Sex-Linked Growth Disorder and Aberrant Pituitary Gene Expression in Nestin-Cre-Mediated Egr1 Conditional Knockout Mice
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
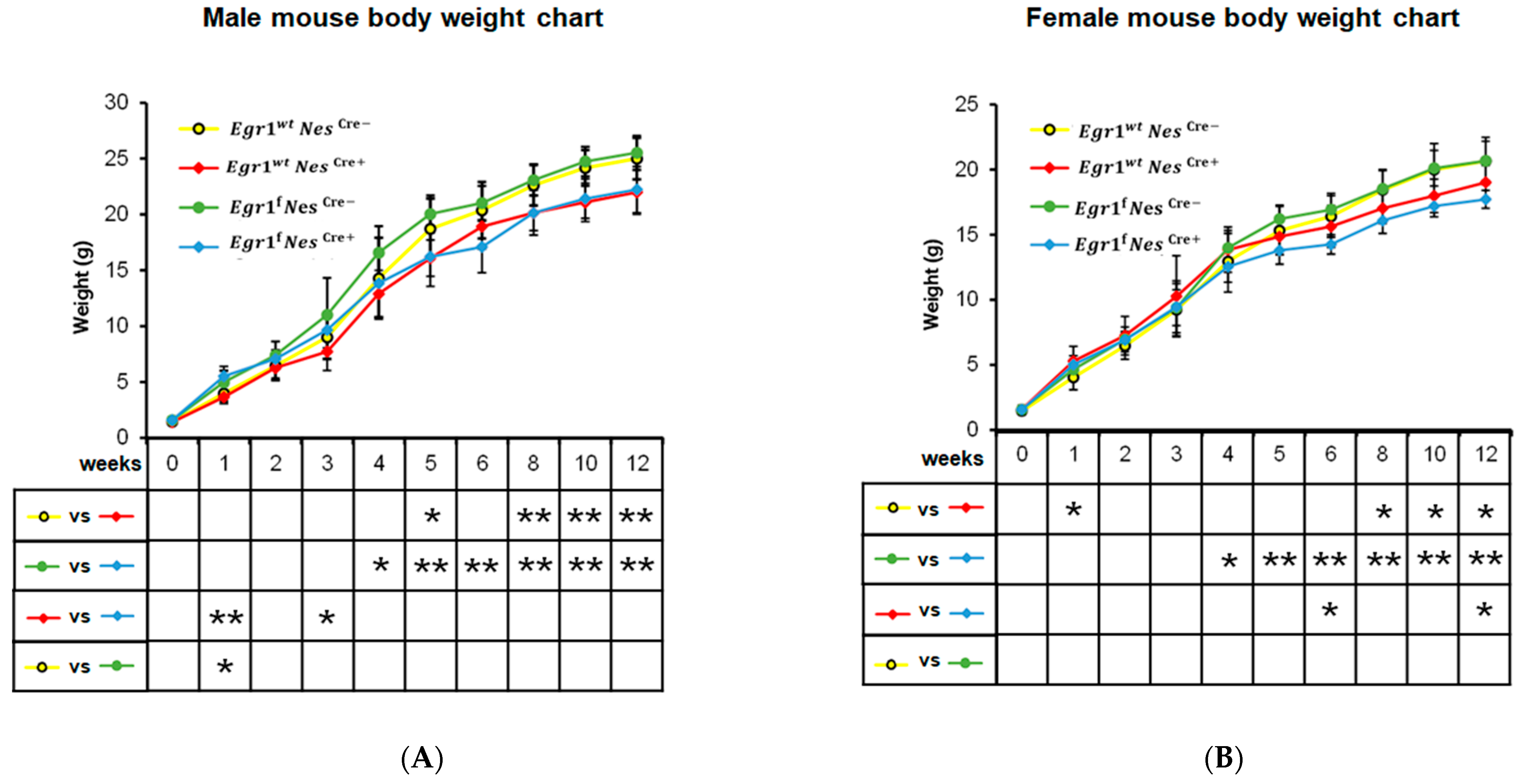
2.2. Measurement of Growth
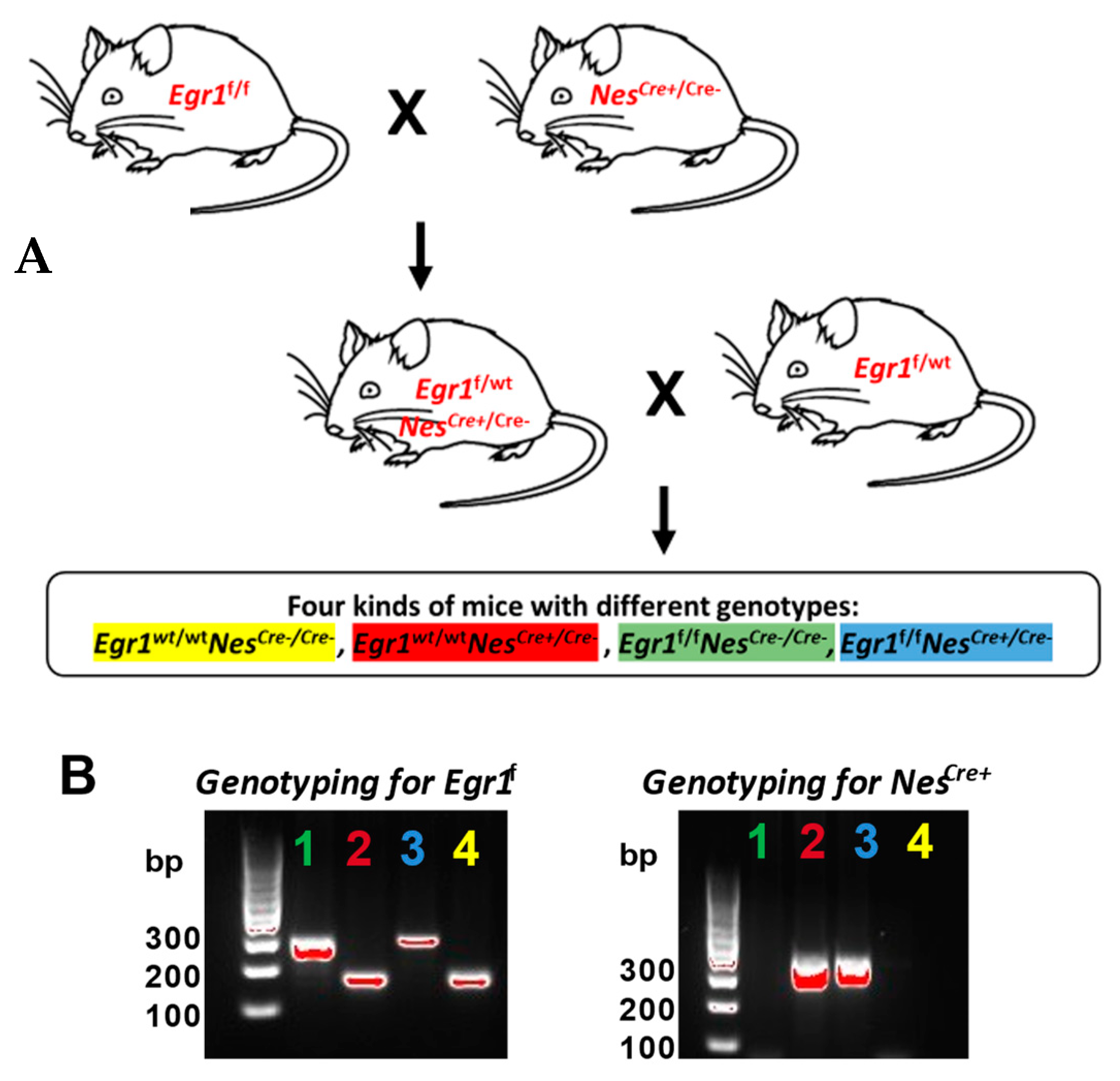
2.3. Genotyping PCR
2.4. Immunohistochemistry (IHC)
2.5. RNAscope
2.6. RNA Extraction, RNA-Seq Library Construction, and Data Analysis
2.7. GO Analysis
2.8. Statistical Analysis
3. Results
3.1. Generation and Characterization of Egr1cKO Mice
3.2. Growth Reduction Observed in Both Nestin-Cre and Egr1cKO Mice
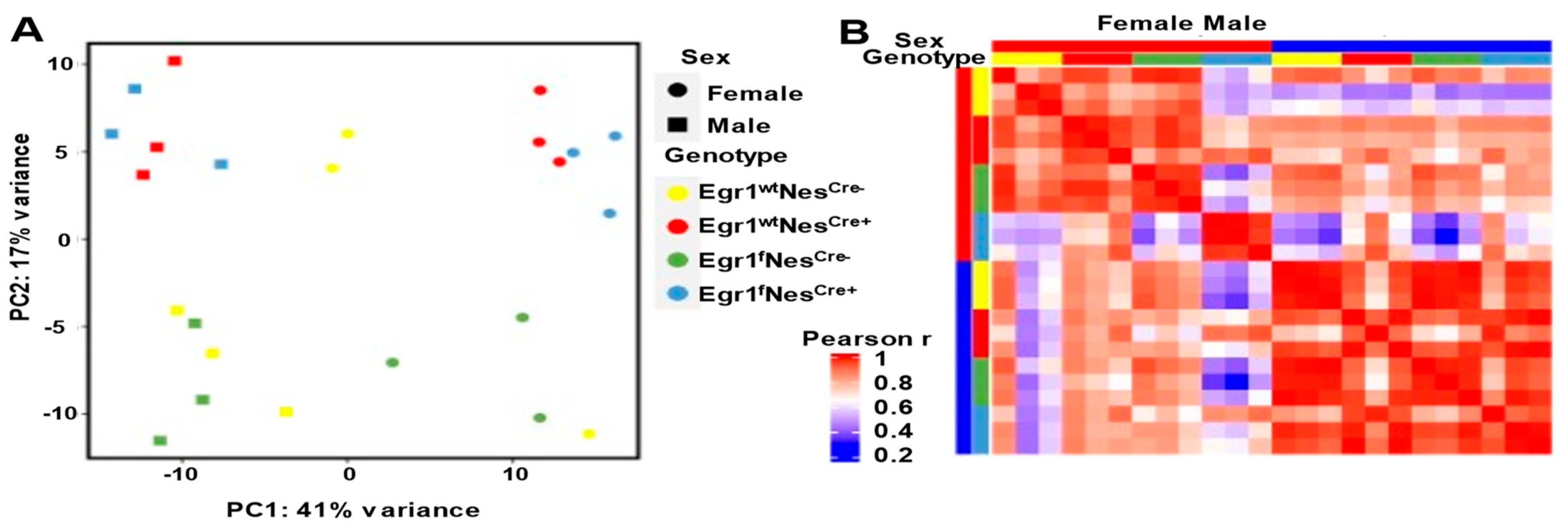
3.3. Sex Differences in Gene Expression Amplified in Nestin-Cre and Egr1cKO Mice
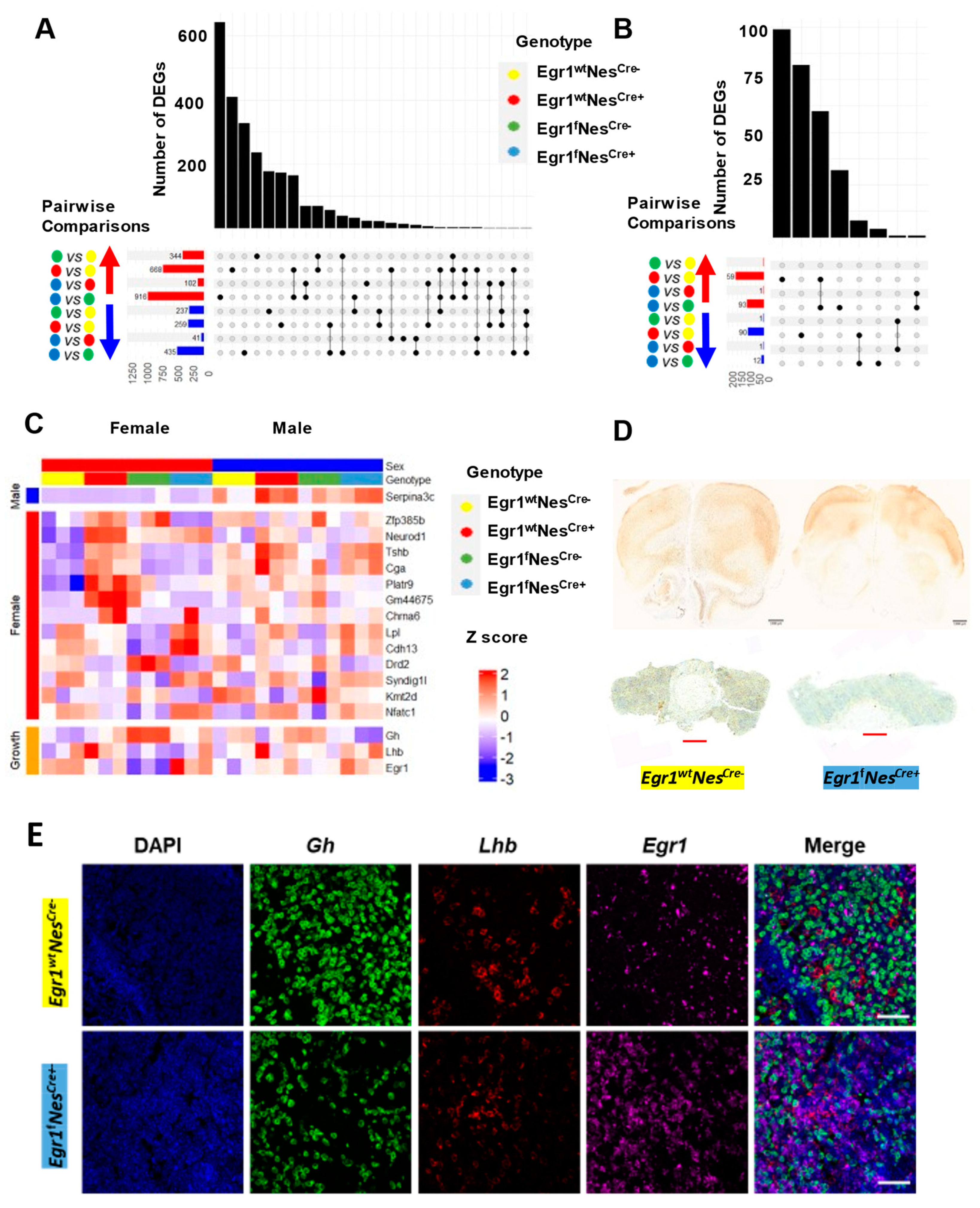
3.4. Aberrant Gene Expression Associated with Nestin-Cre and Egr1cKO Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GH | Growth hormone |
| LH | Luteinizing hormone |
| PRL | Prolactin |
| TSH | Thyroid-stimulating hormone |
| FSH | Follicle-stimulating hormone |
| Egr1 | Early growth response 1 |
| sf1 | Steroidogenic factor 1 |
| Pitx1 | Pituitary homeobox factor 1 |
| EGR1 | Early growth response 1 |
| GnRH | Gonadotropin-releasing hormone |
| PCA | Principal component analysis |
| GO | Gene ontology |
References
- Rizzoti, K. Genetic regulation of murine pituitary development. J. Mol. Endocrinol. 2015, 54, R55–R73. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, P.A.; Smiljanic, K.; Prévide, R.M.; Iben, J.R.; Li, T.; Rokic, M.B.; Sherman, A.; Coon, S.L.; Stojilkovic, S.S. Cell Type- and Sex-Dependent Transcriptome Profiles of Rat Anterior Pituitary Cells. Front. Endocrinol. 2019, 10, 623. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.; Hu, P.; Peel, M.T.; Chen, S.; Camara, P.G.; Epstein, D.J.; Wu, H.; Liebhaber, S.A. Single-cell transcriptomic analysis of adult mouse pituitary reveals sexual dimorphism and physiologic demand-induced cellular plasticity. Protein Cell. 2020, 11, 565–583. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.W.; Call, G.B. Early growth response protein 1 binds to the luteinizing hormone-beta promoter and mediates gonadotropin-releasing hormone-stimulated gene expression. Mol. Endocrinol. 1999, 13, 752–763. [Google Scholar] [PubMed]
- Quirk, C.C.; Lozada, K.L.; Keri, R.A.; Nilson, J.H. A single Pitx1 binding site is essential for activity of the LHbeta promoter in transgenic mice. Mol. Endocrinol. 2001, 15, 734–746. [Google Scholar]
- Jorgensen, J.S.; Quirk, C.C.; Nilson, J.H. Multiple and overlapping combinatorial codes orchestrate hormonal responsiveness and dictate cell-specific expression of the genes encoding luteinizing hormone. Endocr. Rev. 2004, 25, 521–542. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, U.B.; Halvorson, L.M.; Chen, M.T. Sp1, steroidogenic factor 1 (SF-1), and early growth response protein 1 (egr-1) binding sites form a tripartite gonadotropin-releasing hormone response element in the rat luteinizing hormone-beta gene promoter: An integral role for SF-1. Mol. Endocrinol. 2000, 14, 1235–1245. [Google Scholar]
- Tremblay, J.J.; Drouin, J. Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone beta gene transcription. Mol. Cell. Biol. 1999, 19, 2567–2576. [Google Scholar] [CrossRef]
- Hashimoto, H.; Olanrewaju, Y.O.; Zheng, Y.; Wilson, G.G.; Zhang, X.; Cheng, X. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes. Dev. 2014, 28, 2304–2313. [Google Scholar] [CrossRef]
- Veyrac, A.; Besnard, A.; Caboche, J.; Davis, S.; Laroche, S. The transcription factor Zif268/Egr1, brain plasticity, and memory. Progress. Mol. Biol. Transl. Sci. 2014, 122, 89–129. [Google Scholar]
- Li, L.; Carter, J.; Gao, X.; Whitehead, J.; Tourtellotte, W.G. The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors. Mol. Cell. Biol. 2005, 25, 10286–10300. [Google Scholar] [CrossRef]
- Gashler, A.; Sukhatme, V.P. Early growth response protein 1 (Egr-1): Prototype of a zinc-finger family of transcription factors. Prog. Nucleic Acid. Res. Mol. Biol. 1995, 50, 191–224. [Google Scholar] [PubMed]
- Sun, Z.; Xu, X.; He, J.; Murray, A.; Sun, M.-A.; Wei, X.; Wang, X.; McCoig, E.; Xie, E.; Jiang, X.; et al. EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity. Nat. Commun. 2019, 10, 3892. [Google Scholar] [CrossRef] [PubMed]
- Haisenleder, D.J.; Dalkin, A.C.; Ortolano, G.A.; Marshall, J.C.; Shupnik, M.A. A pulsatile gonadotropin-releasing hormone stimulus is required to increase transcription of the gonadotropin subunit genes: Evidence for differential regulation of transcription by pulse frequency in vivo. Endocrinology 1991, 128, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Ruf-Zamojski, F.; Ge, Y.; Nair, V.; Zamojski, M.; Pincas, H.; Toufaily, C.; Tome-Garcia, J.; Stoeckius, M.; Stephenson, W.; Smith, G.R.; et al. Single-cell stabilization method identifies gonadotrope transcriptional dynamics and pituitary cell type heterogeneity. Nucleic Acids Res. 2018, 46, 11370–11380. [Google Scholar] [CrossRef]
- Lee, S.L.; Sadovsky, Y.; Swirnoff, A.H.; Polish, J.A.; Goda, P.; Gavrilina, G.; Milbrandt, J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1). Science 1996, 273, 1219–1221. [Google Scholar] [CrossRef]
- Topilko, P.; Schneider-Maunoury, S.; Levi, G.; Trembleau, A.; Gourdji, D.; Driancourt, M.A.; Rao, C.V.; Charnay, P. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol. Endocrinol. 1998, 12, 107–122. [Google Scholar] [CrossRef]
- Hou, H.; Chan, C.; Yuki, K.E.; Sokolowski, D.; Roy, A.; Qu, R.; Uusküla-Reimand, L.; Faykoo-Martinez, M.; Hudson, M.; Corre, C.; et al. Postnatal developmental trajectory of sex-biased gene expression in the mouse pituitary gland. Biol. Sex. Differ. 2022, 13, 57. [Google Scholar] [CrossRef]
- Knight, C.; Slade, J.P.; Carter, D. The nuclear, 75 kDa form of early growth response protein-1/nerve growth factor-induced A protein is primarily restricted to LH beta-subunit-expressing cells in rat anterior pituitary. Eur. J. Endocrinol. 2000, 143, 817–821. [Google Scholar] [CrossRef]
- Man, P.S.; Wells, T.; Carter, D.A. Egr-1-d2EGFP transgenic rats identify transient populations of neurons and glial cells during postnatal brain development. Gene Expr. Patterns 2007, 7, 872–883. [Google Scholar] [CrossRef]
- Man, P.S.; Wells, T.; Carter, D.A. Cellular distribution of Egr1 transcription in the male rat pituitary gland. J. Mol. Endocrinol. 2014, 53, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Bernal, A.; Arranz, L. Nestin-expressing progenitor cells: Function, identity and therapeutic implications. Cell. Mol. Life Sci. 2018, 75, 2177–2195. [Google Scholar] [CrossRef]
- Tronche, F.; Kellendonk, C.; Kretz, O.; Gass, P.; Anlag, K.; Orban, P.C.; Bock, R.; Klein, R.; Schütz, G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 1999, 23, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Kato, T.; Higuchi, M.; Yako, H.; Chen, M.; Kanno, N.; Ueharu, H.; Kato, Y. Rapid transition of NESTIN-expressing dividing cells from PROP1-positive to PIT1-positive advances prenatal pituitary development. J. Neuroendocrinol. 2013, 25, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Gleiberman, A.S.; Michurina, T.; Encinas, J.M.; Roig, J.L.; Krasnov, P.; Balordi, F.; Fishell, G.; Rosenfeld, M.G.; Enikolopov, G. Genetic approaches identify adult pituitary stem cells. Proc. Natl. Acad. Sci. USA 2008, 105, 6332–6337. [Google Scholar] [CrossRef] [PubMed]
- Araujo, D.M.; Cotman, C.W. Basic FGF in astroglial, microglial, and neuronal cultures: Characterization of binding sites and modulation of release by lymphokines and trophic factors. J. Neurosci. 1992, 12, 1668–1678. [Google Scholar] [CrossRef]
- Friedman, W.J.; Thakur, S.; Seidman, L.; Rabson, A.B. Regulation of nerve growth factor mRNA by interleukin-1 in rat hippocampal astrocytes is mediated by NFkappaB. J. Biol. Chem. 1996, 271, 31115–31120. [Google Scholar] [CrossRef]
- Glaccum, M.B.; Stocking, K.L.; Charrier, K.; Smith, J.L.; Willis, C.R.; Maliszewski, C.; Livingston, D.J.; Peschon, J.J.; Morrissey, P.J. Phenotypic and functional characterization of mice that lack the type I receptor for IL-1. J. Immunol. 1997, 159, 3364–3371. [Google Scholar] [CrossRef]
- Galichet, C.; Lovell-Badge, R.; Rizzoti, K. Nestin-Cre mice are affected by hypopituitarism, which is not due to significant activity of the transgene in the pituitary gland. PLoS ONE 2010, 5, e11443. [Google Scholar] [CrossRef]
- Liang, H.; Hippenmeyer, S.; Ghashghaei, H.T. A Nestin-cre transgenic mouse is insufficient for recombination in early embryonic neural progenitors. Biol. Open 2012, 1, 1200–1203. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swilley, C.; Lin, Y.; Zheng, Y.; Xu, X.; Liu, M.; Zimmerman, K.; Xie, H. Sex-Linked Growth Disorder and Aberrant Pituitary Gene Expression in Nestin-Cre-Mediated Egr1 Conditional Knockout Mice. Biology 2023, 12, 966. https://doi.org/10.3390/biology12070966
Swilley C, Lin Y, Zheng Y, Xu X, Liu M, Zimmerman K, Xie H. Sex-Linked Growth Disorder and Aberrant Pituitary Gene Expression in Nestin-Cre-Mediated Egr1 Conditional Knockout Mice. Biology. 2023; 12(7):966. https://doi.org/10.3390/biology12070966
Chicago/Turabian StyleSwilley, Cody, Yu Lin, Yuze Zheng, Xiguang Xu, Min Liu, Kurt Zimmerman, and Hehuang Xie. 2023. "Sex-Linked Growth Disorder and Aberrant Pituitary Gene Expression in Nestin-Cre-Mediated Egr1 Conditional Knockout Mice" Biology 12, no. 7: 966. https://doi.org/10.3390/biology12070966
APA StyleSwilley, C., Lin, Y., Zheng, Y., Xu, X., Liu, M., Zimmerman, K., & Xie, H. (2023). Sex-Linked Growth Disorder and Aberrant Pituitary Gene Expression in Nestin-Cre-Mediated Egr1 Conditional Knockout Mice. Biology, 12(7), 966. https://doi.org/10.3390/biology12070966





