Detection of Hindwing Landmarks Using Transfer Learning and High-Resolution Networks
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Hardware and Software Environment
2.2. Image and Dataset Preparation
2.3. Network Architecture
3. Results
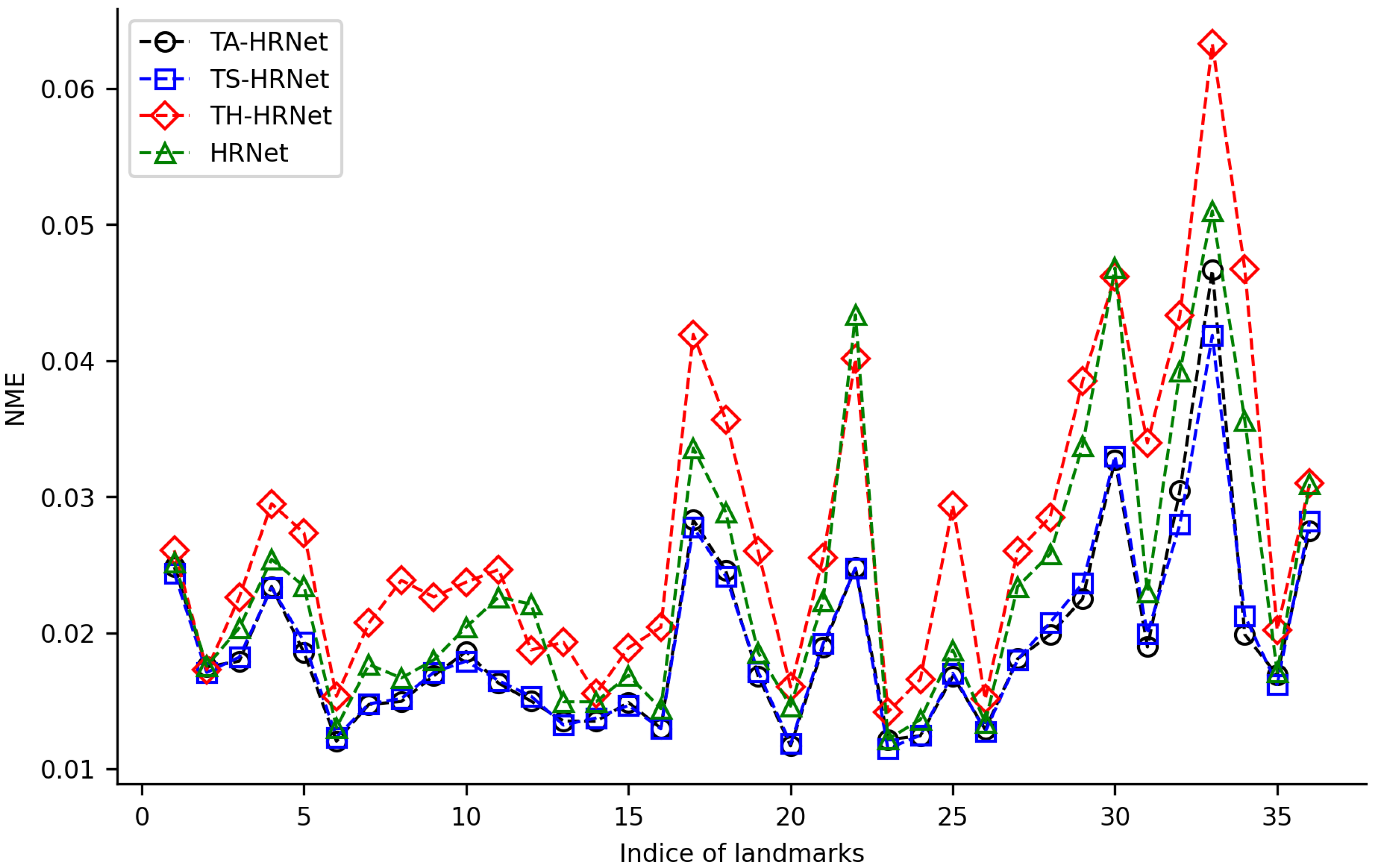
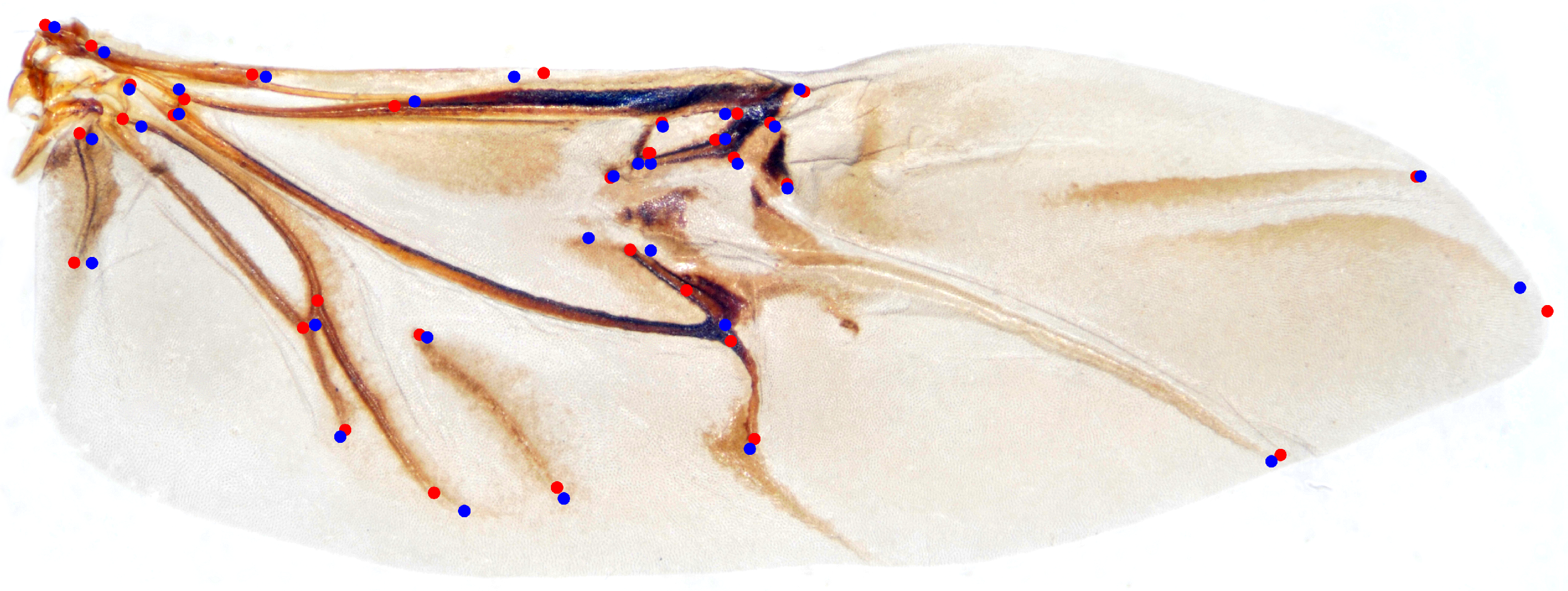
3.1. Performance of Transfer Strategies
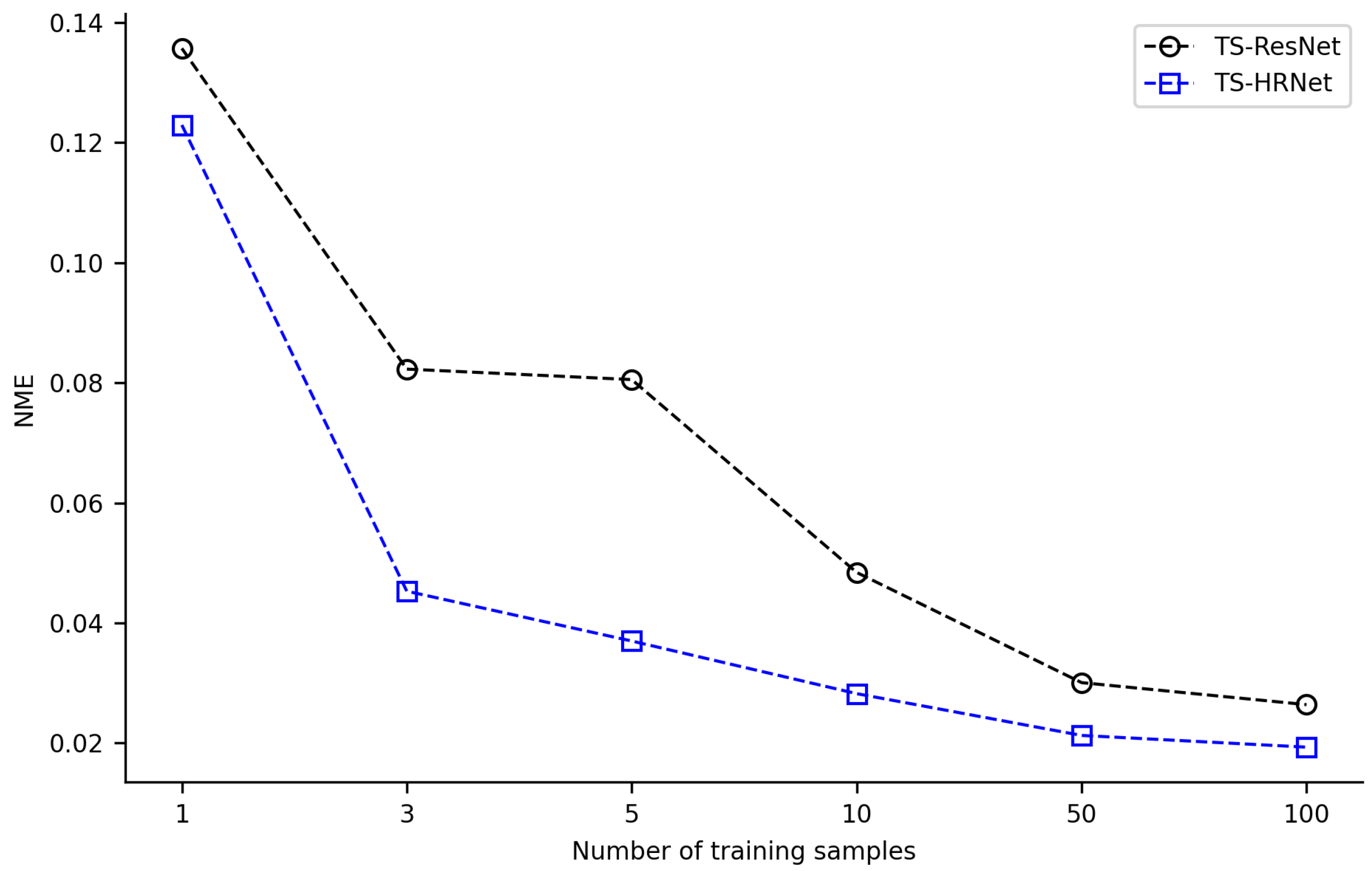
3.2. TS-HRNet Performance on Training Datasets of Varied Sizes
3.3. Performance Comparison of TS-HRNet with TS-ResNet
4. Discussion
4.1. Entomological Significance of Wing Landmark Detection
4.2. Improving Landmark Detection Models with Transfer Learning
4.3. Performance Analysis of TS-HRNet
4.4. Implications for Future Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kukalová-Peck, J. Origin of the Insect Wing and Wing Articulation from the Arthropodan Leg. Can. J. Zool. 1983, 61, 1618–1669. [Google Scholar] [CrossRef]
- Hörnschemeyer, T.; Willkommen, J. The Contribution of Flight System Characters to the Reconstruction of the Phylogeny of the Pterygota. Arthropod Syst. Phylogeny 2007, 65, 15–23. [Google Scholar]
- Changbunjong, T.; Prakaikowit, N.; Maneephan, P.; Kaewwiset, T.; Weluwanarak, T.; Chaiphongpachara, T.; Dujardin, J.P. Landmark Data to Distinguish and Identify Morphologically Close Tabanus Spp. (Diptera: Tabanidae). Insects 2021, 12, 974. [Google Scholar] [CrossRef] [PubMed]
- Simões, R.F.; Wilke, A.B.B.; Chagas, C.R.F.; de Menezes, R.M.T.; Suesdek, L.; Multini, L.C.; Silva, F.S.; Grech, M.G.; Marrelli, M.T.; Kirchgatter, K. Wing Geometric Morphometrics as a Tool for the Identification of Culex Subgenus Mosquitoes of Culex (Diptera: Culicidae). Insects 2020, 11, 567. [Google Scholar] [CrossRef]
- Hamilton, C.A.; Winiger, N.; Rubin, J.J.; Breinholt, J.; Rougerie, R.; Kitching, I.J.; Barber, J.R.; Kawahara, A.Y. Hidden Phylogenomic Signal Helps Elucidate Arsenurine Silkmoth Phylogeny and the Evolution of Body Size and Wing Shape Trade-Offs. Syst. Biol. 2022, 71, 859–874. [Google Scholar] [CrossRef]
- Bai, Y.; Dong, J.; Guan, D.; Xie, J.; Xu, S. Geographic Variation in Wing Size and Shape of the Grasshopper Trilophidia Annulata (Orthoptera: Oedipodidae): Morphological Trait Variations Follow an Ecogeographical Rule. Sci. Rep. 2016, 6, 32680. [Google Scholar] [CrossRef]
- Oliveira-Christe, R.; Wilke, A.B.B.; Marrelli, M.T. Microgeographic Wing-Shape Variation in Aedes Albopictus and Aedes Scapularis (Diptera: Culicidae) Populations. Insects 2020, 11, 862. [Google Scholar] [CrossRef]
- Saito, K.; Nomura, S.; Yamamoto, S.; Niiyama, R.; Okabe, Y. Investigation of Hindwing Folding in Ladybird Beetles by Artificial Elytron Transplantation and Microcomputed Tomography. Proc. Natl. Acad. Sci. USA 2017, 114, 5624–5628. [Google Scholar] [CrossRef]
- Bai, M.; Beutel, R.G.; Song, K.; Liu, W.; Malqin, H.; Li, S.; Hu, X.; Yang, X. Evolutionary Patterns of Hind Wing Morphology in Dung Beetles (Coleoptera: Scarabaeinae). Arthropod Struct. Dev. 2012, 41, 505–513. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, H.Y.; Ge, X.Y.; Yang, Y.X. Evaluating the Significance of Wing Shapes in Inferring Phylogenetic Proximity among the Generic Taxa: An Example of Cantharinae (Coleoptera, Cantharidae). Arthropod Syst. Phylogeny 2023, 81, 303–316. [Google Scholar] [CrossRef]
- Bai, M.; Beutel, R.G.; Shih, C.; Ren, D.; Yang, X. Septiventeridae, a New and Ancestral Fossil Family of Scarabaeoidea (Insecta: Coleoptera) from the Late Jurassic to Early Cretaceous Yixian Formation. J. Syst. Palaeontol. 2013, 11, 359–374. [Google Scholar] [CrossRef]
- Ospina-Garcés, S.M.; Escobar, F.; Baena, M.L.; Davis, A.L.V.; Scholtz, C.H. Do Dung Beetles Show Interrelated Evolutionary Trends in Wing Morphology, Flight Biomechanics and Habitat Preference? Evol. Ecol. 2018, 32, 663–682. [Google Scholar] [CrossRef]
- Tocco, C.; Dacke, M.; Byrne, M. Eye and Wing Structure Closely Reflects the Visual Ecology of Dung Beetles. J. Comp. Physiol. A 2019, 205, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Ariza-Marín, E.R.; Amat-García, G. Morphometric Changes in Wings of Bess Beetles (Coleoptera: Passalidae) Related to Elevation: A Case of Study in the Colombian Andes. Stud. Neotrop. Fauna Environ. 2021, 1–11. [Google Scholar] [CrossRef]
- Meresman, Y.; Husak, J.F.; Ben-Shlomo, R.; Ribak, G. Morphological Diversification Has Led to Inter-Specific Variation in Elastic Wing Deformation during Flight in Scarab Beetles. R. Soc. Open Sci. 2020, 7, 200277. [Google Scholar] [CrossRef]
- Owens, H.L.; Lewis, D.S.; Condamine, F.L.; Kawahara, A.Y.; Guralnick, R.P. Comparative Phylogenetics of Papilio Butterfly Wing Shape and Size Demonstrates Independent Hindwing and Forewing Evolution. Syst. Biol. 2020, 69, 813–819. [Google Scholar] [CrossRef]
- Rohlf, F.J. tpsDig, Digitize Landmarks and Outlines; Department of Ecology and Evolution, State University of New York: New York, NY, USA, 2006. [Google Scholar]
- Venegas, P.; Pérez, N.; Zapata, S.; Mosquera, J.D.; Augot, D.; Rojo-Álvarez, J.L.; Benítez, D. An Approach to Automatic Classification of Culicoides Species by Learning the Wing Morphology. PLoS One 2020, 15, e0241798. [Google Scholar] [CrossRef]
- Bellin, N.; Calzolari, M.; Callegari, E.; Bonilauri, P.; Grisendi, A.; Dottori, M.; Rossi, V. Geometric Morphometrics and Machine Learning as Tools for the Identification of Sibling Mosquito Species of the Maculipennis Complex (Anopheles). Infect. Genet. Evol. 2021, 95, 105034. [Google Scholar] [CrossRef]
- Valan, M.; Makonyi, K.; Maki, A.; Vondráček, D.; Ronquist, F. Automated Taxonomic Identification of Insects with Expert-Level Accuracy Using Effective Feature Transfer from Convolutional Networks. Syst. Biol. 2019, 68, 876–895. [Google Scholar] [CrossRef]
- Rodrigues, P.J.; Gomes, W.; Pinto, M.A. DeepWings©: Automatic Wing Geometric Morphometrics Classification of Honey Bee (Apis mellifera) Subspecies Using Deep Learning for Detecting Landmarks. Big Data Cogn. Comput. 2022, 6, 70. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Ho, B.H.; Lai, H.P.; Tran, H.T.; Bañuls, A.L.; Prudhomme, J.; Le, H.T. A Lightweight Keypoint Matching Framework for Insect Wing Morphometric Landmark Detection. Ecol. Inform. 2022, 70, 101694. [Google Scholar] [CrossRef]
- Sun, K.; Xiao, B.; Liu, D.; Wang, J. Deep High-Resolution Representation Learning for Human Pose Estimation. In Proceedings of the Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, 15–20 June 2019; pp. 5693–5703. [CrossRef]
- Arroyo, J.C.T. Coleoptera Classification Using Convolutional Neural Network and Transfer Learning. IJETT 2021, 69, 1–5. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 10, 1345–1359. [Google Scholar] [CrossRef]
- Ribani, R.; Marengoni, M. A survey of transfer learning for convolutional neural networks. In Proceedings of the 2019 32nd SIBGRAPI Conference on Graphics, Patterns and Images Tutorials (SIBGRAPI-T), Rio de Janeiro, Brazil, 28–31 October 2019; pp. 47–57. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- He, K.; Girshick, R.; Dollar, P. Rethinking ImageNet pre-training. In Proceedings of the Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 4918–4927.
- Kingma, D.P.; Ba, J.L. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Xiao, B.; Wu, H.; Wei, Y. Simple baselines for human pose estimation and tracking. In Proceedings of the Computer Vision—ECCV, Munich, Germany, 8–14 September 2018; Springer: Cham, Switzerland, 2018; pp. 472–487. [Google Scholar] [CrossRef]
- Wang, B.; Sun, R.; Yang, X.; Niu, B.; Zhang, T.; Zhao, Y.; Zhang, Y.; Zhang, Y.; Han, J. Recognition of Rare Microfossils Using Transfer Learning and Deep Residual Networks. Biology 2023, 12, 16. [Google Scholar] [CrossRef]
- Ren, J.; Bai, M.; Yang, X.K.; Zhang, R.Z.; Ge, S.Q. Geometric Morphometrics Analysis of the Hind Wing of Leaf Beetles: Proximal and Distal Parts Are Separate Modules. Zookeys 2017, 685, 131–149. [Google Scholar] [CrossRef]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common Objects in Context. In Proceedings of the European Conference on Computer Vision; Lecture Notes in Computer Science; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer: Zurich, Switzerland, 2014; pp. 740–755. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Oquab, M.; Bottou, L.; Laptev, I.; Sivic, J. Learning and Transferring Mid-Level Image Representations Using Convolutional Neural Networks. In Proceedings of the Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1717–1724.
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A survey on deep transfer learning. In Proceedings of the Artificial Neural Networks and Machine Learning—ICANN, Rhodes, Greece, 4–7 October 2018; Lecture Notes in Computer Science. Kůrková, V., Manolopoulos, Y., Hammer, B., Iliadis, L., Maglogiannis, I., Eds.; Springer: Cham, Switzerland, 2018; pp. 270–279. [Google Scholar] [CrossRef]
- Lan, X.; Hu, Q.; Cheng, J. ATF: An Alternating Training Framework for Weakly Supervised Face Alignment. IEEE Trans. Multimed. 2022, 25, 1798–1809. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, H.; Li, C.; Kim, J.; Wei, F. ADNet: Leveraging error-bias towards normal direction in face alignment. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 3060–3070. [Google Scholar] [CrossRef]
- Shimmi, O.; Matsuda, S.; Hatakeyama, M. Insights into the Molecular Mechanisms Underlying Diversified Wing Venation among Insects. Proc. R. Soc. B Biol. Sci. 2014, 281, 20140264. [Google Scholar] [CrossRef]
- Lawrence, J.F.; Zhou, Y.L.Z.; Lemann, C.; Sinclair, B.; Ślipiński, A. The Hind Wing of Coleoptera (Insecta): Morphology, Nomenclature and Phylogenetic Significance. Part 1. General Discussion and Archostemata–Elateroidea. Ann. Zool. 2021, 71, 421–606. [Google Scholar] [CrossRef]
- Lawrence, J.F.; Zhou, Y.L.Z.; Lemann, C.; Sinclair, B.; Ślipiński, A. The Hind Wing of Coleoptera (Insecta): Morphology, Nomenclature and Phylogenetic Significance: Part 2. Further Discussion, Histeroidea, Bostrichoidea to Curculionoidea. Ann. Zool. 2022, 72, 433–755. [Google Scholar] [CrossRef]
- Soule, A.J.; Decker, L.E.; Hunter, M.D. Effects of Diet and Temperature on Monarch Butterfly Wing Morphology and Flight Ability. J. Insect. Conserv. 2020, 24, 961–975. [Google Scholar] [CrossRef]
- Henriques, D.; Chávez-Galarza, J.; Teixeira, J.S.G.; Ferreira, H.; J. Neves, C.; Francoy, T.M.; Pinto, M.A. Wing Geometric Morphometrics of Workers and Drones and Single Nucleotide Polymorphisms Provide Similar Genetic Structure in the Iberian Honey Bee (Apis Mellifera Iberiensis). Insects 2020, 11, 89. [Google Scholar] [CrossRef]
- Song, F.; Yan, Y.; Sun, J. Review of Insect-Inspired Wing Micro Air Vehicle. Arthropod Struct. Dev. 2023, 72, 101225. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.V.; Park, H.C. Mechanisms of Collision Recovery in Flying Beetles and Flapping-Wing Robots. Science 2020, 370, 1214–1219. [Google Scholar] [CrossRef]
- Zeng, Y.; O’Malley, C.; Singhal, S.; Rahim, F.; Park, S.; Chen, X.; Dudley, R. A Tale of Winglets: Evolution of Flight Morphology in Stick Insects. Front. Ecol. Evol. 2020, 8, 121. [Google Scholar] [CrossRef]
- Dudley, R. The Biomechanics of Insect Flight: Form, Function, Evolution; Princeton University Press: Princeton, NJ, USA, 2002. [Google Scholar]
- Sun, J.; Wu, W.; Ling, M.; Tong, J.; Ren, L. Fluid Analysis of Vein of Beetle Hindwing during Unfolding Action. Int. J. Heat Mass Transf. 2016, 101, 379–386. [Google Scholar] [CrossRef]
- Nakata, T.; Liu, H. Aerodynamic Performance of a Hovering Hawkmoth with Flexible Wings: A Computational Approach. Proc. R. Soc. B Biol. Sci. 2011, 279, 722–731. [Google Scholar] [CrossRef]
- Song, Z.; Tong, J.; Pfleging, W.; Sun, J. A Review: Learning from the Flight of Beetles. Comput. Biol. Med. 2021, 133, 104397. [Google Scholar] [CrossRef]
- Cannet, A.; Simon-Chane, C.; Akhoundi, M.; Histace, A.; Romain, O.; Souchaud, M.; Jacob, P.; Delaunay, P.; Sereno, D.; Bousses, P.; et al. Wing Interferential Patterns (WIPs) and Machine Learning, a Step toward Automatized Tsetse (Glossina spp.) Identification. Sci. Rep. 2022, 12, 20086. [Google Scholar] [CrossRef]
- Park, J.; Kim, D.I.; Choi, B.; Kang, W.; Kwon, H.W. Classification and Morphological Analysis of Vector Mosquitoes Using Deep Convolutional Neural Networks. Sci. Rep. 2020, 10, 1012. [Google Scholar] [CrossRef]
- Buschbacher, K.; Ahrens, D.; Espeland, M.; Steinhage, V. Image-Based Species Identification of Wild Bees Using Convolutional Neural Networks. Ecol. Inform. 2020, 55, 101017. [Google Scholar] [CrossRef]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; Curran Associates, Inc.: New York, NY, USA, 2014; Volume 27. [Google Scholar]
- Zhu, X.; Lei, Z.; Liu, X.; Shi, H.; Li, S.Z. Face alignment across large poses: A 3D solution. In Proceedings of the Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 146–155.
- Murty, N.A.R.; Bashivan, P.; Abate, A.; DiCarlo, J.J.; Kanwisher, N. Computational Models of Category-Selective Brain Regions Enable High-Throughput Tests of Selectivity. Nat. Commun. 2021, 12, 5540. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, C.P.; McIntyre, G.S. Geometric Morphometrics of Developmental Instability: Analyzing Patterns of Fluctuating Asymmetry with Procrustes Methods. Evolution 1998, 52, 1363–1375. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, C.P.; Zaklan, S.D. Morphological Integration between Developmental Compartments in the Drosophila Wing. Evolution 2000, 54, 1273–1285. [Google Scholar] [CrossRef] [PubMed]
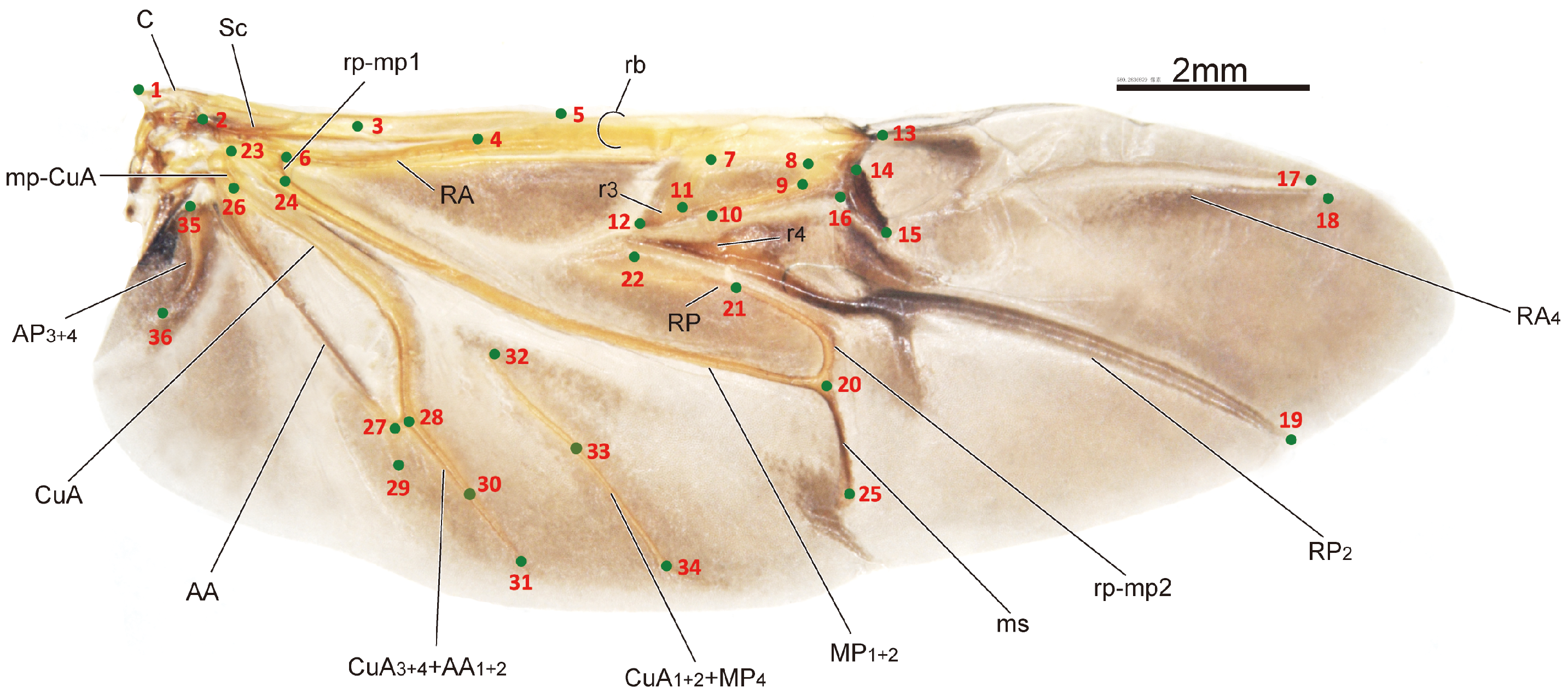
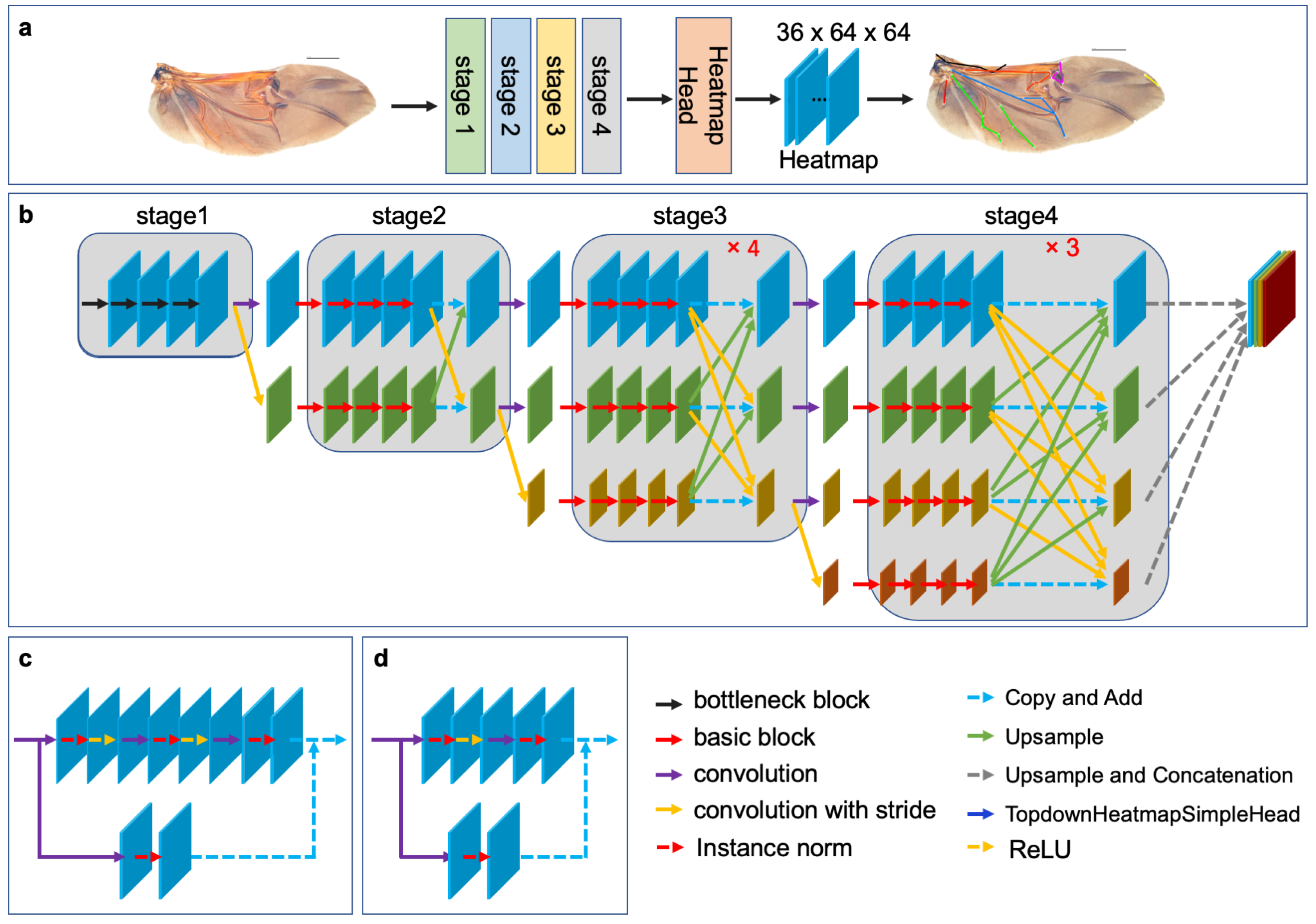

| Landmark Index | Position Description |
|---|---|
| 1 | Proximal anterior point of humeral plate (HP) |
| 2 | The crossing point of BSc and Sc |
| 3 | The point of Sc getting to bifurcate into ScA and ScP |
| 4 | The crossing point of ScP and RA |
| 5 | The crossing point of ScA and RA |
| 6 | The crossing point of rp-m1 and RA |
| 7 | Proximal anterior point of radial cell |
| 8 | Distal anterior point of radial cell |
| 9 | Distal posterior point of radial cell |
| 10 | Anterior point of r4 (or the crossing point of r4 and radial cell) |
| 11 | Proximal posterior point of radial cell |
| 12 | Proximal point of r3 |
| 13 | Apical hinge |
| 14 | The anterior point of triangular area of radial cell’s distal side |
| 15 | The posterior point of triangular area of radial cell’s distal side |
| 16 | The proximal point of triangular area of radial cell’s distal side |
| 17 | The distal point of RA_4 |
| 18 | The distal point of RA_1 |
| 19 | The distal point of RP_2 |
| 20 | The point of MP1+2 getting to bifurcate |
| 21 | The posterior point of r4, or the crossing point of r4 and rp-mp2 |
| 22 | The proximal point of RP |
| 23 | Anterior point of mp-cua |
| 24 | The crossing point of rm-mp1and MP |
| 25 | The posterior of medial spur |
| 26 | Posterior point of mp-cua |
| 27 | The point of AA getting to bifurcate |
| 28 | The point of AA1+2 getting to fuse with CuA3+4 |
| 29 | The posterior or distal point of AA3+4 |
| 30 | The proximal point of cv |
| 31 | The posterior or distal point of AA1+2+CuA3+4 |
| 32 | Anterior point of CuA1+2+MP4 |
| 33 | The distal point of cv |
| 34 | Posterior point of CuA1+2+MP_4 |
| 35 | The base point of AP3+4 |
| 36 | The posterior point of AP3+4 |
| Strategies | Stage1 | Stage2 | Stage3 | Stage4 | Hh (Heatmap Head) |
|---|---|---|---|---|---|
| TH-HRNet | ✕ | ✕ | ✕ | ✕ | ✓ |
| ✕ | ✕ | ✕ | ✓ | ✓ | |
| TS-HRNet | ✕ | ✕ | ✓ | ✓ | ✓ |
| ✕ | ✓ | ✓ | ✓ | ✓ | |
| TA-HRNet | ✓ | ✓ | ✓ | ✓ | ✓ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Liu, X.; Li, W.; Li, C.; Ma, G.; Yang, G.; Ren, J.; Ge, S. Detection of Hindwing Landmarks Using Transfer Learning and High-Resolution Networks. Biology 2023, 12, 1006. https://doi.org/10.3390/biology12071006
Yang Y, Liu X, Li W, Li C, Ma G, Yang G, Ren J, Ge S. Detection of Hindwing Landmarks Using Transfer Learning and High-Resolution Networks. Biology. 2023; 12(7):1006. https://doi.org/10.3390/biology12071006
Chicago/Turabian StyleYang, Yi, Xiaokun Liu, Wenjie Li, Congqiao Li, Ge Ma, Guangqin Yang, Jing Ren, and Siqin Ge. 2023. "Detection of Hindwing Landmarks Using Transfer Learning and High-Resolution Networks" Biology 12, no. 7: 1006. https://doi.org/10.3390/biology12071006
APA StyleYang, Y., Liu, X., Li, W., Li, C., Ma, G., Yang, G., Ren, J., & Ge, S. (2023). Detection of Hindwing Landmarks Using Transfer Learning and High-Resolution Networks. Biology, 12(7), 1006. https://doi.org/10.3390/biology12071006






