Glucose Inhibits Yeast AMPK (Snf1) by Three Independent Mechanisms
Abstract
Simple Summary
Abstract
1. Introduction
2. Glucose Sensing Pathways in S. cerevisiae
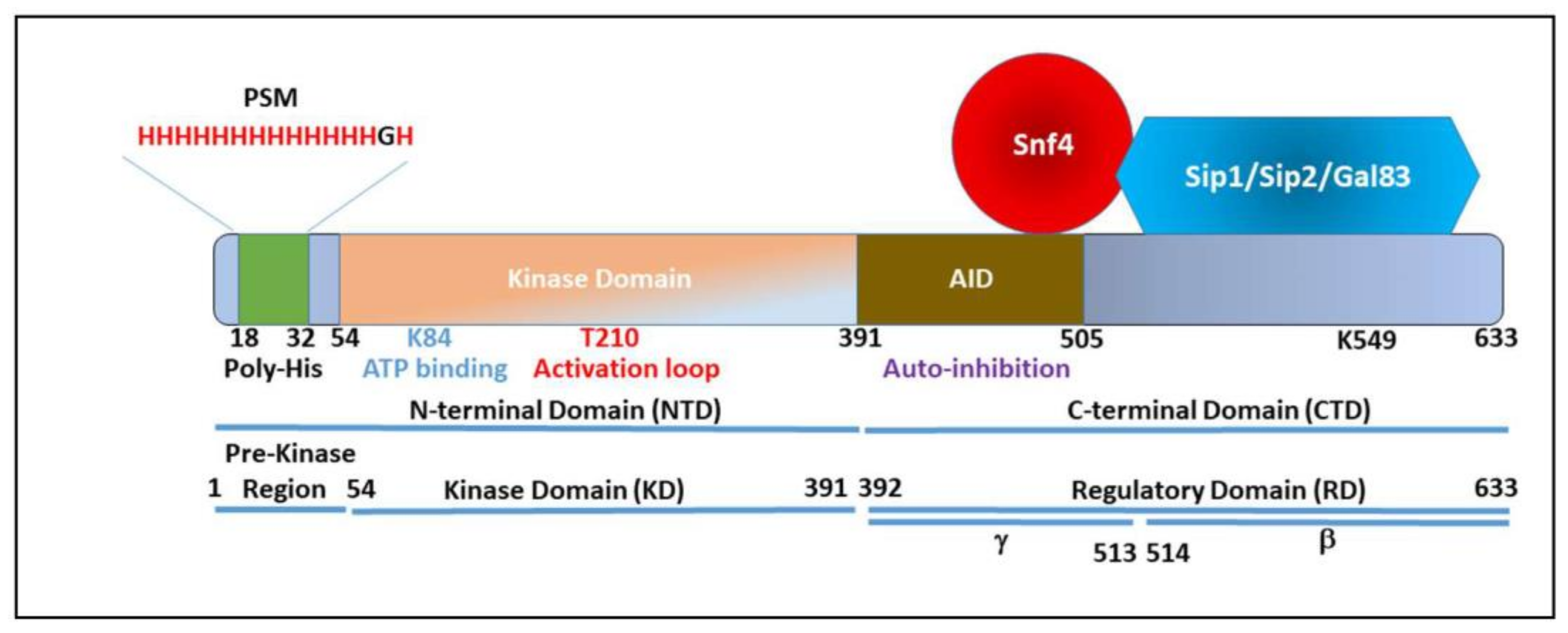
3. Snf1 Structure
4. Regulation of Snf1
4.1. Phosphorylation
4.2. SUMOylation
4.3. Regulation of Snf1 by pH
5. The Polyhistidine Tract Is a Multi-Tool
6. Other Functions of Polyhistidine
7. Relationship between the Snf1-Activating Pathways
8. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roustan, V.; Jain, A.; Teige, M.; Ebersberger, I.; Weckwerth, W. An evolutionary perspective of AMPK-TOR signaling in the three domains of life. J. Exp. Bot. 2016, 67, 3897–3907. [Google Scholar] [CrossRef] [PubMed]
- Vincent, O.; Carlson, M. Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 1998, 17, 7002–7008. [Google Scholar] [CrossRef] [PubMed]
- Treitel, M.A.; Kuchin, S.; Carlson, M. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998, 18, 6273–6280. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Chen, Y.; Siewers, V.; Nielsen, J. Improving production of malonyl coenzyme A-derived metabolites by abolishing Snf1-dependent regulation of Acc1. mBio 2014, 5, e01130-14. [Google Scholar] [CrossRef]
- Benanti, J.A.; Cheung, S.K.; Brady, M.C.; Toczyski, D.P. A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat. Cell Biol. 2007, 9, 1184–1191. [Google Scholar] [CrossRef]
- Becuwe, M.; Vieira, N.; Lara, D.; Gomes-Rezende, J.; Soares-Cunha, C.; Casal, M.; Haguenauer-Tsapis, R.; Vincent, O.; Paiva, S.; Léon, S. A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J. Cell Biol. 2012, 196, 247–259. [Google Scholar] [CrossRef]
- Fujita, S.; Sato, D.; Kasai, H.; Ohashi, M.; Tsukue, S.; Takekoshi, Y.; Gomi, K.; Shintani, T. The C-terminal region of the yeast monocarboxylate transporter Jen1 acts as a glucose signal-responding degron recognized by the α-arrestin Rod1. J. Biol. Chem. 2018, 293, 10926–10936. [Google Scholar] [CrossRef]
- Nicastro, R.; Tripodi, F.; Gaggini, M.; Castoldi, A.; Reghellin, V.; Nonnis, S.; Tedeschi, G.; Coccetti, P. Snf1 Phosphorylates Adenylate Cyclase and Negatively Regulates Protein Kinase A-dependent Transcription in Saccharomyces cerevisiae*. J. Biol. Chem. 2015, 290, 24715–24726. [Google Scholar] [CrossRef]
- Ozcan, S.; Dover, J.; Johnston, M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998, 17, 2566–2573. [Google Scholar] [CrossRef]
- Moriya, H.; Johnston, M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. USA 2004, 101, 1572–1577. [Google Scholar] [CrossRef] [PubMed]
- Flick, K.M.; Spielewoy, N.; Kalashnikova, T.I.; Guaderrama, M.; Zhu, Q.; Chang, H.-C.; Wittenberg, C. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Biol. Cell 2003, 14, 3230–3241. [Google Scholar] [CrossRef]
- Polish, J.A.; Kim, J.-H.; Johnston, M. How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose. Genetics 2005, 169, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Jouandot, D.; Cho, K.H.; Kim, J.-H. Understanding the mechanism of glucose-induced relief of Rgt1-mediated repression in yeast. FEBS Open Bio 2014, 4, 105–111. [Google Scholar] [CrossRef]
- Kim, J.-H.; Brachet, V.; Moriya, H.; Johnston, M. Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot. Cell 2006, 5, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Gadura, N.; Robinson, L.C.; Michels, C.A. Glc7-Reg1 phosphatase signals to Yck1,2 casein kinase 1 to regulate transport activity and glucose-induced inactivation of Saccharomyces maltose permease. Genetics 2006, 172, 1427–1439. [Google Scholar] [CrossRef]
- Simpson-Lavy, K.J.; Johnston, M. SUMOylation regulates the SNF1 protein kinase. Proc. Natl. Acad. Sci. USA 2013, 110, 17432–17437. [Google Scholar] [CrossRef] [PubMed]
- Peeters, T.; Louwet, W.; Geladé, R.; Nauwelaers, D.; Thevelein, J.M.; Versele, M. Kelch-repeat proteins interacting with the Galpha protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc. Natl. Acad. Sci. USA 2006, 103, 13034–13039. [Google Scholar] [CrossRef] [PubMed]
- Kraakman, L.; Lemaire, K.; Ma, P.; Teunissen, A.W.; Donaton, M.C.; van Dijck, P.; Winderickx, J.; de Winde, J.H.; Thevelein, J.M. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 1999, 32, 1002–1012. [Google Scholar] [CrossRef]
- Yun, C.W.; Tamaki, H.; Nakayama, R.; Yamamoto, K.; Kumagai, H. Gpr1p, a putative G-protein coupled receptor, regulates glucose-dependent cellular cAMP level in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1998, 252, 29–33. [Google Scholar] [CrossRef]
- Toda, T.; Cameron, S.; Sass, P.; Zoller, M.; Scott, J.D.; McMullen, B.; Hurwitz, M.; Krebs, E.G.; Wigler, M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987, 7, 1371–1377. [Google Scholar] [CrossRef]
- Toda, T.; Cameron, S.; Sass, P.; Zoller, M.; Wigler, M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 1987, 50, 277–287. [Google Scholar] [CrossRef]
- Lee, P.; Cho, B.-R.; Joo, H.-S.; Hahn, J.-S. Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol. Microbiol. 2008, 70, 882–895. [Google Scholar] [CrossRef] [PubMed]
- Isom, D.G.; Page, S.C.; Collins, L.B.; Kapolka, N.J.; Taghon, G.J.; Dohlman, H.G. Coordinated regulation of intracellular pH by two glucose-sensing pathways in yeast. J. Biol. Chem. 2018, 293, 2318–2329. [Google Scholar] [CrossRef] [PubMed]
- Dechant, R.; Binda, M.; Lee, S.S.; Pelet, S.; Winderickx, J.; Peter, M. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J. 2010, 29, 2515–2526. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Zhao, C.; Chen, D.; Yun, C.; Li, H.; Bai, L. Structure and activation mechanism of the hexameric plasma membrane H+-ATPase. Nat. Commun. 2021, 12, 6439. [Google Scholar] [CrossRef]
- Martínez-Muñoz, G.A.; Kane, P. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J. Biol. Chem. 2008, 283, 20309–20319. [Google Scholar] [CrossRef]
- Mazón, M.J.; Eraso, P.; Portillo, F. Specific phosphoantibodies reveal two phosphorylation sites in yeast Pma1 in response to glucose. FEMS Yeast Res. 2015, 15, fov030. [Google Scholar] [CrossRef] [PubMed]
- Piper, P.W.; Ortiz-Calderon, C.; Holyoak, C.; Coote, P.; Cole, M. Hsp30, the integral plasma membrane heat shock protein of Saccharmyces cerevisiae, is a stress-inducible regulator of plasma membrane H+-ATPase. Cell Stress Chaper. 1997, 2, 12. [Google Scholar] [CrossRef]
- Kane, P.M. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J. Biol. Chem. 1995, 270, 17025–17032. [Google Scholar] [CrossRef]
- Orij, R.; Postmus, J.; ter Beek, A.; Brul, S.; Smits, G.J. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 2009, 155, 268–278. [Google Scholar] [CrossRef]
- Petrovska, I.; Nüske, E.; Munder, M.C.; Kulasegaran, G.; Malinovska, L.; Kroschwald, S.; Richter, D.; Fahmy, K.; Gibson, K.; Verbavatz, J.-M.; et al. Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife 2014, 3, e02409. [Google Scholar] [CrossRef]
- Young, B.P.; Shin, J.J.H.; Orij, R.; Chao, J.T.; Li, S.C.; Guan, X.L.; Khong, A.; Jan, E.; Wenk, M.R.; Prinz, W.A.; et al. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 2010, 329, 1085–1088. [Google Scholar] [CrossRef] [PubMed]
- Geva, Y.; Crissman, J.; Arakel, E.C.; Gómez-Navarro, N.; Chuartzman, S.G.; Stahmer, K.R.; Schwappach, B.; Miller, E.A.; Schuldiner, M. Two novel effectors of trafficking and maturation of the yeast plasma membrane H+-ATPase. Traffic 2017, 18, 672–682. [Google Scholar] [CrossRef]
- Eraso, P.; Mazón, M.J.; Portillo, F. Yeast protein kinase Ptk2 localizes at the plasma membrane and phosphorylates in vitro the C-terminal peptide of the H+-ATPase. Biochim. Biophys. Acta 2006, 1758, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Goossens, A.; de La Fuente, N.; Forment, J.; Serrano, R.; Portillo, F. Regulation of yeast H+-ATPase by protein kinases belonging to a family dedicated to activation of plasma membrane transporters. Mol. Cell. Biol. 2000, 20, 7654–7661. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.B.; Kardos, T.B.; Monk, B.C. Regulation and pH-dependent expression of a bilaterally truncated yeast plasma membrane H+-ATPase. Biochim. Biophys. Acta 1998, 1372, 261–271. [Google Scholar] [CrossRef]
- Jiang, R.; Carlson, M. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 1996, 10, 3105–3115. [Google Scholar] [CrossRef]
- Jiang, R.; Carlson, M. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 1997, 17, 2099–2106. [Google Scholar] [CrossRef]
- Vincent, O.; Townley, R.; Kuchin, S.; Carlson, M. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 2001, 15, 1104–1114. [Google Scholar] [CrossRef]
- Meng, L.; Liu, H.-L.; Lin, X.; Hu, X.-P.; Teng, K.-R.; Liu, S.-X. Enhanced multi-stress tolerance and glucose utilization of Saccharomyces cerevisiae by overexpression of the SNF1 gene and varied beta isoform of Snf1 dominates in stresses. Microb. Cell Fact. 2020, 19, 134. [Google Scholar] [CrossRef]
- Calabrese, M.F.; Rajamohan, F.; Harris, M.S.; Caspers, N.L.; Magyar, R.; Withka, J.M.; Wang, H.; Borzilleri, K.A.; Sahasrabudhe, P.V.; Hoth, L.R.; et al. Structural basis for AMPK activation: Natural and synthetic ligands regulate kinase activity from opposite poles by different molecular mechanisms. Structure 2014, 22, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Bridges, M.D.; Yan, Y.; de Waal, P.W.; Zhou, X.E.; Suino-Powell, K.M.; Xu, H.E.; Hubbell, W.L.; Melcher, K. Conformational heterogeneity of the allosteric drug and metabolite (ADaM) site in AMP-activated protein kinase (AMPK). J. Biol. Chem. 2018, 293, 16994–17007. [Google Scholar] [CrossRef] [PubMed]
- Simpson-Lavy, K.J.; Kupiec, M. Regulation of yeast Snf1 (AMPK) by a polyhistidine containing pH sensing module. iScience 2022, 25, 105083. [Google Scholar] [CrossRef] [PubMed]
- Amodeo, G.A.; Rudolph, M.J.; Tong, L. Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature 2007, 449, 492–495. [Google Scholar] [CrossRef]
- Leech, A.; Nath, N.; McCartney, R.R.; Schmidt, M.C. Isolation of mutations in the catalytic domain of the snf1 kinase that render its activity independent of the snf4 subunit. Eukaryot. Cell 2003, 2, 265–273. [Google Scholar] [CrossRef]
- Elbing, K.; Rubenstein, E.M.; McCartney, R.R.; Schmidt, M.C. Subunits of the Snf1 kinase heterotrimer show interdependence for association and activity. J. Biol. Chem. 2006, 281, 26170–26180. [Google Scholar] [CrossRef]
- Schnell, H.M.; Jochem, M.; Micoogullari, Y.; Riggs, C.L.; Ivanov, P.; Welsch, H.; Ravindran, R.; Anderson, P.; Robinson, L.C.; Tatchell, K.; et al. Reg1 and Snf1 regulate stress-induced relocalization of protein phosphatase-1 to cytoplasmic granules. FEBS J. 2021, 288, 4833–4848. [Google Scholar] [CrossRef]
- Pérez-Sampietro, M.; Casas, C.; Herrero, E. The AMPK family member Snf1 protects Saccharomyces cerevisiae cells upon glutathione oxidation. PLoS ONE 2013, 8, e58283. [Google Scholar] [CrossRef]
- Portillo, F.; Mulet, J.M.; Serrano, R. A role for the non-phosphorylated form of yeast Snf1: Tolerance to toxic cations and activation of potassium transport. FEBS Lett. 2005, 579, 512–516. [Google Scholar] [CrossRef]
- Hong, S.-P.; Carlson, M. Regulation of Snf1 Protein Kinase in Response to Environmental Stress. J. Biol. Chem. 2007, 282, 16838–16845. [Google Scholar] [CrossRef]
- Dubacq, C.; Chevalier, A.; Mann, C. The protein kinase Snf1 is required for tolerance to the ribonucleotide reductase inhibitor hydroxyurea. Mol. Cell. Biol. 2004, 24, 2560–2572. [Google Scholar] [CrossRef]
- Casamayor, A.; Serrano, R.; Platara, M.; Casado, C.; Ruiz, A.; Ariño, J. The role of the Snf1 kinase in the adaptive response of Saccharomyces cerevisiae to alkaline pH stress. Biochem. J. 2012, 444, 39–49. [Google Scholar] [CrossRef]
- Hedbacker, K.; Hong, S.-P.; Carlson, M. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol. Cell. Biol. 2004, 24, 8255–8263. [Google Scholar] [CrossRef]
- Hong, S.-P.; Leiper, F.C.; Woods, A.; Carling, D.; Carlson, M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 2003, 100, 8839–8843. [Google Scholar] [CrossRef]
- Kim, M.-D.; Hong, S.-P.; Carlson, M. Role of Tos3, a Snf1 protein kinase kinase, during growth of Saccharomyces cerevisiae on nonfermentable carbon sources. Eukaryot. Cell 2005, 4, 861–866. [Google Scholar] [CrossRef]
- Barrett, L.; Orlova, M.; Maziarz, M.; Kuchin, S. Protein kinase A contributes to the negative control of Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot. Cell 2012, 11, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Milanesi, R.; Tripodi, F.; Vertemara, J.; Tisi, R.; Coccetti, P. AMPK Phosphorylation Is Controlled by Glucose Transport Rate in a PKA-Independent Manner. Int. J. Mol. Sci. 2021, 22, 9483. [Google Scholar] [CrossRef]
- Estruch, F.; Treitel, M.A.; Yang, X.; Carlson, M. N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 1992, 132, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Ludin, K.; Jiang, R.; Carlson, M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1998, 95, 6245–6250. [Google Scholar] [CrossRef]
- Frederick, D.L.; Tatchell, K. The REG2 gene of Saccharomyces cerevisiae encodes a type 1 protein phosphatase-binding protein that functions with Reg1p and the Snf1 protein kinase to regulate growth. Mol. Cell. Biol. 1996, 16, 2922–2931. [Google Scholar] [CrossRef] [PubMed]
- Maziarz, M.; Shevade, A.; Barrett, L.; Kuchin, S. Springing into Action: Reg2 Negatively Regulates Snf1 Protein Kinase and Facilitates Recovery from Prolonged Glucose Starvation in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2016, 82, 3875–3885. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.; Liu, Y.; Xu, X.; Carlson, M. Heterotrimer-independent regulation of activation-loop phosphorylation of Snf1 protein kinase involves two protein phosphatases. Proc. Natl. Acad. Sci. USA 2012, 109, 8652–8657. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.; Xu, X.; Carlson, M. Ptc1 protein phosphatase 2C contributes to glucose regulation of SNF1/AMP-activated protein kinase (AMPK) in Saccharomyces cerevisiae. J. Biol. Chem. 2013, 288, 31052–31058. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, D.; Albacar, M.; Zhang, C.; Calafí, C.; López-Malo, M.; Torres-Torronteras, J.; Martí, R.; Kovalchuk, S.I.; Pinson, B.; Jensen, O.N.; et al. Yeast Ppz1 protein phosphatase toxicity involves the alteration of multiple cellular targets. Sci. Rep. 2020, 10, 15613. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.V.; Heath, R.; Underwood, E.; Sanders, M.J.; Carmena, D.; McCartney, R.R.; Leiper, F.C.; Xiao, B.; Jing, C.; Walker, P.A.; et al. ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell Metab. 2011, 14, 707–714. [Google Scholar] [CrossRef]
- Sanders, M.J.; Grondin, P.O.; Hegarty, B.D.; Snowden, M.A.; Carling, D. Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem. J. 2007, 403, 139–148. [Google Scholar] [CrossRef]
- Chandrashekarappa, D.G.; McCartney, R.R.; Schmidt, M.C. Ligand binding to the AMP-activated protein kinase active site mediates protection of the activation loop from dephosphorylation. J. Biol. Chem. 2013, 288, 89–98. [Google Scholar] [CrossRef]
- Psakhye, I.; Jentsch, S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 2012, 151, 807–820. [Google Scholar] [CrossRef]
- Xie, Y.; Kerscher, O.; Kroetz, M.B.; McConchie, H.F.; Sung, P.; Hochstrasser, M. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 2007, 282, 34176–34184. [Google Scholar] [CrossRef]
- Simpson-Lavy, K.J.; Bronstein, A.; Kupiec, M.; Johnston, M. Cross-Talk between Carbon Metabolism and the DNA Damage Response in S. cerevisiae. Cell Rep. 2015, 12, 1865–1875. [Google Scholar] [CrossRef]
- Wilson, M.A.; Koutelou, E.; Hirsch, C.; Akdemir, K.; Schibler, A.; Barton, M.C.; Dent, S.Y.R. Ubp8 and SAGA regulate Snf1 AMP kinase activity. Mol. Cell. Biol. 2011, 31, 3126–3135. [Google Scholar] [CrossRef]
- Crozet, P.; Margalha, L.; Butowt, R.; Fernandes, N.; Elias, C.A.; Orosa, B.; Tomanov, K.; Teige, M.; Bachmair, A.; Sadanandom, A.; et al. SUMOylation represses SnRK1 signaling in Arabidopsis. Plant J. 2016, 85, 120–133. [Google Scholar] [CrossRef]
- Yan, Y.; Ollila, S.; Wong, I.P.L.; Vallenius, T.; Palvimo, J.J.; Vaahtomeri, K.; Mäkelä, T.P. SUMOylation of AMPKα1 by PIAS4 specifically regulates mTORC1 signalling. Nat. Commun. 2015, 6, 8979. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.-M.; Du, Q.-S.; Meng, J.-Z.; Pang, Z.-W.; Huang, R.-B. The multiple roles of histidine in protein interactions. Chem. Cent. J. 2013, 7, 44. [Google Scholar] [CrossRef]
- Whitten, S.T.; García-Moreno, E.B.; Hilser, V.J. Local conformational fluctuations can modulate the coupling between proton binding and global structural transitions in proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 4282–4287. [Google Scholar] [CrossRef] [PubMed]
- Celenza, J.L.; Carlson, M. Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol. Cell. Biol. 1989, 9, 5034–5044. [Google Scholar] [CrossRef] [PubMed]
- Simpson-Lavy, K.J.; Kupiec, M. The polyHIS Tract of Yeast AMPK Coordinates Carbon Metabolism with Iron Availability. Int. J. Mol. Sci. 2023, 24, 1368. [Google Scholar] [CrossRef]
- Engel, S.R.; Dietrich, F.S.; Fisk, D.G.; Binkley, G.; Balakrishnan, R.; Costanzo, M.C.; Dwight, S.S.; Hitz, B.C.; Karra, K.; Nash, R.S.; et al. The reference genome sequence of Saccharomyces cerevisiae: Then and now. G3 Genes Genomes Genet. 2014, 4, 389–398. [Google Scholar] [CrossRef]
- Salichs, E.; Ledda, A.; Mularoni, L.; Albà, M.M.; de la Luna, S. Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLoS Genet. 2009, 5, e1000397. [Google Scholar] [CrossRef]
- Pertea, M.; Shumate, A.; Pertea, G.; Varabyou, A.; Breitwieser, F.P.; Chang, Y.-C.; Madugundu, A.K.; Pandey, A.; Salzberg, S.L. CHESS: A new human gene catalog curated from thousands of large-scale RNA sequencing experiments reveals extensive transcriptional noise. Genome Biol. 2018, 19, 208. [Google Scholar] [CrossRef]
- Cherny, V.V.; Morgan, D.; Thomas, S.; Smith, S.M.E.; DeCoursey, T.E. Histidine168 is crucial for ΔpH-dependent gating of the human voltage-gated proton channel, hHV1. J. Gen. Physiol. 2018, 150, 851–862. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, C.; Li, S.J. The pH-sensitive structure of the C-terminal domain of voltage-gated proton channel and the thermodynamic characteristics of Zn2+ binding to this domain. Biochem. Biophys. Res. Commun. 2015, 456, 207–212. [Google Scholar] [CrossRef]
- Gutierrez, J.I.; Brittingham, G.P.; Karadeniz, Y.; Tran, K.D.; Dutta, A.; Holehouse, A.S.; Peterson, C.L.; Holt, L.J. SWI/SNF senses carbon starvation with a pH-sensitive low-complexity sequence. eLife 2022, 11, e70344. [Google Scholar] [CrossRef]
- Vercoulen, Y.; Kondo, Y.; Iwig, J.S.; Janssen, A.B.; White, K.A.; Amini, M.; Barber, D.L.; Kuriyan, J.; Roose, J.P. A Histidine pH sensor regulates activation of the Ras-specific guanine nucleotide exchange factor RasGRP1. eLife 2017, 6, e29002. [Google Scholar] [CrossRef]
- Hoque, M.; Young, T.M.; Lee, C.-G.; Serrero, G.; Mathews, M.B.; Pe’ery, T. The growth factor granulin interacts with cyclin T1 and modulates P-TEFb-dependent transcription. Mol. Cell. Biol. 2003, 23, 1688–1702. [Google Scholar] [CrossRef]
- Wang, W.; Qiao, S.; Li, G.; Cheng, J.; Yang, C.; Zhong, C.; Stovall, D.B.; Shi, J.; Teng, C.; Li, D.; et al. A histidine cluster determines YY1-compartmentalized coactivators and chromatin elements in phase-separated enhancer clusters. Nucleic Acids Res. 2022, 50, 4917–4937. [Google Scholar] [CrossRef]
- Hecel, A.; Wątły, J.; Rowińska-Żyrek, M.; Świątek-Kozłowska, J.; Kozłowski, H. Histidine tracts in human transcription factors: Insight into metal ion coordination ability. J. Biol. Inorg. Chem. 2018, 23, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Bellotti, D.; Sinigaglia, A.; Guerrini, R.; Marzola, E.; Rowińska-Żyrek, M.; Remelli, M. The N-terminal domain of Helicobacter pylori’s Hpn protein: The role of multiple histidine residues. J. Inorg. Biochem. 2021, 214, 111304. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Scott, D.R.; Vagin, O.; Tokhtaeva, E.; Marcus, E.A.; Sachs, G. Measurement of Internal pH in Helicobacter pylori by Using Green Fluorescent Protein Fluorimetry. J. Bacteriol. 2018, 200, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, A.; Rao, N.N.; Ault-Riché, D. Inorganic polyphosphate: A molecule of many functions. Annu. Rev. Biochem. 1999, 68, 89–125. [Google Scholar] [CrossRef]
- Neville, N.; Lehotsky, K.; Yang, Z.; Klupt, K.A.; Denoncourt, A.; Downey, M.; Jia, Z. Ionic polyphosphorylation of histidine repeat proteins by inorganic polyphosphate. bioRxiv 2023. [Google Scholar] [CrossRef]
- Hedbacker, K.; Townley, R.; Carlson, M. Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol. Cell. Biol. 2004, 24, 1836–1843. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simpson-Lavy, K.; Kupiec, M. Glucose Inhibits Yeast AMPK (Snf1) by Three Independent Mechanisms. Biology 2023, 12, 1007. https://doi.org/10.3390/biology12071007
Simpson-Lavy K, Kupiec M. Glucose Inhibits Yeast AMPK (Snf1) by Three Independent Mechanisms. Biology. 2023; 12(7):1007. https://doi.org/10.3390/biology12071007
Chicago/Turabian StyleSimpson-Lavy, Kobi, and Martin Kupiec. 2023. "Glucose Inhibits Yeast AMPK (Snf1) by Three Independent Mechanisms" Biology 12, no. 7: 1007. https://doi.org/10.3390/biology12071007
APA StyleSimpson-Lavy, K., & Kupiec, M. (2023). Glucose Inhibits Yeast AMPK (Snf1) by Three Independent Mechanisms. Biology, 12(7), 1007. https://doi.org/10.3390/biology12071007





