Cotton RSG2 Mediates Plant Resistance against Verticillium dahliae by miR482b Regulation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of GhCNL Disease Resistance Family
2.2. Plant Materials and Growth Condition
2.3. Pathogen Culture, Inoculation, Activation and Disease Detection
2.4. Fungal Recovery Culture and DNA Abundance Detection
2.5. RNA Extraction and cDNA Synthesis
2.6. Analysis of Gene Expression by RT-qPCR
2.7. Gene Amplification, Vector Construction and Agrobacterium-Mediated Transformation
2.8. Activation, Resuspension and Injection of Agrobacterium
2.9. GUS Histochemical Staining Analysis
3. Results
3.1. Characterization Analysis of Cotton miR482b and CNLs
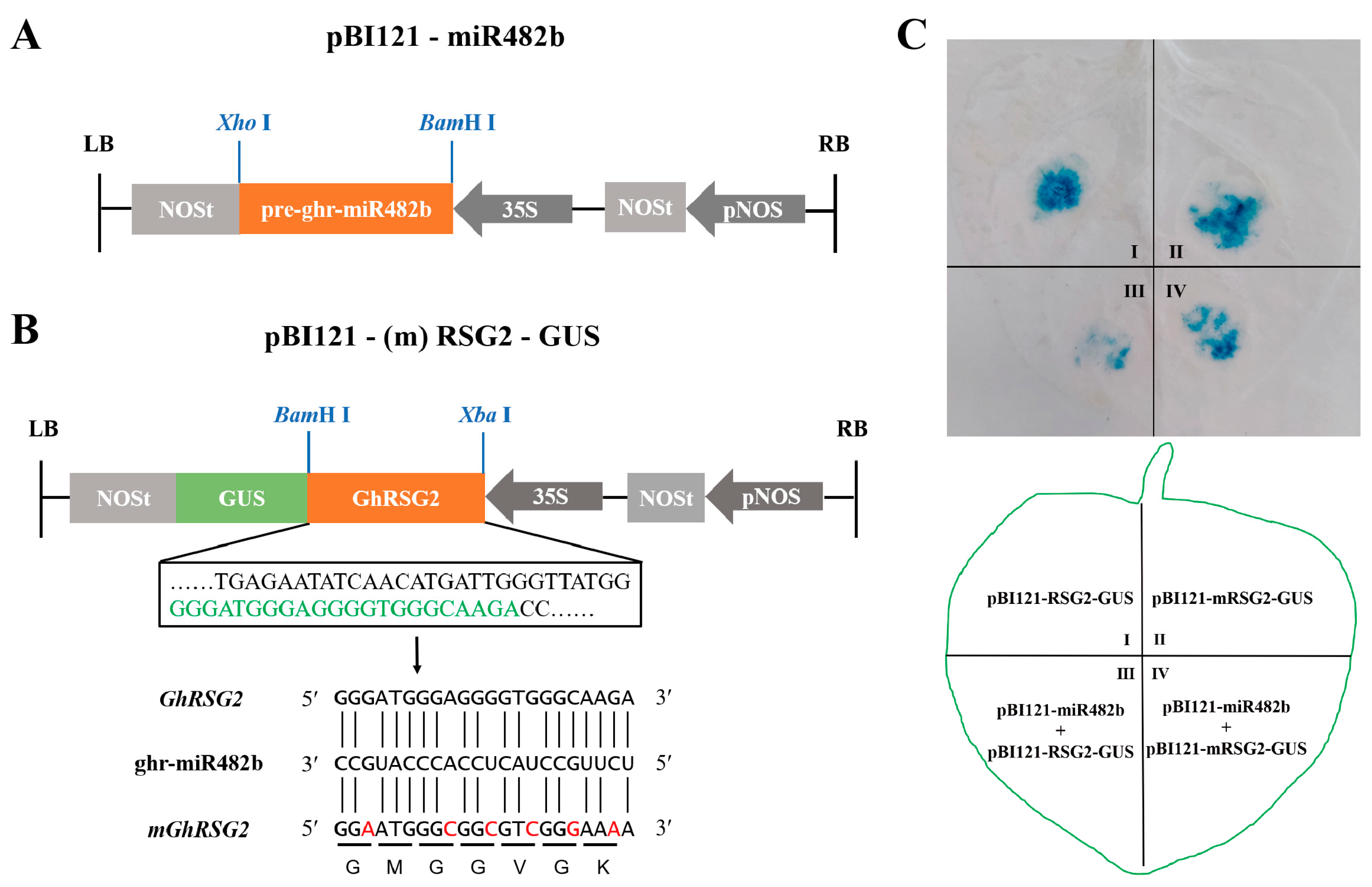
3.2. The GhRSG2 mRNA Degradation Directed by ghr-miR482b
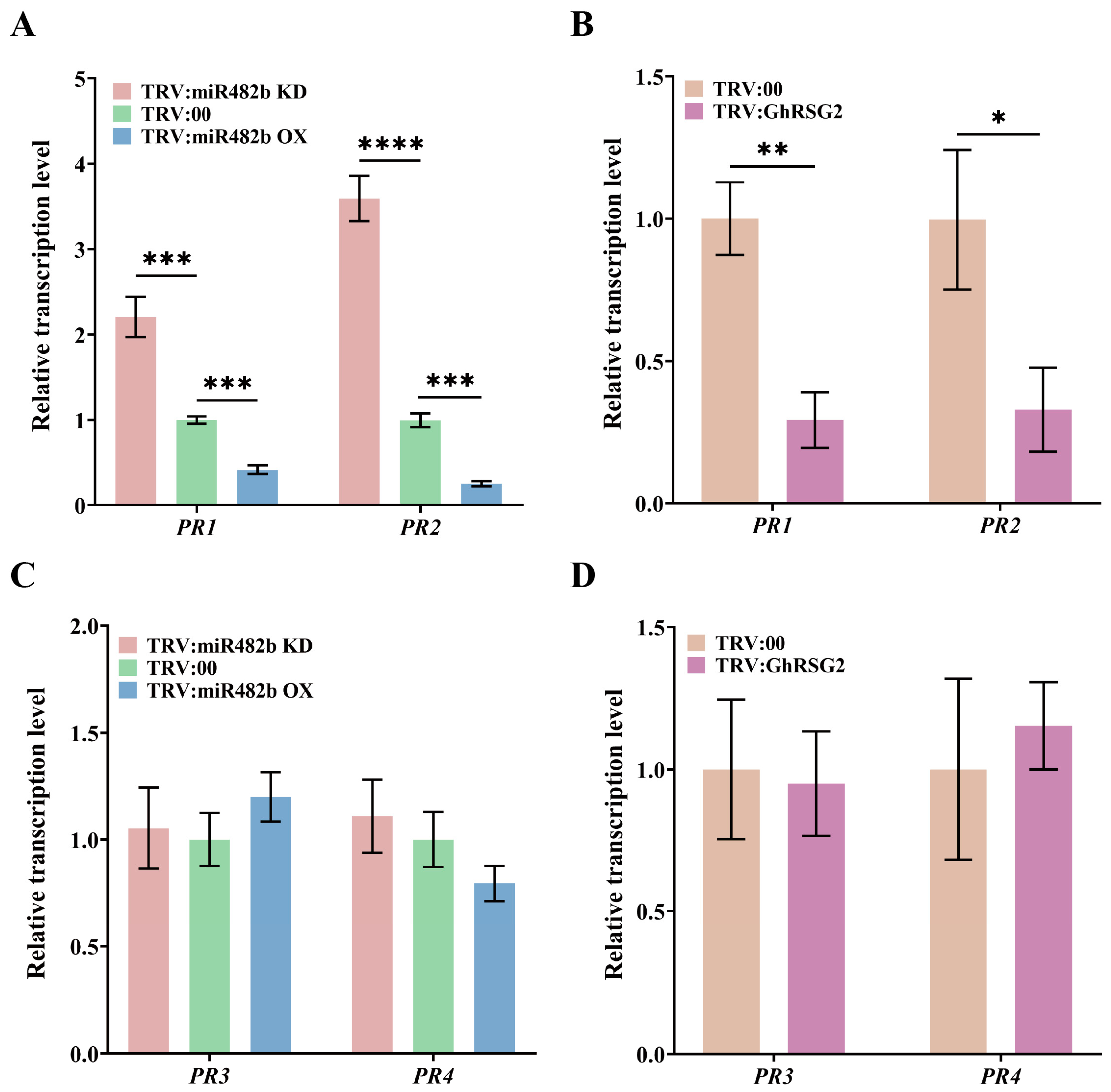
3.3. The ghr-miR482b Negatively Regulated Plant Resistance to V. dahliae Infection
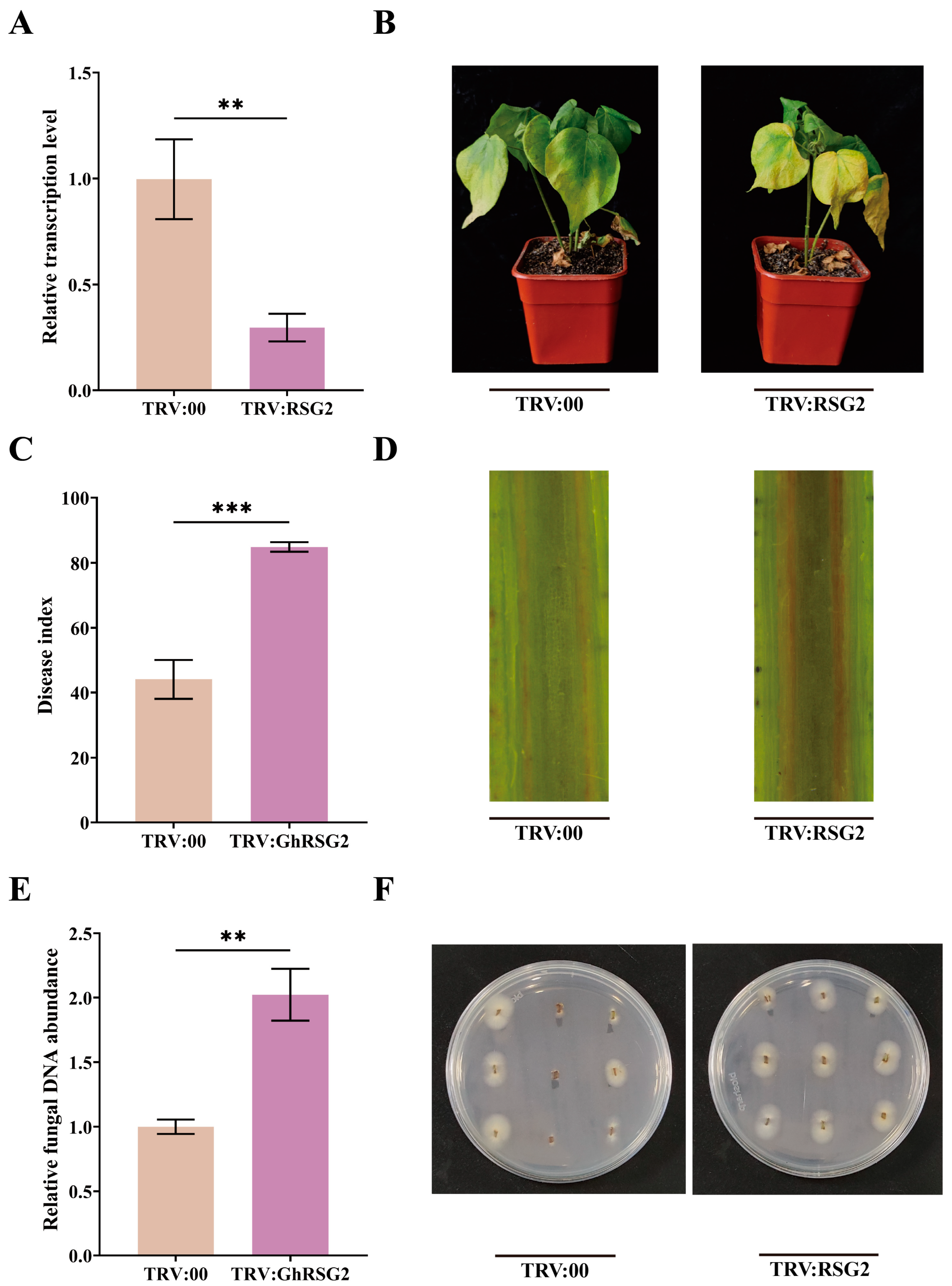
3.4. The Silence of GhRSG2 Reduced Plant Resistance to V. dahliae Infection
3.5. The ghr-miR482b-GhRSG2 Module Mediates Plant Resistance to V. dahliae Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef]
- D’Ario, M.; Griffiths-Jones, S.; Kim, M. Small RNAs: Big impact on plant development. Trends Plant Sci. 2017, 22, 1056–1068. [Google Scholar] [CrossRef]
- Chen, C.; Zeng, Z.; Liu, Z.; Xia, R. Small RNAs, emerging regulators critical for the development of horticultural traits. Hortic. Res. 2018, 5, 63. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, L.; Yang, Y.; Schmid, M.; Wang, Y. MiRNA mediated regulation and interaction between plants and pathogens. Int. J. Mol. Sci. 2021, 22, 2913. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Djami-Tchatchou, A.T.; Dubery, I.A. MiR393 regulation of lectin receptor-like kinases associated with LPS perception in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2019, 513, 88–92. [Google Scholar] [CrossRef]
- Shi, G.; Wang, S.; Wang, P.; Zhan, J.; Tang, Y.; Zhao, G.; Li, F.; Ge, X.; Wu, J. Cotton miR393-TIR1 module regulates plant defense against Verticillium dahliae via auxin perception and signaling. Front. Plant Sci. 2022, 13, 888703. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xia, Y.; Lin, S.; Wang, Y.; Guo, B.; Song, X.; Ding, S.; Zheng, L.; Feng, R.; Chen, S.; et al. Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Plant J. 2018, 95, 584–597. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Lei, Y.; Liu, J.; Hao, M.; Zhang, Z.; Tang, Y.; Chen, A.; Wu, J. The ghr-miR164 and GhNAC100 modulate cotton plant resistance against Verticillium dahlia. Plant Sci. 2020, 293, 110438. [Google Scholar] [CrossRef]
- Salvador-Guirao, R.; Baldrich, P.; Weigel, D.; Rubio-Somoza, I.; San Segundo, B. The microRNA miR773 is involved in the Arabidopsis immune response to fungal pathogens. Mol. Plant Microbe Interact. 2018, 31, 249–259. [Google Scholar] [CrossRef]
- Yu, X.; Hou, Y.; Chen, W.; Wang, S.; Wang, P.; Qu, S. Malus hupehensis miR168 targets to ARGONAUTE1 and contributes to the resistance against Botryosphaeria dothidea infection by altering defense responses. Plant Cell Physiol. 2017, 58, 1541–1557. [Google Scholar] [CrossRef]
- Feng, H.; Xu, M.; Gao, Y.; Liang, J.; Guo, F.; Guo, Y.; Huang, L. Vm-milR37 contributes to pathogenicity by regulating glutathione peroxidase gene VmGP in Valsa mali. Mol. Plant Pathol. 2021, 22, 243–254. [Google Scholar] [CrossRef]
- Hu, G.; Hao, M.; Wang, L.; Liu, J.; Zhang, Z.; Tang, Y.; Peng, Q.; Yang, Z.; Wu, J. The cotton miR477-CBP60A module participates in plant defense against Verticillium dahlia. Mol. Plant Microbe. Interact. 2020, 33, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Tang, Y.; Jia, P.; Zeng, Y.; Wang, B.; Wu, P.; Quan, Y.; Chen, A.; Li, Y.; Wu, J. A cotton lignin biosynthesis gene, GhLAC4, fine-tuned by ghr-miR397 modulates plant resistance against Verticillium dahliae. Front. Plant Sci. 2021, 12, 743795. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Tang, Y.; Hu, G.; Quan, Y.; Chen, A.; Zhong, N.; Peng, Q.; Wu, J. Cotton miR319b-targeted TCP4-like enhances plant defense against Verticillium dahliae by activating GhICS1 transcription expression. Front. Plant Sci. 2022, 13, 870882. [Google Scholar] [CrossRef]
- Liao, L.; Xie, B.; Guan, P.; Jiang, N.; Cui, J. New insight into the molecular mechanism of miR482/2118 during plant resistance to pathogens. Front. Plant Sci. 2022, 13, 1026762. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Sun, Y.H.; Shi, R.; Clark, C.; Li, L.; Chiang, V.L. Novel and mechanical stress-responsive MicroRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 2005, 17, 2186–2203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Waseem, M.; Zeng, Z.; Xu, J.; Chen, C.; Liu, Y.; Zhai, J.; Xia, R. MicroRNA482/2118, a miRNA superfamily essential for both disease resistance and plant development. New Phytol. 2022, 233, 2047–2057. [Google Scholar] [CrossRef]
- Xia, R.; Xu, J.; Arikit, S.; Meyers, B.C. Extensive families of miRNAs and PHAS loci in Norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol. Biol. Evol. 2015, 32, 2905–2918. [Google Scholar] [CrossRef]
- Shen, E.; Chen, T.; Zhu, X.; Fan, L.; Sun, J.; Llewellyn, D.J.; Wilson, I.; Zhu, Q.H. Expansion of MIR482/2118 by a class-II transposable element in cotton. Plant J. 2020, 103, 2084–2099. [Google Scholar] [CrossRef]
- Shivaprasad, P.V.; Chen, H.M.; Patel, K.; Bond, D.M.; Santos, B.A.; Baulcombe, D.C. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef]
- Zhai, J.; Jeong, D.H.; De Paoli, E.; Park, S.; Rosen, B.D.; Li, Y.; Gonzalez, A.J.; Yan, Z.; Kitto, S.L.; Grusak, M.A.; et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011, 25, 2540–2553. [Google Scholar] [CrossRef]
- Canto-Pastor, A.; Santos, B.; Valli, A.A.; Summers, W.; Schornack, S.; Baulcombe, D.C. Enhanced resistance to bacterial and oomycete pathogens by short tandem target mimic RNAs in tomato. Proc. Natl. Acad. Sci. USA 2019, 116, 2755–2760. [Google Scholar] [CrossRef]
- Permar, V.; Singh, A.; Pandey, V.; Alatar, A.A.; Faisal, M.; Jain, R.K.; Praveen, S. Tospo viral infection instigates necrosis and premature senescence by micro RNA controlled programmed cell death in Vigna unguiculata. Physiol. Mol. Plant Pathol. 2014, 88, 77–84. [Google Scholar] [CrossRef]
- Zhao, C.; Li, T.; Zhao, Y.; Zhang, B.; Li, A.; Zhao, S.; Hou, L.; Xia, H.; Fan, S.; Qiu, J.; et al. Integrated small RNA and mRNA expression profiles reveal miRNAs and their target genes in response to Aspergillus flavus growth in peanut seeds. BMC Plant Biol. 2020, 20, 215. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Fan, L.; Liu, Y.; Xu, H.; Llewellyn, D.; Wilson, I. MiR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS ONE 2013, 8, e84390. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, W.; Chen, J.; Liu, J.; Xia, M.; Shen, F. Identification of miRNAs and their targets in cotton inoculated with Verticillium dahliae by high-throughput sequencing and degradome analysis. Int. J. Mol. Sci. 2015, 16, 14749–14768. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Jin, S.; Yuan, Y.; Liu, Q.; Zhang, X.; Wilson, I. CRISPR/Cas9-mediated saturated mutagenesis of the cotton MIR482 family for dissecting the functionality of individual members in disease response. Plant Direct 2022, 6, e410. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Mu, X.; Liu, C.; Cai, J.; Shi, K.; Zhu, W.; Yang, Q. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Integr. Plant Biol. 2015, 57, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Deng, Y.; Wu, T.; Subramanian, S.; Yu, O. Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 2010, 153, 1759–1770. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Dickerman, A.W.; Michelmore, R.W.; Sivaramakrishnan, S.; Sobral, B.W.; Young, N.D. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 1999, 20, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef]
- Fei, Q.; Zhang, Y.; Xia, R.; Meyers, B.C. Small RNAs add zing to the Zig-Zag-Zig model of plant defenses. Mol. Plant Microbe Interact. 2016, 29, 165–169. [Google Scholar] [CrossRef]
- Han, G.Z. Origin and evolution of the plant immune system. New Phytol. 2019, 222, 70–83. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Jiang, N.; Cui, J.; Yang, G.; He, X.; Meng, J.; Luan, Y. Comparative transcriptome analysis shows the defense response networks regulated by miR482b. Plant Cell Rep. 2019, 38, 1–13. [Google Scholar] [CrossRef]
- Liu, W.; Cui, J.; Luan, Y. SlmiR482e-5p regulates tomato resistance to Phytophthora infestans infection along with slmiR482e-3p via sllncRNA39298-mediated inhibition. Physiol. Mol. Plant Pathol. 2022, 121, 101875. [Google Scholar] [CrossRef]
- Long, L.; Xu, F.C.; Zhao, J.R.; Li, B.; Xu, L.; Gao, W. GbMPK3 overexpression increases cotton sensitivity to Verticillium dahliae by regulating salicylic acid signaling. Plant Sci. 2020, 292, 110374. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.P.; Sun, S.C.; Zhang, X.Y.; Li, Y.J.; Liu, F.; Zhu, Q.H.; Xue, F.; Sun, J. GhWRKY70D13 regulates resistance to Verticillium dahliae in cotton through the ethylene and jasmonic acid signaling pathways. Front. Plant Sci. 2020, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019, 51, 224–229. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, A.V.; Jiang, T.; Keating, A.E.; Berger, B. Paircoil2: Improved prediction of coiled coils from sequence. Bioinformatics 2006, 22, 356–358. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Hunter, S.; Jones, P.; Mitchell, A.; Apweiler, R.; Attwood, T.K.; Bateman, A.; Bernard, T.; Binns, D.; Bork, P.; Burge, S.; et al. InterPro in 2011: New developments in the family and domain prediction database. Nucleic Acids Res. 2012, 40, D306–D312. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Gao, F.; Zhou, B.J.; Li, G.Y.; Jia, P.S.; Li, H.; Zhao, Y.L.; Zhao, P.; Xia, G.X.; Guo, H.S. A glutamic acid-rich protein identified in Verticillium dahliae from an insertional mutagenesis affects microsclerotial formation and pathogenicity. PLoS ONE 2010, 5, e15319. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, J.; Ding, L.; Zou, L.; Li, Y.; Chen, G.; Zhang, T. Constitutive expression of a novel antimicrobial protein, Hcm1, confers resistance to both Verticillium and Fusarium wilts in cotton. Sci. Rep. 2016, 6, 20773. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhang, Z.; Lei, Y.; Hu, G.; Liu, J.; Hao, M.; Chen, A.; Peng, Q.; Wu, J. Cotton WATs modulate SA biosynthesis and local lignin deposition participating in plant resistance against Verticillium dahliae. Front. Plant Sci. 2019, 10, 526. [Google Scholar] [CrossRef]
- Wang, L.; Wu, S.M.; Zhu, Y.; Fan, Q.; Zhang, Z.N.; Hu, G.; Peng, Q.Z.; Wu, J.H. Functional characterization of a novel jasmonate ZIM-domain interactor (NINJA) from upland cotton (Gossypium hirsutum). Plant Physiol. Biochem. 2017, 112, 152–160. [Google Scholar] [CrossRef]
- Xiong, X.P.; Sun, S.C.; Zhu, Q.H.; Zhang, X.Y.; Liu, F.; Li, Y.J.; Xue, F.; Sun, J. Transcriptome analysis and RNA interference reveal GhGDH2 regulating cotton resistance to Verticillium wilt by JA and SA signaling pathways. Front. Plant Sci. 2021, 12, 654676. [Google Scholar] [CrossRef]
- Yan, J.; Gu, Y.; Jia, X.; Kang, W.; Pan, S.; Tang, X.; Chen, X.; Tang, G. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 2012, 24, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Boccara, M.; Sarazin, A.; Thiébeauld, O.; Jay, F.; Voinnet, O.; Navarro, L.; Colot, V. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 2014, 10, e1003883. [Google Scholar] [CrossRef] [PubMed]
- DeYoung, B.J.; Innes, R.W. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 2006, 7, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Caplan, J.; Padmanabhan, M.; Dinesh-Kumar, S.P. Plant NB-LRR immune receptors: From recognition to transcriptional reprogramming. Cell Host Microbe 2008, 3, 126–135. [Google Scholar] [CrossRef]
- Zhu, X.; Lu, C.; Du, L.; Ye, X.; Liu, X.; Coules, A.; Zhang, Z. The wheat NB-LRR gene TaRCR1 is required for host defence response to the necrotrophic fungal pathogen Rhizoctonia cerealis. Plant Biotechnol. J. 2017, 15, 674–687. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, H.; Cai, T.; Deng, Y.; Zhuang, R.; Zhang, N.; Zeng, Y.; Zheng, Y.; Tang, R.; Pan, R.; et al. Overexpression of a novel peanut NBS-LRR gene AhRRS5 enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Biotechnol. J. 2017, 15, 39–55. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, P.; Lu, C.; Wang, B.; Zhang, F.; Shi, L.; Xu, Y.; Chen, A.; Si, H.; Su, J.; Wu, J. Cotton RSG2 Mediates Plant Resistance against Verticillium dahliae by miR482b Regulation. Biology 2023, 12, 898. https://doi.org/10.3390/biology12070898
Wu P, Lu C, Wang B, Zhang F, Shi L, Xu Y, Chen A, Si H, Su J, Wu J. Cotton RSG2 Mediates Plant Resistance against Verticillium dahliae by miR482b Regulation. Biology. 2023; 12(7):898. https://doi.org/10.3390/biology12070898
Chicago/Turabian StyleWu, Pan, Chengzhe Lu, Bingting Wang, Feiyan Zhang, Linfang Shi, Yunjiao Xu, Aimin Chen, Huaijun Si, Junji Su, and Jiahe Wu. 2023. "Cotton RSG2 Mediates Plant Resistance against Verticillium dahliae by miR482b Regulation" Biology 12, no. 7: 898. https://doi.org/10.3390/biology12070898
APA StyleWu, P., Lu, C., Wang, B., Zhang, F., Shi, L., Xu, Y., Chen, A., Si, H., Su, J., & Wu, J. (2023). Cotton RSG2 Mediates Plant Resistance against Verticillium dahliae by miR482b Regulation. Biology, 12(7), 898. https://doi.org/10.3390/biology12070898






