An Updated Overview of Existing Cancer Databases and Identified Needs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
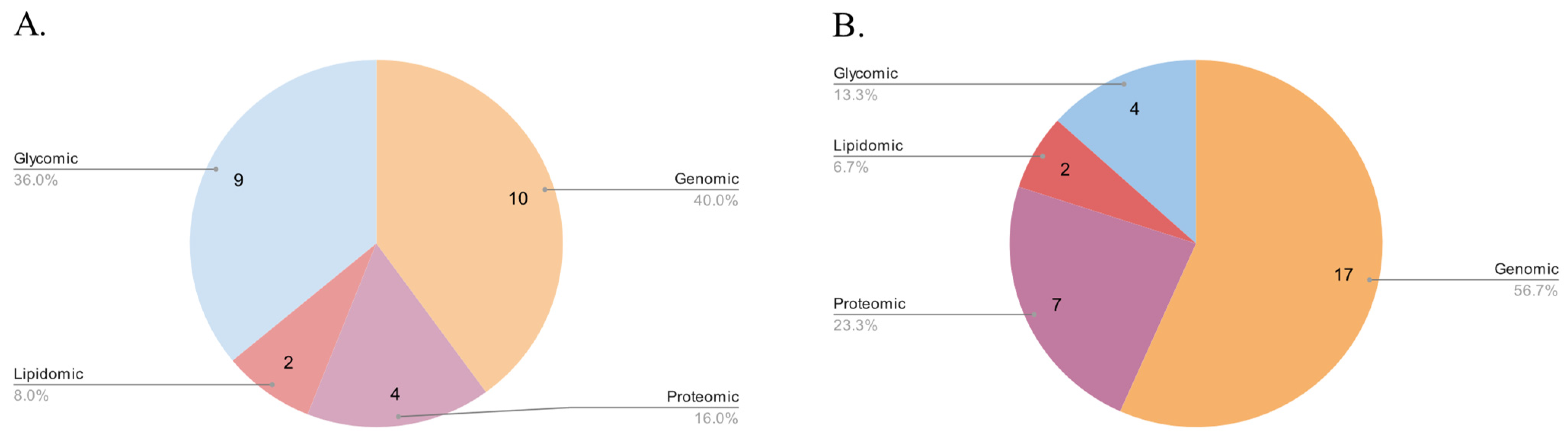
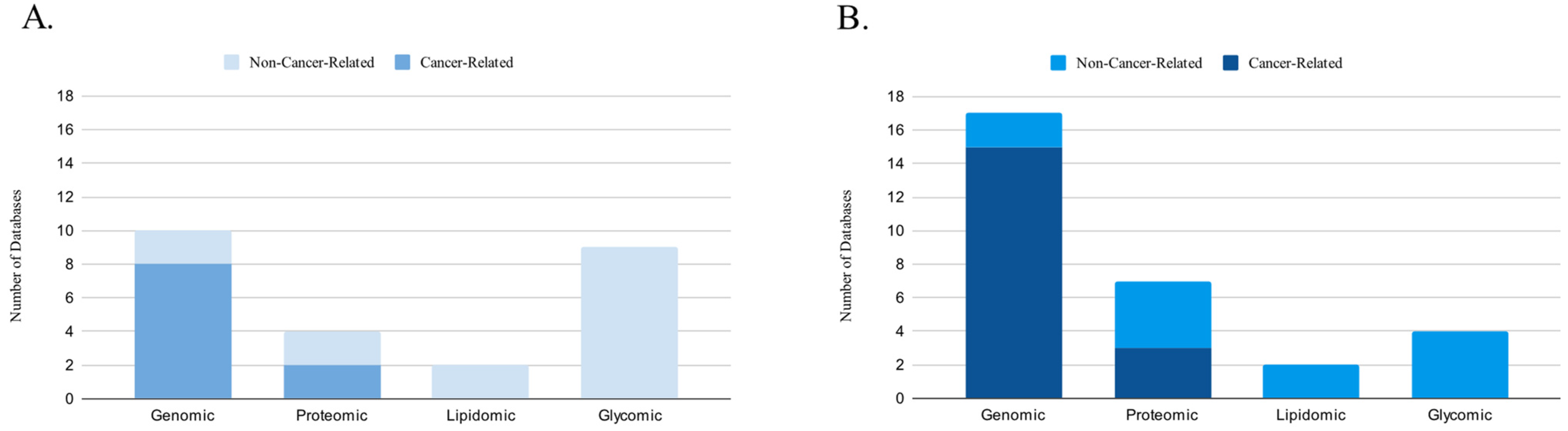
3. Results
3.1. Genomic Databases
Cancer Specific Databases
3.2. Proteomic Databases
3.3. Lipidomic Databases
3.4. Glycomic Databases
3.5. Clinical Trial Databases
3.6. Other Cancer Databases
Retired Databases
3.7. Web-Based Servers
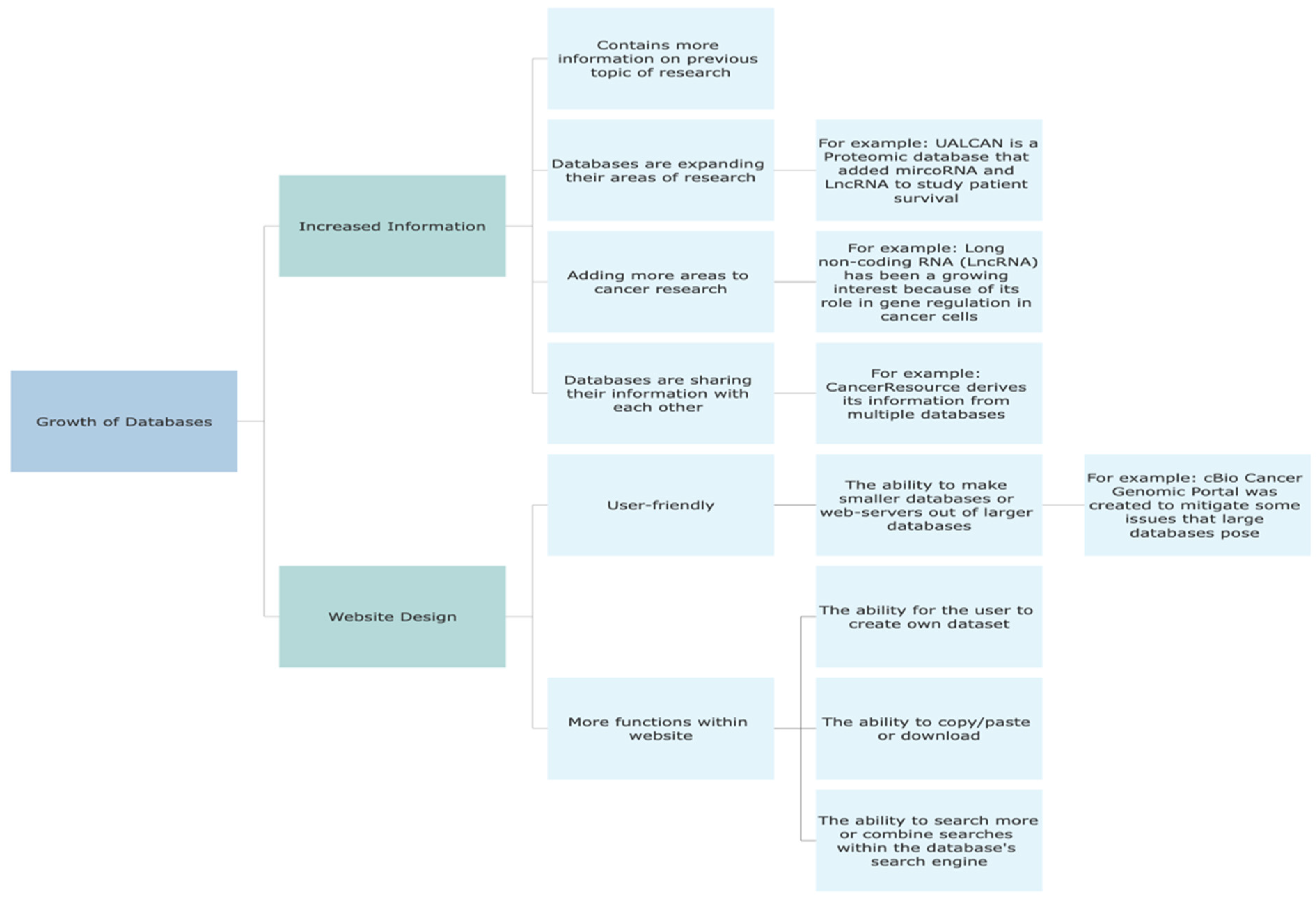
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Faguet, G.B. A brief history of cancer: Age-old milestones underlying our current knowledge database. Int. J. Cancer 2015, 136, 2022–2036. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, I.B.; Case, K. The History of Cancer Research: Introducing an AACR Centennial Series. Cancer Res. 2008, 68, 6861–6862. [Google Scholar] [CrossRef] [PubMed]
- SEER Training Modules. War Facts and the War on Cancer; National Cancer Institute: Bethesda, MA, USA, 2023. [Google Scholar]
- SEER Training Modules. Brief History of Cancer Registration; National Cancer Institute: Bethesda, MA, USA, 2023. [Google Scholar]
- Ursin, G. Cancer registration in the era of modern oncology and GDPR. Acta Oncol. 2019, 58, 1547–1548. [Google Scholar] [CrossRef] [PubMed]
- Pavlopoulou, A.; Spandidos, D.A.; Michalopoulos, I. Human cancer databases (Review). Oncol. Rep. 2015, 33, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. Review The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 2015, 68–77. [Google Scholar] [CrossRef]
- Sarver, A.L.; Sarver, A.E.; Yuan, C.; Subramanian, S. OMCD: OncomiR Cancer Database. BMC Cancer 2018, 18, 1223. [Google Scholar] [CrossRef]
- Mei, S.; Meyer, C.A.; Zheng, R.; Qin, Q.; Wu, Q.; Jiang, P.; Li, B.; Shi, X.; Wang, B.; Fan, J.; et al. Cistrome cancer: A web resource for integrative gene regulation modeling in cancer. Cancer Res. 2017, 77, e19–e22. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Zhang, J.; Bajari, R.; Andric, D.; Gerthoffert, F.; Lepsa, A.; Nahal-Bose, H.; Stein, L.D.; Ferretti, V. The International Cancer Genome Consortium Data Portal. Nat. Biotechnol. 2019, 37, 367–369. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- The Human Genome Browser at UCSC. Available online: https://genome.cshlp.org/content/12/6/996.short (accessed on 6 February 2023).
- Clough, E.; Barrett, T. The Gene Expression Omnibus database. Methods Mol. Biol. 2016, 1418, 93–110. [Google Scholar] [PubMed]
- Flicek, P.; Amode, M.R.; Barrell, D.; Beal, K.; Billis, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fitzgerald, S.; et al. Ensembl 2014. Nucleic Acids Res. 2014, 42, D749–D755. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Girón, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef]
- Martin, F.J.; Amode, M.R.; Aneja, A.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2023. Nucleic Acids Res. 2023, 51, D933–D941. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.A.; Ferretti, V.; Grossman, R.L.; Staudt, L.M. The NCI Genomic Data Commons as an engine for precision medicine. Blood 2017, 130, 453–459. [Google Scholar] [CrossRef] [PubMed]
- GDC. Available online: https://portal.gdc.cancer.gov/ (accessed on 15 February 2023).
- Cappelli, E.; Cumbo, F.; Bernasconi, A.; Canakoglu, A.; Ceri, S.; Masseroli, M.; Weitschek, E. OpenGDC: Unifying, Modeling, Integrating Cancer Genomic Data and Clinical Metadata. Appl. Sci. 2020, 10, 6367. [Google Scholar] [CrossRef]
- Futreal, P.A.; Coin, L.; Marshall, M.; Down, T.; Hubbard, T.; Wooster, R.; Rahman, N.; Stratton, M.R. A census of human cancer genes. Nat. Rev. Cancer 2004, 4, 177–183. [Google Scholar] [CrossRef]
- Repana, D.; Nulsen, J.; Dressler, L.; Bortolomeazzi, M.; Venkata, S.K.; Tourna, A.; Yakovleva, A.; Palmieri, T. The Network of Cancer Genes (NCG): A comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens 06 Biological Sciences 0604 Genetics 11 Medical and Health Sciences 1112 Oncology and Carcinogenesis 06 Biological Sciences 0601 Biochemistry and Cell Biology. Genome Biol. 2019, 20, 1–20. [Google Scholar]
- Zhang, D.; Huo, D.; Xie, H.; Wu, L.; Zhang, J.; Liu, L.; Jin, Q.; Chen, X. CHG: A Systematically Integrated Database of Cancer Hallmark Genes. Front. Genet. 2020, 11, 29. [Google Scholar] [CrossRef]
- Bamford, S.; Dawson, E.; Forbes, S.; Clements, J.; Pettett, R.; Dogan, A.; Flanagan, A.; Teague, J.; Futreal, A.P.; Stratton, M.R.; et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 2004, 91, 355–358. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.-L.; Li, M.; Goncearenco, A.; Panchenko, A.R. Finding driver mutations in cancer: Elucidating the role of background mutational processes. PLoS Comput. Biol. 2019, 15, e1006981. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Carrio-Cordo, P.; Gao, B.; Paloots, R.; Baudis, M. The Progenetix oncogenomic resource in 2021. Database 2021, 2021, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Progenetix. Available online: https://progenetix.org/ (accessed on 15 February 2023).
- Ping, J.; Oyebamiji, O.; Yu, H.; Ness, S.; Chien, J.; Ye, F.; Kang, H.; Samuels, D.; Ivanov, S.; Chen, D.; et al. MutEx: A multifaceted gateway for exploring integrative pan-cancer genomic data. Briefings Bioinform. 2020, 21, 1479–1486. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pander, A.; Chinnaiyan, A.M. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform 1. 2004. Available online: www.oncomine.org (accessed on 6 February 2023).
- Rhodes, D.R.; Kalyana-Sundaram, S.; Mahavisno, V.; Varambally, R.; Yu, J.; Briggs, B.B.; Barrette, T.R.; Anstet, M.J.; Kincead-Beal, C.; Kulkarni, P.; et al. Oncomine 3.0: Genes, Pathways, and Networks in a Collection of 18,000 Cancer Gene Expression Profiles. Neoplasia 2007, 9, 166–180. [Google Scholar] [CrossRef]
- Vestergaard, L.K.; Oliveira, D.N.P.; Poulsen, T.S.; Høgdall, C.K.; Høgdall, E.V. OncomineTM comprehensive assay v3 vs. OncomineTM comprehensive assay plus. Cancers 2021, 13, 5230. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Lin, S.; Girard, L.; Zhou, Y.; Yang, L.; Ci, B.; Zhou, Q.; Luo, D.; Yao, B.; Tang, H.; et al. LCE: An open web portal to explore gene expression and clinical associations in lung cancer. Oncogene 2018, 38, 2551–2564. [Google Scholar] [CrossRef]
- Koshkin, V.S.; Patel, V.G.; Ali, A.; Bilen, M.A.; Ravindranathan, D.; Park, J.J.; Kellezi, O.; Cieslik, M.; Shaya, J.; Cabal, A.; et al. PROMISE: A real-world clinical-genomic database to address knowledge gaps in prostate cancer. Prostate Cancer Prostatic Dis. 2021, 25, 388–396. [Google Scholar] [CrossRef]
- Lian, Q.; Wang, S.; Zhang, G.; Wang, D.; Luo, G.; Tang, J.; Chen, L.; Gu, J. HCCDB: A Database of Hepatocellular Carcinoma Expression Atlas. Genom. Proteom. Bioinform. 2018, 16, 269–275. [Google Scholar] [CrossRef]
- Edwards, N.J.; Oberti, M.; Thangudu, R.R.; Cai, S.; McGarvey, P.B.; Jacob, S.; Madhavan, S.; Ketchum, K.A. The CPTAC Data Portal: A Resource for Cancer Proteomics Research. J. Proteome Res. 2015, 14, 2707–2713. [Google Scholar] [CrossRef]
- Clinical Proteomic Tumor Analysis Consortium (CPTAC)|NCI Genomic Data Commons. Available online: https://gdc.cancer.gov/about-gdc/contributed-genomic-data-cancer-research/clinical-proteomic-tumor-analysis-consortium-cptac (accessed on 6 February 2023).
- Lindgren, C.M.; Adams, D.W.; Kimball, B.; Boekweg, H.; Tayler, S.; Pugh, S.L.; Payne, S.H. Simplified and Unified Access to Cancer Proteogenomic Data. J. Proteome Res. 2021, 20, 1902–1910. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. Correction to ‘The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets’. Nucleic Acids Res. 2021, 49, 10800. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, B.; Xu, J.; Wang, X.; Xie, L.; Zhang, B.; Li, Y.; Li, J. CanProVar 2.0: An Updated Database of Human Cancer Proteome Variation. J. Proteome Res. 2017, 16, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Rose, P.W.; Prlić, A.; Bi, C.; Bluhm, W.F.; Christie, C.H.; Dutta, S.; Green, R.K.; Goodsell, D.S.; Westbrook, J.D.; Woo, J.; et al. The RCSB Protein Data Bank: Views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015, 43, D345–D356. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.U. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2014, 42, 7486. [Google Scholar] [CrossRef]
- Bateman, A; UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res 2019, 47, D506–D515. [CrossRef]
- Orsburn, B.C. Proteome Discoverer—A Community Enhanced Data Processing Suite for Protein Informatics. Proteomes 2021, 9, 15. [Google Scholar] [CrossRef]
- O’Donovan, C.; Martin, M.J.; Gattiker, A.; Gasteiger, E.; Bairoch, A.; Apweiler, R. High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Briefings Bioinform. 2002, 3, 275–284. [Google Scholar] [CrossRef]
- Moriya, Y.; Kawano, S.; Okuda, S.; Watanabe, Y.; Matsumoto, M.; Takami, T.; Kobayashi, D.; Yamanouchi, Y.; Araki, N.; Yoshizawa, A.C.; et al. The jPOST environment: An integrated proteomics data repository and database. Nucleic Acids Res. 2019, 47, D1218–D1224. [Google Scholar] [CrossRef]
- Shao, X.; Taha, I.N.; Clauser, K.; Gao, Y.; Naba, A. MatrisomeDB: The ECM-protein knowledge database. Nucleic Acids Res. 2020, 48, D1136–D1144. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Zhao, H.; Zeng, Y. Lipidomics: A promising cancer biomarker. Clin. Transl. Med. 2018, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Buszewska-Forajta, M.; Pomastowski, P.; Monedeiro, F.; Walczak-Skierska, J.; Markuszewski, M.; Matuszewski, M.; Markuszewski, M.J.; Buszewski, B. Lipidomics as a Diagnostic Tool for Prostate Cancer. Cancers 2021, 13, 2000. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Huang, Y.; Kong, X.; Jia, B.; Lu, X.; Chen, Y.; Huang, Z.; Li, Y.-Y.; Dai, W. DBLiPro: A Database for Lipids and Proteins in Human Lipid Metabolism. Phenomics 2023, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cotter, D.; Maer, A.; Guda, C.; Saunders, B.; Subramaniam, S. LMPD: LIPID MAPS proteome database. Nucleic Acids Res. 2006, 34 (Suppl. S1), D507–D510. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35 (Suppl. S1), D527–D532. [Google Scholar] [CrossRef]
- Liebisch, G.; Fahy, E.; Aoki, J.; Dennis, E.A.; Durand, T.; Ejsing, C.S.; Fedorova, M.; Feussner, I.; Griffiths, W.J.; Köfeler, H.; et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 2020, 61, 1539–1555. [Google Scholar] [CrossRef]
- Blair, B.B.; Funkhouser, A.T.; Goodwin, J.L.; Strigenz, A.M.; Chaballout, B.H.; Martin, J.C.; Arthur, C.M.; Funk, C.R.; Edenfield, W.J.; Blenda, A.V. Increased Circulating Levels of Galectin Proteins in Patients with Breast, Colon, and Lung Cancer. Cancers 2021, 13, 4819. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.-T.; Stowell, S.R. The role of galectins in immunity and infection. Nat. Rev. Immunol. 2023, 23, 1–16. [Google Scholar] [CrossRef]
- Funkhouser, A.T.; Strigenz, A.M.; Blair, B.B.; Miller, A.P.; Shealy, J.C.; Ewing, J.A.; Martin, J.C.; Funk, C.R.; Edenfield, W.J.; Blenda, A.V. KIT Mutations Correlate with Higher Galectin Levels and Brain Metastasis in Breast and Non-Small Cell Lung Cancer. Cancers 2022, 14, 2781. [Google Scholar] [CrossRef] [PubMed]
- Hizal, D.B.; Wolozny, D.; Colao, J.; Jacobson, E.; Tian, Y.; Krag, S.S.; Betenbaugh, M.J.; Zhang, H. Glycoproteomic and glycomic databases. Clin. Proteom. 2014, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhang, H. Glycoproteomics and clinical applications. Proteom.-Clin. Appl. 2010, 4, 124–132. [Google Scholar] [CrossRef]
- Kim, E.H.; Misek, D.E. Glycoproteomics-Based Identification of Cancer Biomarkers. Int. J. Proteom. 2011, 2011, 2010–2166. [Google Scholar] [CrossRef]
- Pan, S.; Chen, R.; Aebersold, R.; Brentnall, T.A. Mass Spectrometry Based Glycoproteomics—From a Proteomics Perspective. Mol. Cell. Proteom. 2011, 10, R110.003251. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.A.; Relvas-Santos, M.; Peixoto, A.; Silva, A.M.; Santos, L.L. Glycoproteogenomics: Setting the Course for Next-generation Cancer Neoantigen Discovery for Cancer Vaccines. Genom. Proteom. Bioinform. 2021, 19, 25–43. [Google Scholar] [CrossRef]
- Cooper, C.A.; Harrison, M.J.; Wilkins, M.R.; Packer, N.H. GlycoSuiteDB: A new curated relational database of glycoprotein glycan structures and their biological sources. Nucleic Acids Res. 2001, 29, 332–335. [Google Scholar] [CrossRef]
- Hayes, C.A.; Karlsson, N.G.; Struwe, W.B.; Lisacek, F.; Rudd, P.M.; Packer, N.H.; Campbell, M.P. UniCarb-DB: A database resource for glycomic discovery. Bioinformatics 2011, 27, 1343–1344. [Google Scholar] [CrossRef]
- von der Lieth, C.-W.; Freire, A.A.; Blank, D.; Campbell, M.P.; Ceroni, A.; Damerell, D.R.; Dell, A.; Dwek, R.A.; Ernst, B.; Fogh, R.; et al. EUROCarbDB: An open-access platform for glycoinformatics. Glycobiology 2011, 21, 493–502. [Google Scholar] [CrossRef]
- Zhang, H.; Loriaux, P.; Eng, J.; Campbell, D.; Keller, A.; Moss, P.; Bonneau, R.; Zhang, N.; Zhou, Y.; Wollscheid, B.; et al. UniPep—a database for human N-linked glycosites: A resource for biomarker discovery. Genome Biol. 2006, 7, R73. [Google Scholar] [CrossRef]
- Togayachi, A.; Dae, K.-Y.; Shikanai, T.; Narimatsu, H. A Database System for Glycogenes (GGDB). Exp. Glycosci. 2008, 423–425. [Google Scholar] [CrossRef]
- Ranzinger, R.; Frank, M.; von der Lieth, C.-W.; Herget, S. Glycome-DB.org: A portal for querying across the digital world of carbohydrate sequences. Glycobiology 2009, 19, 1563–1567. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.P.; Royle, L.; Radcliffe, C.M.; Dwek, R.A.; Rudd, P.M. GlycoBase and autoGU: Tools for HPLC-based glycan analysis. Bioinformatics 2008, 24, 1214–1216. [Google Scholar] [CrossRef]
- Zhao, S.; Walsh, I.; Abrahams, J.L.; Royle, L.; Nguyen-Khuong, T.; Spencer, D.; Fernandes, D.L.; Packer, N.H.; Rudd, P.M.; Campbell, M.P. GlycoStore: A database of retention properties for glycan analysis. Bioinformatics 2018, 34, 3231–3232. [Google Scholar] [CrossRef] [PubMed]
- Ranzinger, R.; Aoki-Kinoshita, K.F.; Campbell, M.P.; Kawano, S.; Lütteke, T.; Okuda, S.; Shinmachi, D.; Shikanai, T.; Sawaki, H.; Toukach, P.; et al. GlycoRDF: An ontology to standardize glycomics data in RDF. Bioinformatics 2015, 31, 919–925. [Google Scholar] [CrossRef]
- Weatherly, D.B.; Arpinar, F.S.; Porterfield, M.; Tiemeyer, M.; York, W.S.; Ranzinger, R. GRITS Toolbox—A freely available software for processing, annotating and archiving glycomics mass spectrometry data. Glycobiology 2019, 29, 452–460. [Google Scholar] [CrossRef]
- Tiemeyer, M.; Aoki, K.; Paulson, J.; Cummings, R.D.; York, W.S.; Karlsson, N.G.; Lisacek, F.; Packer, N.H.; Campbell, M.P.; Aoki, N.P.; et al. GlyTouCan: An accessible glycan structure repository. Glycobiology 2017, 27, 915–919. [Google Scholar] [CrossRef]
- Hirabayashi, J.; Tateno, H.; Shikanai, T.; Aoki-Kinoshita, K.F.; Narimatsu, H. The Lectin Frontier Database (LfDB), and Data Generation Based on Frontal Affinity Chromatography. Molecules 2015, 20, 951–973. [Google Scholar] [CrossRef]
- Toukach, P.V.; Shirkovskaya, A.I. Carbohydrate Structure Database and Other Glycan Databases as an Important Element of Glycoinformatics. Russ. J. Bioorg. Chem. 2022, 48, 457–466. [Google Scholar] [CrossRef]
- Solomon, B.D.; Nguyen, A.-D.; Bear, K.A.; Wolfsberg, T.G. Clinical Genomic Database. Proc. Natl. Acad. Sci. 2013, 110, 9851–9855. [Google Scholar] [CrossRef]
- Hartmaier, R.J.; Albacker, L.A.; Chmielecki, J.; Bailey, M.; He, J.; Goldberg, M.E.; Ramkissoon, S.; Suh, J.; Elvin, J.A.; Chiacchia, S.; et al. High-throughput genomic profiling of adult solid tumors reveals novel insights into cancer pathogenesis. Cancer Res. 2017, 77, 2464–2475. [Google Scholar] [CrossRef] [PubMed]
- Mudaranthakam, D.P.; Thompson, J.; Hu, J.; Pei, D.; Chintala, S.R.; Park, M.; Fridley, B.L.; Gajewski, B.; Koestler, D.C.; Mayo, M.S. A Curated Cancer Clinical Outcomes Database (C3OD) for accelerating patient recruitment in cancer clinical trials. JAMIA Open 2018, 1, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Overgaard, J.; Jovanovic, A.; Godballe, C.; Eriksen, J.G. The Danish Head and Neck Cancer database. Clin. Epidemiol. 2016, 8, 491–496. [Google Scholar] [CrossRef] [PubMed]
- McCabe, R.M. National Cancer Database: The Past, Present, and Future of the Cancer Registry and Its Efforts to Improve the Quality of Cancer Care. Semin. Radiat. Oncol. 2019, 29, 323–325. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.C.; Paquette, I.M. Surveillance, Epidemiology, and End Results (SEER) and SEER-Medicare Databases: Use in Clinical Research for Improving Colorectal Cancer Outcomes. Clin. Colon Rectal Surg. 2019, 32, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Kattman, B.L. ClinVar at five years: Delivering on the promise. Hum. Mutat. 2018, 39, 1623–1630. [Google Scholar] [CrossRef]
- Nanda, J.S.; Kumar, R.; Raghava, G.P.S. dbEM: A database of epigenetic modifiers curated from cancerous and normal genomes. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Ullah, S.; Ullah, F.; Rahman, W.; Karras, A.D.; Ullah, A.; Ahmad, G.; Ijaz, M.; Gao, T. The Cancer Research Database (CRDB): Integrated Platform to Gain Statistical Insight Into the Correlation between Cancer and COVID-19. JMIR Cancer 2022, 8. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, G.; Zhang, L.; Wang, Q.; Li, H.; Han, Y.; Xie, L.; Yan, Z.; Li, Y.; An, Y.; et al. Comprehensive Review of Web Servers and Bioinformatics Tools for Cancer Prognosis Analysis. Front. Oncol. 2020, 10, 68. [Google Scholar] [CrossRef]
- Goswami, C.P.; Nakshatri, H. PROGgeneV2: Enhancements on the existing database. BMC Cancer 2014, 14, 1–6. [Google Scholar] [CrossRef]
- Kumar, R.; Chaudhary, K.; Gupta, S.; Singh, H.; Kumar, S.; Gautam, A.; Kapoor, P.; Raghava, G.P.S. CancerDR: Cancer Drug Resistance Database. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-H.; Shen, P.-C.; Chen, C.-Y.; Hsu, A.-N.; Cho, Y.-C.; Lai, Y.-L.; Chen, F.-H.; Li, C.-Y.; Wang, S.-C.; Chen, M.; et al. DriverDBv3: A multi-omics database for cancer driver gene research. Nucleic Acids Res. 2020, 48, D863–D870. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Wang, P.; Tian, R.; Wang, S.; Guo, Q.; Luo, M.; Zhou, W.; Liu, G.; Jiang, H.; Jiang, Q. LncRNA2Target v2.0: A comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2019, 47, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Shang, S.; Guo, S.; Li, X.; Zhou, H.; Liu, H.; Sun, Y.; Wang, J.; Wang, P.; Zhi, H.; et al. Lnc2Cancer 3.0: An updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 2021, 49, D1251–D1258. [Google Scholar] [CrossRef] [PubMed]
- Carithers, L.J.; Moore, H.M. The Genotype-Tissue Expression (GTEx) Project. Biopreservation Biobanking 2015, 13, 307–308. [Google Scholar] [CrossRef]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2021. Nucleic Acids Res. 2021, 49, D1138–D1143. [Google Scholar] [CrossRef]
- Chen, X.; Ji, Z.L.; Chen, Y.Z. TTD: Therapeutic Target Database. Nucleic Acids Res. 2002, 30, 412–415. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S.; Li, F.; Zhou, Y.; Zhang, Y.; Wang, Z.; Zhang, R.; Zhu, J.; Ren, Y.; Tan, Y.; et al. Therapeutic target database 2020: Enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020, 48, D1031–D1041. [Google Scholar] [CrossRef]
- Thorn, C.F.; Klein, T.E.; Altman, R.B. PharmGKB: The pharmacogenomics knowledge base. Methods Mol. Biol. 2013, 1015, 311–320. [Google Scholar]
- Gong, L.; Whirl-Carrillo, M.; Klein, T.E. PharmGKB, an Integrated Resource of Pharmacogenomic Knowledge. Curr. Protoc. 2021, 1, e226. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, J.; Meinel, T.; Dunkel, M.; Murgueitio, M.S.; Adams, R.; Blasse, C.; Eckert, A.; Preissner, S.; Preissner, R. CancerResource: A comprehensive database of cancer-relevant proteins and compound interactions supported by experimental knowledge. Nucleic Acids Res. 2011, 39, D960–D967. [Google Scholar] [CrossRef] [PubMed]
- Gohlke, B.-O.; Nickel, J.; Otto, R.; Dunkel, M.; Preissner, R. CancerResource—updated database of cancer-relevant proteins, mutations and interacting drugs. Nucleic Acids Res. 2016, 44, D932–D937. [Google Scholar] [CrossRef]
- Küntzer, J.; Maisel, D.; Lenhof, H.-P.; Klostermann, S.; Burtscher, H. The Roche Cancer Genome Database 2.0. BMC Med Genom. 2011, 4, 43. [Google Scholar] [CrossRef]
- Higgins, M.E.; Claremont, M.; Major, J.E.; Sander, C.; Lash, A.E. CancerGenes: A gene selection resource for cancer genome projects. Nucleic Acids Res. 2007, 35 (Suppl. S1), D721–D726. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-J.; Hu, F.-F.; Xia, M.-X.; Han, L.; Zhang, Q.; Guo, A.-Y. GSCALite: A web server for gene set cancer analysis. Bioinformatics 2018, 34, 3771–3772. [Google Scholar] [CrossRef]
- Hamosh, A.; Amberger, J.S.; Bocchini, C.; Scott, A.F.; Rasmussen, S.A. Online Mendelian Inheritance in Man (OMIM®): Victor McKusick’s magnum opus. Am. J. Med Genet. Part A 2021, 185, 3259–3265. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Wen, B.; Wang, X.; Zhang, B. PepQuery enables fast, accurate, and convenient proteomic validation of novel genomic alterations. Genome Res. 2019, 29, 485–493. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 1–9. [Google Scholar] [CrossRef]
| Name | Content/Functionality | Web Service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| The Cancer Genome Atlas Cases = 11,315 | Genome sequencing across 33 tumor types | Yes | Yes | Yes | F, A, I, R | https://www.cancer.gov/ccg/research/genome-sequencing/tcga (accessed on 14 March 2023) |
| OncomiR Cancer Database OMCD Cases = 9500 | Comparative genomic analysis of miRNA data sequencing | Yes | N/A | Yes | F, A, I | http://www.oncomir.org/cgi-bin/dbSearch.cgi (accessed on 14 March 2023) |
| cBio Cancer Genomic Portal | Genomic analysis of cancer-related genes | Yes | Yes | Yes | F, A, I, R | https://www.cbioportal.org/ (accessed on 14 March 2023) |
| International Cancer Genome Consortium (ICGC) Donors ~24,500 | Catalog of mutational abnormalities in major tumor types | Yes | Yes | Yes | F, A, I, R | https://dcc.icgc.org/ (accessed on 14 March 2023) |
| Human Genome Browser at USCS | Genomic data | Yes | Yes | F, A, R | https://genome.ucsc.edu/index.html (accessed on 14 March 2023) | |
| Gene Expression Omnibus Database (GEO) | Gene expression data | Yes | Yes | F, A, R | https://www.ncbi.nlm.nih.gov/geo/ (accessed on 14 March 2023) | |
| Ensembl | Genomic analysis | Yes | Yes | F, A, R | https://www.ensembl.org/index.html (accessed on 14 March 2023) | |
| National Cancer Institute Genomic Commons (GDC) Cases = 22,000 | Storage, analysis, and sharing of clinical data of patients | Yes | Yes | Yes | F, A, I, R | https://portal.gdc.cancer.gov/ (accessed on 14 March 2023) |
| Network of Cancer Genes | Cancer genes, healthy drivers and their properties | Yes | Yes | Yes | F, A, I, R | http://ncg.kcl.ac.uk/index.php (accessed on 14 March 2023) |
| Catalogue of Somatic Mutation in Cancer (COSMIC) | Somatic mutations in human cancer | Yes | Yes | Yes | F, A, I, R | https://cancer.sanger.ac.uk/cosmic (accessed on 14 March 2023) |
| Mutagene | Mutational profiles in 37 cancer types | Yes | Yes | Yes | F, A, I, R | https://www.ncbi.nlm.nih.gov/research/mutagene/ (accessed on 14 March 2023) |
| Progenetix Samples = 142,063 | Cancer copy number abnormalities (CNA) | Yes | Yes | Yes | F, A, I, R | https://progenetix.org/ (accessed on 14 March 2023) |
| MutEx | Stores and explores the relationships between gene expression, somatic mutation,mutational burden and and survival | Yes | Yes | Yes | F, A, I, R | http://www.innovebioinfo.com/Databases/Mutationdb_About.php (accessed on 14 March 2023) |
| Oncomine | Precision oncology through next-generation sequencing | Yes | Yes | Yes | F, A, I, R | https://www.oncomine.com/ (accessed on 14 March 2023) |
| Lung Cancer Explorer (LCE) Entries = 356 | Enables the exploration of lung cancer in various analyses | Yes | Yes | F, A, R | https://lce.biohpc.swmed.edu/lungcancer/imageset_tcga.php (accessed on 14 March 2023) | |
| Prostate Cancer Precision Medicine Multi-Institutional Collaborative Effort PROMISE | Analyzes prostate cancer genes and patient outcomes | Yes | Yes | F, A, I | https://www.prostatecancerpromise.org/research/ (accessed on 14 March 2023) | |
| HCCDb | Information about hepatocellular carcinoma | Yes | Yes | F, A, R | http://lifeome.net/database/hccdb/home.html (accessed on 14 March 2023) |
| Name | Content/Functionality | Web Service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| Clinical Proteomic Tumor Analysis Consortium (CPTAC) | Application of large-scale proteome and genome analysis | Yes | Yes | Yes | F, A, I, R | https://proteomics.cancer.gov/programs/cptac (accessed on 14 March 2023) |
| String Database | Protein-protein interactions, functional enrichment analysis | Yes | Yes | Yes | F, A, I, R | https://string-db.org/ (accessed on 14 March 2023) |
| Ualcan | Analyzes cancer transcriptome, proteome, and patient survival data | Yes | N/A | Yes | F, A, I | https://ualcan.path.uab.edu/ (accessed on 14 March 2023) |
| CanProVar | Proteomic variations | Yes | Yes | F, A, R | http://119.3.70.71/CanProVar/index.html (accessed on 14 March 2023) | |
| RCSB Protein Data Bank * | Works with UniProt and analyzes protein structures | Yes | Yes | Yes | F, A, I, R | https://www.rcsb.org/ (accessed on 14 March 2023) |
| Universal Protein Resource (UniProt) * | Information about protein structures and interactions | Yes | Yes | Yes | F, A, I, R | https://www.uniprot.org/ (accessed on 14 March 2023) |
| Proteome Discover * | Not free to access (attempted to access on 14 March 2023) | |||||
| Swiss-Prot and TrEMBL * | A part of the UniProt database | Yes | Yes | Yes | F, A, I, R | https://www.uniprot.org/uniprotkb (accessed on 14 March 2023) |
| jPOST * | Post-translational modifications on proteins | Yes | Yes | Yes | F, A, I, R | https://globe.jpostdb.org/ (accessed on 14 March 2023) |
| MatrisomeDB * | Proteomic data from studies of ECM ** | Yes | Yes | F, A, R | https://matrisomedb.org/ (accessed on 14 March 2023) |
| Databases | Content/Functionality | Web Service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| DBLiPro * | Focuses on human lipid metabolism and provides lipidome-centric analysis | Yes | Yes | Yes | F, A, I, R | http://lipid.cloudna.cn/home (accessed on 14 March 2023) |
| Lipid Maps * | Lipid structures | Yes | Yes | Yes | F, A, I, R | https://www.lipidmaps.org/ (accessed on 14 March 2023) |
| Name | Content/Functionality | Web Service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| UniCarb-DB * | Carbohydrates characterized by LC-MS | Yes | Yes | Yes | F, A, I, R | https://unicarb-db.expasy.org/about (accessed on 14 March 2023) |
| UniPep * | N-linked glycosites for proteomic analyses | Yes | Yes | F, A, R | https://unipep.systemsbiology.net/ (accessed on 14 March 2023) | |
| GlycoGene (GGDB) * | Information about glycogenes | Yes | Yes | F, A, R | https://www.glycogene.com/ (accessed on 14 March 2023) | |
| Glycome-DB * | A part of the GlyTouCan database | http://www.glycome-db.org/ (accessed on 14 March 2023) | ||||
| GlycoRDF * | Glycomics data | Yes | Yes | F, A, R | https://github.com/glycoinfo/GlycoRDF/wiki (accessed on 14 March 2023) | |
| GRITs Toolbox* | Processing, annotating and archiving of glycomics data with a focus on MS data | Yes | Yes | Yes | F, A, I, R | http://www.grits-toolbox.org/ (accessed on 14 March 2023) |
| GlyTouCan * | Glycan structure repository | Yes | Yes | F, A, R | https://glytoucan.org/ (accessed on 14 March 2023) | |
| The Lectin Frontier Database (LfDB) * | Lectin-standard oligosaccharide interactions | Yes | F, A | https://acgg.asia/lfdb2/ (accessed on 14 March 2023) | ||
| Carbohydrate Structure Database (CSDB) * | Manually curated natural carbohydrate structures, taxonomy, bibliography, NMR, and other data | Yes | Yes, this is done by an external source | F, A, I, R | http://csdb.glycoscience.ru/database/index.html (accessed on 14 March 2023) |
| Name | Content/Functionality | Web Service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| Clinical Genomic Database (GCD) | Genetic information pertaining to patient care | Yes | Yes | F, A, R | https://research.nhgri.nih.gov/CGD/ (accessed on 14 March 2023) | |
| Foundation Medicine Adult-Cancer-Clinical Dataset | Clinical relevance of rare alterations and diseases | Yes | Yes | Yes | F, A, I, R | https://gdc.cancer.gov/about-gdc/contributed-genomic-data-cancer-research/foundation-medicine/foundation-medicine (accessed on 14 March 2023) |
| A Curated Cancer Clinical Outcome Database (C3OD) | Cannot access (14 March 2023) | |||||
| Danish Head and Neck Cancer Database | National guidelines, clinical studies for improved treatment | Yes | Yes | F, A, R | https://www.dahanca.dk/IndexPage (accessed on 14 March 2023) | |
| National Cancer Database (NCDB) | Requires login access | F | https://www.facs.org/quality-programs/cancer-programs/national-cancer-database/ (accessed on 14 March 2023) | |||
| Surveillance Epidemiology and End Results (SEER) * | Information on cancer statistics | Yes | F, A | https://seer.cancer.gov/ (accessed on 14 March 2023) | ||
| ClinVar * | Information about genomic variations and their relationship to human health. | Yes | Yes | F, A, R | https://www.ncbi.nlm.nih.gov/clinvar/ (accessed on 14 March 2023) |
| Name | Content/Functionality | Web Service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| Database of Epigenetics Modifiers (dbEM) * | Genomic information about epigenetic modifiers from cancerous and normal genomes | Yes | F, A | https://webs.iiitd.edu.in/raghava/dbem/index.php (accessed on 14 March 2023) | ||
| Cancer Research Database (CRDB) | Analyses of other cancer research databases | Yes | F, A | https://www.habdsk.org/crdb (accessed on 14 March 2023) | ||
| PROGgene | Pan Cancer Prognostics | Yes | Yes | F, A, I | http://www.progtools.net/gene/index.php (accessed on 14 March 2023) | |
| Cancer Drug Resistance (CancerDR) | Information about anticancer drugs and their effectiveness against cancer cell lines | Yes | Yes | F, A, R | https://webs.iiitd.edu.in/raghava/cancerdr/index.html (accessed on 14 March 2023) | |
| DriverDBv3 | Cancer driver genes and mutations | Yes | Yes | Yes | F, A, I, R | http://driverdb.tms.cmu.edu.tw/ (accessed on 14 March 2023) |
| LncRNA2Target 2.0 * | Server unreachable (14 March 2023) | |||||
| Lnc2Cancers 3.0 | Server unreachable (14 March 2023) | |||||
| Genotype Expression Project (GTEx) * | Gene expression data, QTLs, and histology images | Yes | Yes | F, A, R | https://www.gtexportal.org/home/ (accessed on 14 March 2023) | |
| Comparative Toxicogenomic Database (CTD) * | Chemical–gene/protein interactions, chemical–disease and gene–disease relationships | Yes | Yes | Yes | F, A, I, R | http://ctdbase.org/ (accessed on 14 March 2023) |
| Therapeutic Target Database (TTD) | Therapeutic protein and nucleic acid targets, the targeted disease, pathway information and the corresponding drugs | Yes | Yes | Yes | F, A, I, R | https://db.idrblab.net/ttd/ (accessed on 14 March 2023) |
| Pharmacogenomics Knowledge Base (PharmGKB) | Knowledge about the impact of genetic variation on drug response | Yes | Yes | F, A, I, R | https://www.pharmgkb.org/ (accessed on 14 March 2023) | |
| DrugBank | Information about drugs, mechanisms of action, and interactions | Yes | Yes | F, A, I, R | https://go.drugbank.com/ (accessed on 14 March 2023) |
| Name | Content/Functionality | Web Service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| Gene Set Cancer Analysis (GSCALite) | Analysis platform for gene set cancer analysis | Yes | F, A | http://bioinfo.life.hust.edu.cn/web/GSCALite/ (accessed on 14 March 2023) | ||
| Online Mendelian Inheritance in Man (OMIM) | Online catalog of human genes and genetic disorders | Yes | Yes | F, A, R | https://www.omim.org/ (accessed on 14 March 2023) | |
| Gene Expression Profiling Interactive Analysis (GEPIA) * | Analysis of RNA sequencing expression data | Yes | Yes | Yes | F, A, I, R | http://gepia.cancer-pku.cn/ (accessed on 14 March 2023) |
| PepQuery * | Universal targeted peptide search engine | Yes | Yes | F, A, R | http://www.pepquery.org/ (accessed on 14 March 2023) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Austin, B.K.; Firooz, A.; Valafar, H.; Blenda, A.V. An Updated Overview of Existing Cancer Databases and Identified Needs. Biology 2023, 12, 1152. https://doi.org/10.3390/biology12081152
Austin BK, Firooz A, Valafar H, Blenda AV. An Updated Overview of Existing Cancer Databases and Identified Needs. Biology. 2023; 12(8):1152. https://doi.org/10.3390/biology12081152
Chicago/Turabian StyleAustin, Brittany K., Ali Firooz, Homayoun Valafar, and Anna V. Blenda. 2023. "An Updated Overview of Existing Cancer Databases and Identified Needs" Biology 12, no. 8: 1152. https://doi.org/10.3390/biology12081152
APA StyleAustin, B. K., Firooz, A., Valafar, H., & Blenda, A. V. (2023). An Updated Overview of Existing Cancer Databases and Identified Needs. Biology, 12(8), 1152. https://doi.org/10.3390/biology12081152








