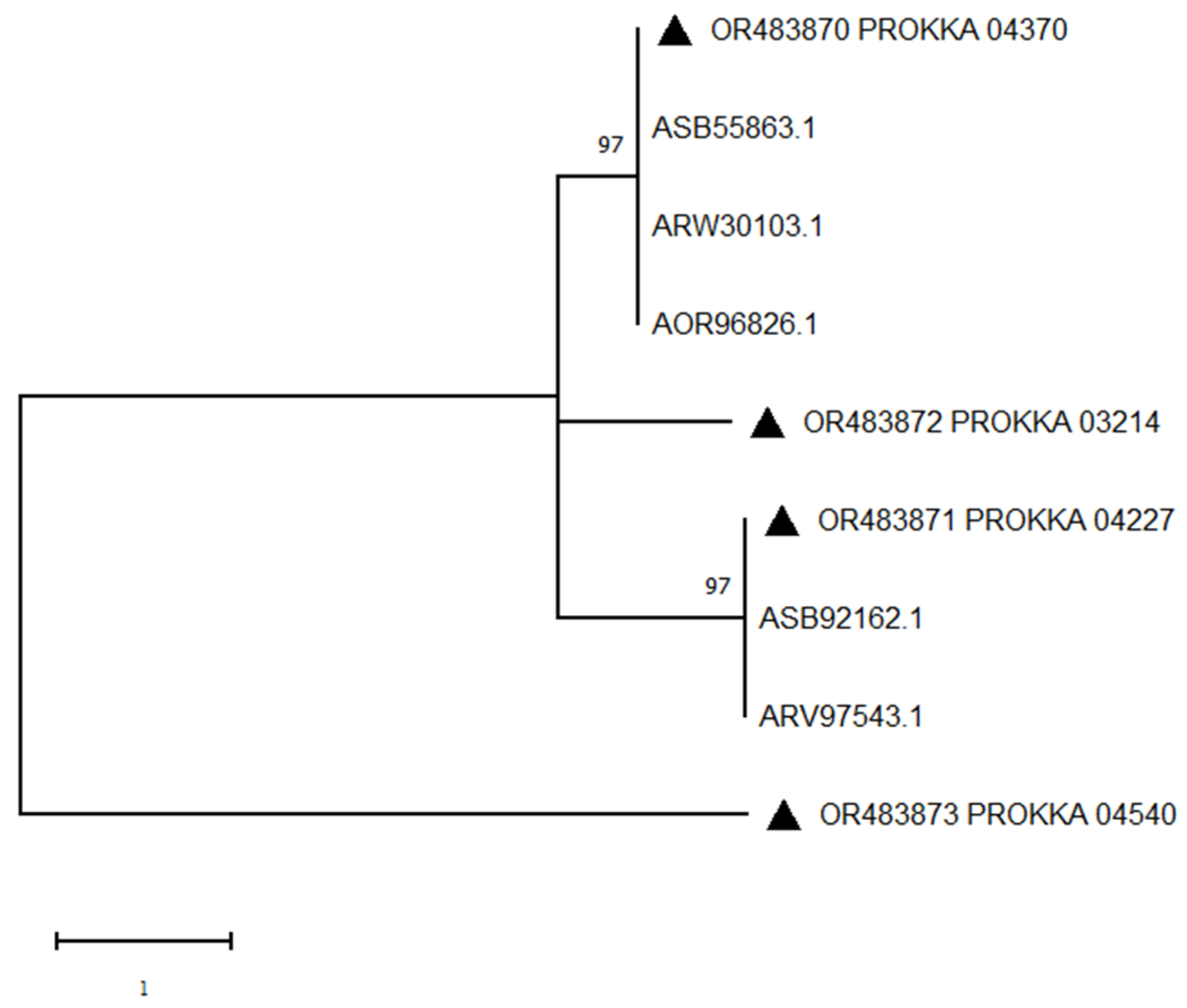
3.9. Mining Potential AFT-Degrading Enzymes of Strain B. subtilis ZJ20
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gizachew, D.; Chang, C.H.; Szonyi, B.; De La Torre, S.; Ting, W.E. Aflatoxin B1 (AFB1) production by Aspergillus flavus and Aspergillus parasiticus on ground Nyjer seeds: The effect of water activity and temperature. Int. J. Food Microbiol. 2019, 296, 8–13. [Google Scholar] [CrossRef]
- Wang, P.; Ma, L.; Jin, J.; Zheng, M.; Pan, L.; Zhao, Y.; Sun, X.; Liu, Y.; Xing, F. The anti-aflatoxigenic mechanism of cinnamaldehyde in Aspergillus flavus. Sci. Rep. 2019, 9, 10499. [Google Scholar] [CrossRef] [PubMed]
- Kensler, T.W.; Roebuck, B.D.; Wogan, G.N.; Groopman, J.D. Aflatoxin: A 50-year odyssey of mechanistic and translational toxicology. Toxicol. Sci. Off. J. Soc. Toxicol. 2011, 120 (Suppl. 1), S28–S48. [Google Scholar] [CrossRef]
- Emmanuel, K.T.; Els, V.P.; Bart, H.; Evelyne, D.; Els, V.H.; Els, D. Carry-over of some Fusarium mycotoxins in tissues and eggs of chickens fed experimentally mycotoxin-contaminated diets. Food Chem. Toxicol. 2020, 145, 111715. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, F.; Na, L.; Jia, M.; Ishfaq, M.; Zhang, Y.; Liu, M.; Wu, C. Ferulic acid alleviates AFB1-induced duodenal barrier damage in rats via up-regulating tight junction proteins, down-regulating ROCK, competing CYP450 enzyme and activating GST. Ecotoxicol. Environ. Saf. 2022, 241, 113805. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Chen, Q.; Wang, L.; Gao, X.; Zhu, W.; Mu, P.; Deng, Y. Aflatoxin B1 Induces Neurotoxicity through Reactive Oxygen Species Generation, DNA Damage, Apoptosis, and S-Phase Cell Cycle Arrest. Int. J. Mol. Sci. 2020, 21, 6517. [Google Scholar] [CrossRef]
- Guindon-Kezis, K.A.; Mulder, J.E.; Massey, T.E. In vivo treatment with aflatoxin B1 increases DNA oxidation, base excision repair activity and 8-oxoguanine DNA glycosylase 1 levels in mouse lung. Toxicology 2014, 321, 21–26. [Google Scholar] [CrossRef]
- Zhu, Q.; Ma, Y.; Liang, J.; Wei, Z.; Li, M.; Zhang, Y.; Liu, M.; He, H.; Qu, C.; Cai, J.; et al. AHR mediates the aflatoxin B1 toxicity associated with hepatocellular carcinoma. Signal Transduct. Target. Ther. 2021, 6, 299. [Google Scholar] [CrossRef]
- Abdel-Daim, M.M.; Abdeen, A.; Jalouli, M.; Abdelkader, A.; Megahed, A.; Alkahtane, A.; Almeer, R.; Alhoshani, N.M.; Al-Johani, N.S.; Alkahtani, S.; et al. Fucoidan supplementation modulates hepato-renal oxidative stress and DNA damage induced by aflatoxin B1 intoxication in rats. Sci. Total Environ. 2021, 768, 144781. [Google Scholar] [CrossRef]
- Eshelli, M.; Qader, M.M.; Jambi, E.J.; Hursthouse, A.S.; Rateb, M.E. Current Status and Future Opportunities of Omics Tools in Mycotoxin Research. Toxins 2018, 10, 433. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, H.; Hamamoto, R.; Schaffer, D.; Duan, S. Functional genomics, genetics, and bioinformatics. BioMed Res. Int. 2015, 2015, 184824. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Feng, K.; Ding, T.; Huang, K.; Yan, H.; Liu, X.; Zhang, Z. Complete genome sequence of Bacillus licheniformis BL-010. Microb. Pathog. 2018, 118, 199–201. [Google Scholar] [CrossRef]
- De Vos, W.M.; Bron, P.A.; Kleerebezem, M. Post-genomics of lactic acid bacteria and other food-grade bacteria to discover gut functionality. Curr. Opin. Biotechnol. 2004, 15, 86–93. [Google Scholar] [CrossRef]
- Bian, L.; Zheng, M.; Chang, T.; Zhou, J.; Zhang, C. Degradation of Aflatoxin B1 by recombinant laccase extracellular produced from Escherichia coli. Ecotoxicol. Environ. Saf. 2022, 244, 114062. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Qin, X.; Hao, Z.; Luo, H.; Yao, B.; Su, X. Degradation of Four Major Mycotoxins by Eight Manganese Peroxidases in Presence of a Dicarboxylic Acid. Toxins 2019, 11, 566. [Google Scholar] [CrossRef]
- Qin, X.; Su, X.; Tu, T.; Zhang, J.; Wang, X.; Wang, Y.; Wang, Y.; Bai, Y.; Yao, B.; Luo, H.; et al. Enzymatic Degradation of Multiple Major Mycotoxins by Dye-Decolorizing Peroxidase from Bacillus subtilis. Toxins 2021, 13, 429. [Google Scholar] [CrossRef]
- Xing, F.; Wang, L.; Liu, X.; Selvaraj, J.N.; Wang, Y.; Zhao, Y.; Liu, Y. Aflatoxin B1 inhibition in Aspergillus flavus by Aspergillus niger through down-regulating expression of major biosynthetic genes and AFB1 degradation by atoxigenic A. flavus. Int. J. Food Microbiol. 2017, 256, 1–10. [Google Scholar] [CrossRef]
- Xie, X.H.; Fu, X.; Yan, X.Y.; Peng, W.F.; Kang, L.X. A Broad-Specificity Chitinase from Penicillium oxalicum k10 Exhibits Antifungal Activity and Biodegradation Properties of Chitin. Mar. Drugs 2021, 19, 356. [Google Scholar] [CrossRef]
- González Pereyra, M.L.; Martínez, M.P.; Cavaglieri, L.R. Presence of aiiA homologue genes encoding for N-Acyl homoserine lactone-degrading enzyme in aflatoxin B1-decontaminating Bacillus strains with potential use as feed additives. Food Chem. Toxicol. 2019, 124, 316–323. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Boetzer, M.; Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol. 2012, 13, R56. [Google Scholar] [CrossRef] [PubMed]
- Massouras, A.; Hens, K.; Gubelmann, C.; Uplekar, S.; Decouttere, F.; Rougemont, J.; Cole, S.T.; Deplancke, B. Primer-initiated sequence synthesis to detect and assemble structural variants. Nat. Methods 2010, 7, 485–486. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Zheng, J.; Hu, B.; Zhang, X.; Ge, Q.; Yan, Y.; Akresi, J.; Piyush, V.; Huang, L.; Yin, Y. dbCAN-seq update: CAZyme gene clusters and substrates in microbiomes. Nucleic Acids Res. 2023, 51, D557–D563. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Swain, B.K.; Johri, T.S. Effect of supplemental methionine, choline and their combinations on the performance and immune response of broilers. Br. Poult. Sci. 2000, 41, 83–88. [Google Scholar] [CrossRef]
- Domenichini, A.; Adamska, A.; Falasca, M. ABC transporters as cancer drivers: Potential functions in cancer development. Biochim. Biophys. Acta. Gen. Subj. 2019, 1863, 52–60. [Google Scholar] [CrossRef]
- Metzler-Zebeli, B.U.; Koger, S.; Sharma, S.; Sener-Aydemir, A.; Ruczizka, U.; Kreutzmann, H.; Ladinig, A. Short-Chain Fatty Acids Modulate Permeability, Motility and Gene Expression in the Porcine Fetal Jejunum Ex Vivo. Nutrients 2022, 14, 2524. [Google Scholar] [CrossRef]
- Kuroishi, T. Regulation of immunological and inflammatory functions by biotin. Can. J. Physiol. Pharmacol. 2015, 93, 1091–1096. [Google Scholar] [CrossRef]
- Nelson, M.L.; Levy, S.B. The history of the tetracyclines. Ann. N. Y. Acad. Sci. 2011, 1241, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Wardman, J.F.; Bains, R.K.; Rahfeld, P.; Withers, S.G. Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol. 2022, 20, 542–556. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Fleites, C.; Proctor, M.; Roberts, S.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Insights into the synthesis of lipopolysaccharide and antibiotics through the structures of two retaining glycosyltransferases from family GT4. Chem. Biol. 2006, 13, 1143–1152. [Google Scholar] [CrossRef]
- Kleessen, B.; Hartmann, L.; Blaut, M. Oligofructose and long-chain inulin: Influence on the gut microbial ecology of rats associated with a human faecal flora. Br. J. Nutr. 2001, 86, 291–300. [Google Scholar] [CrossRef]
- Sidar, A.; Albuquerque, E.D.; Voshol, G.P.; Ram, A.F.J.; Vijgenboom, E.; Punt, P.J. Carbohydrate Binding Modules: Diversity of Domain Architecture in Amylases and Cellulases from Filamentous Microorganisms. Front. Bioeng. Biotechnol. 2020, 8, 871. [Google Scholar] [CrossRef]
- Hanif, A.; Zhang, F.; Li, P.; Li, C.; Xu, Y.; Zubair, M.; Zhang, M.; Jia, D.; Zhao, X.; Liang, J.; et al. Fengycin Produced by Bacillus amyloliquefaciens FZB42 Inhibits Fusarium graminearum Growth and Mycotoxins Biosynthesis. Toxins 2019, 11, 295. [Google Scholar] [CrossRef]
- Krishnan, N.; Velramar, B.; Velu, R.K. Investigation of antifungal activity of surfactin against mycotoxigenic phytopathogenic fungus Fusarium moniliforme and its impact in seed germination and mycotoxicosis. Pestic. Biochem. Physiol. 2019, 155, 101–107. [Google Scholar] [CrossRef]
- Delves-Broughton, J.; Blackburn, P.; Evans, R.J.; Hugenholtz, J. Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 1996, 69, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Shelburne, C.E.; An, F.Y.; Dholpe, V.; Ramamoorthy, A.; Lopatin, D.E.; Lantz, M.S. The spectrum of antimicrobial activity of the bacteriocin subtilosin A. J. Antimicrob. Chemother. 2007, 59, 297–300. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, M.P.; Abraham, W.R. Antimicrobial and biofilm inhibiting diketopiperazines. Curr. Med. Chem. 2012, 19, 3564–3577. [Google Scholar] [CrossRef] [PubMed]
- González-Burgos, E.; Gómez-Serranillos, M.P. Terpene compounds in nature: A review of their potential antioxidant activity. Curr. Med. Chem. 2012, 19, 5319–5341. [Google Scholar] [CrossRef] [PubMed]
- Kuttan, G.; Pratheeshkumar, P.; Manu, K.A.; Kuttan, R. Inhibition of tumor progression by naturally occurring terpenoids. Pharm. Biol. 2011, 49, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Sweeney-Jones, A.M.; Gagaring, K.; Antonova-Koch, J.; Zhou, H.; Mojib, N.; Soapi, K.; Skolnick, J.; McNamara, C.W.; Kubanek, J. Antimalarial Peptide and Polyketide Natural Products from the Fijian Marine Cyanobacterium Moorea producens. Mar. Drugs 2020, 18, 167. [Google Scholar] [CrossRef]
- Von Wirén, N.; Khodr, H.; Hider, R.C. Hydroxylated phytosiderophore species possess an enhanced chelate stability and affinity for iron (III). Plant Physiol. 2000, 124, 1149–1158. [Google Scholar] [CrossRef] [PubMed][Green Version]


| Strain Number | AFB1 Degradation Rate (%) | Strain Number | AFB1 Degradation Rate (%) |
|---|---|---|---|
| ZJ3 | 40.26 ± 0.22 | ZJ16 | 45.86 ± 1.36 |
| ZJ6 | 52.09 ± 1.23 | ZJ20 | 84.23 ± 0.13 |
| ZJ8 | 42.86 ± 2.16 | ZJ22 | 38.73 ± 0.32 |
| ZJ10 | 52.06 ± 1.16 | ZJ27 | 54.28 ± 2.03 |
| ZJ11 | 35.86 ± 1.14 | ZJ31 | 41.49 ± 0.12 |
| ZJ13 | 68.47 ± 0.18 | ZJ32 | 53.29 ± 1.24 |
| Class | Number |
|---|---|
| Size (base) | 4,326,240 |
| G + C content (%) | 42.9 |
| Protein Coding Genes | 4659 |
| Min length (base) | 45 |
| Max length (base) | 10,806 |
| Average length (base) | 817.69 |
| Total coding gene (base) | 3,809,638 |
| Coding ratio (%) | 88.06 |
| tRNA | 85 |
| rRNA | 11 |
| Repeat Region | |
| Repeat Region Count | 0 |
| Total Repeat Region (base) | 0 |
| Repeat Ratio (%) | 0 |
| COG Code | Number | Proportion (%) | Description |
|---|---|---|---|
| C | 164 | 5.35 | Energy production and conversion |
| D | 37 | 1.21 | Cell cycle control, cell division, and chromosome partitioning |
| E | 248 | 8.09 | Amino acid transport and metabolism |
| F | 82 | 2.68 | Nucleotide transport and metabolism |
| G | 260 | 8.48 | Carbohydrate transport and metabolism |
| H | 125 | 4.08 | Coenzyme transport and metabolism |
| I | 72 | 2.35 | Lipid transport and metabolism |
| J | 168 | 5.48 | Translation, ribosomal structure, and biogenesis |
| K | 257 | 8.38 | Transcription |
| L | 133 | 4.34 | Replication, recombination, and repair |
| M | 185 | 6.04 | Cell wall/membrane/envelope biogenesis |
| N | 24 | 0.78 | Cell motility |
| O | 91 | 2.97 | Posttranslational modification, protein turnover, chaperones |
| P | 171 | 5.58 | Inorganic ion transport and metabolism |
| Q | 60 | 1.96 | Secondary metabolite biosynthesis, transport, and catabolism |
| R | 400 | 13.05 | General function prediction only |
| S | 356 | 11.62 | Function unknown |
| T | 129 | 4.21 | Signal transduction mechanisms |
| U | 45 | 1.47 | Intracellular trafficking, secretion, and vesicular transport |
| V | 58 | 1.89 | Defense mechanisms |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, M.; Guo, J.; Jia, Y.; Liao, C.; He, L.; Li, J.; Wei, Y.; Chen, S.; Chen, J.; Shang, K.; et al. A Bacillus subtilis Strain ZJ20 with AFB1 Detoxification Ability: A Comprehensive Analysis. Biology 2023, 12, 1195. https://doi.org/10.3390/biology12091195
Huang M, Guo J, Jia Y, Liao C, He L, Li J, Wei Y, Chen S, Chen J, Shang K, et al. A Bacillus subtilis Strain ZJ20 with AFB1 Detoxification Ability: A Comprehensive Analysis. Biology. 2023; 12(9):1195. https://doi.org/10.3390/biology12091195
Chicago/Turabian StyleHuang, Meixue, Jing Guo, Yanyan Jia, Chengshui Liao, Lei He, Jing Li, Ying Wei, Songbiao Chen, Jian Chen, Ke Shang, and et al. 2023. "A Bacillus subtilis Strain ZJ20 with AFB1 Detoxification Ability: A Comprehensive Analysis" Biology 12, no. 9: 1195. https://doi.org/10.3390/biology12091195
APA StyleHuang, M., Guo, J., Jia, Y., Liao, C., He, L., Li, J., Wei, Y., Chen, S., Chen, J., Shang, K., Guo, R., Ding, K., & Yu, Z. (2023). A Bacillus subtilis Strain ZJ20 with AFB1 Detoxification Ability: A Comprehensive Analysis. Biology, 12(9), 1195. https://doi.org/10.3390/biology12091195






