Sialylation: A Cloak for Tumors to Trick the Immune System in the Microenvironment
Abstract
Simple Summary
Abstract
1. Introduction
2. Sialylation Accumulates in Cancer Tissues and Promotes Tumor Development
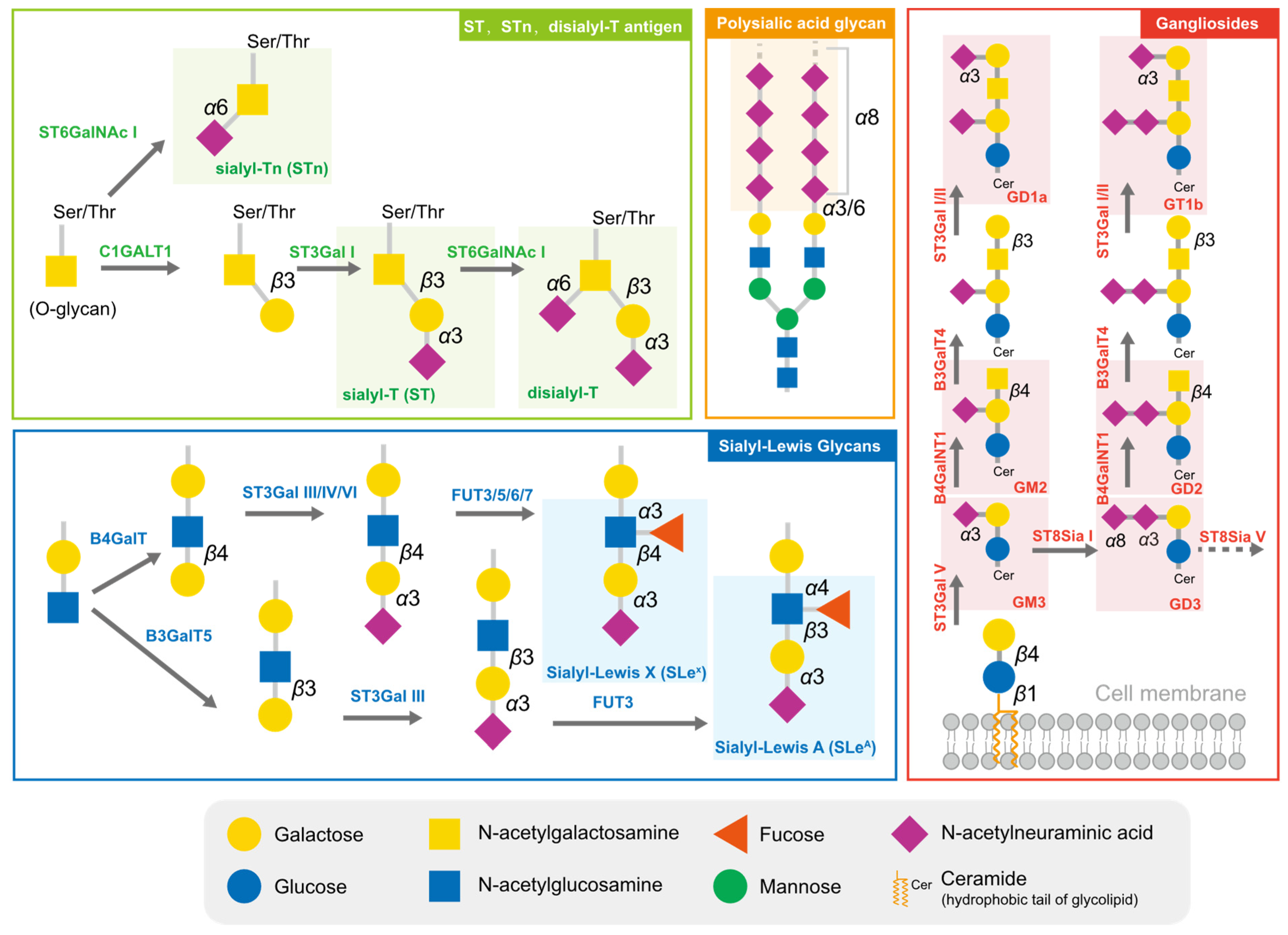
2.1. Typical Sialylated Glycans in Tumors
2.2. Sialyltransferases Are Critical Enzymes for Hypersialylation
2.3. The Function of Sialidases in Tumor Sialylation Regulation
3. Sialylation Reshapes the Tumor Microenvironment
3.1. Sialylation Serves as a Camouflage for the Passive Protection of Tumor Cells
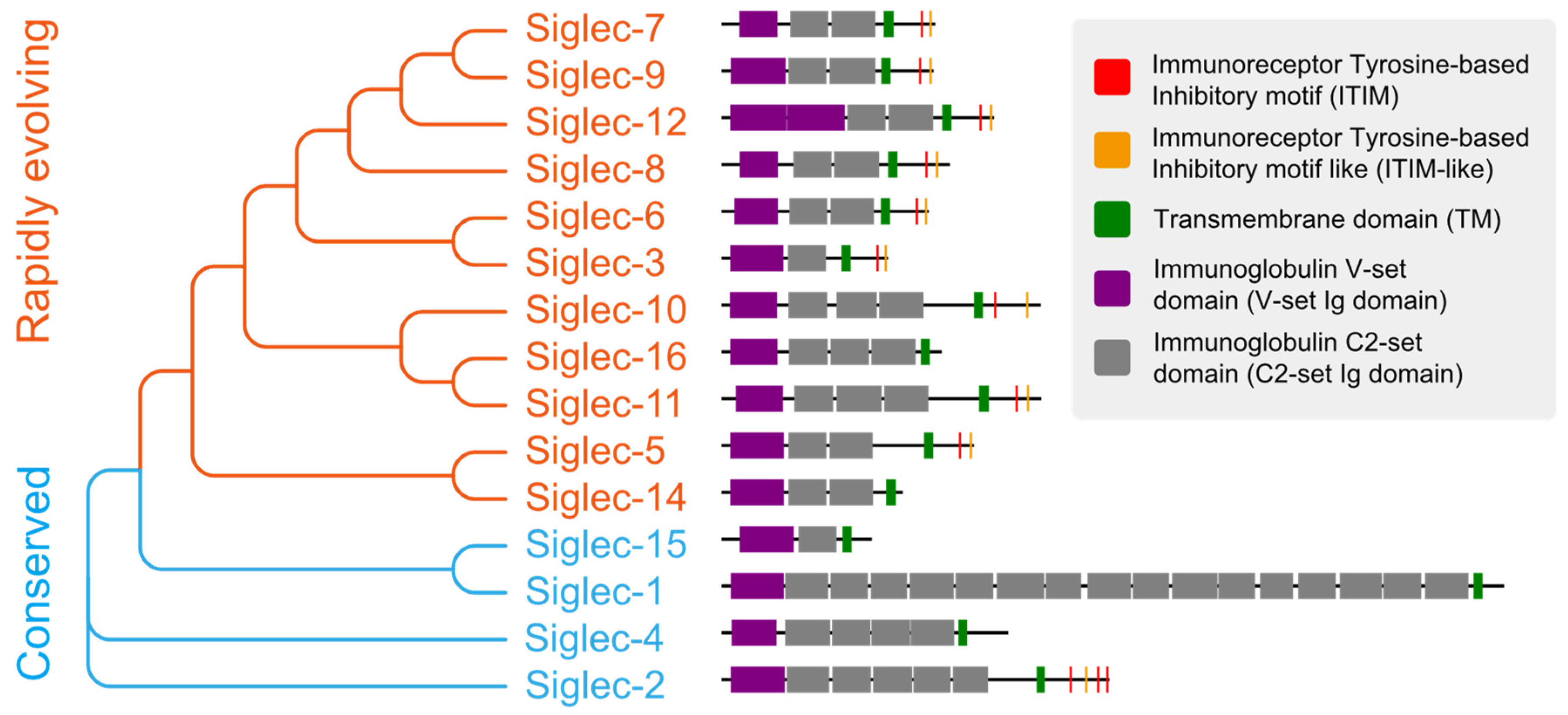
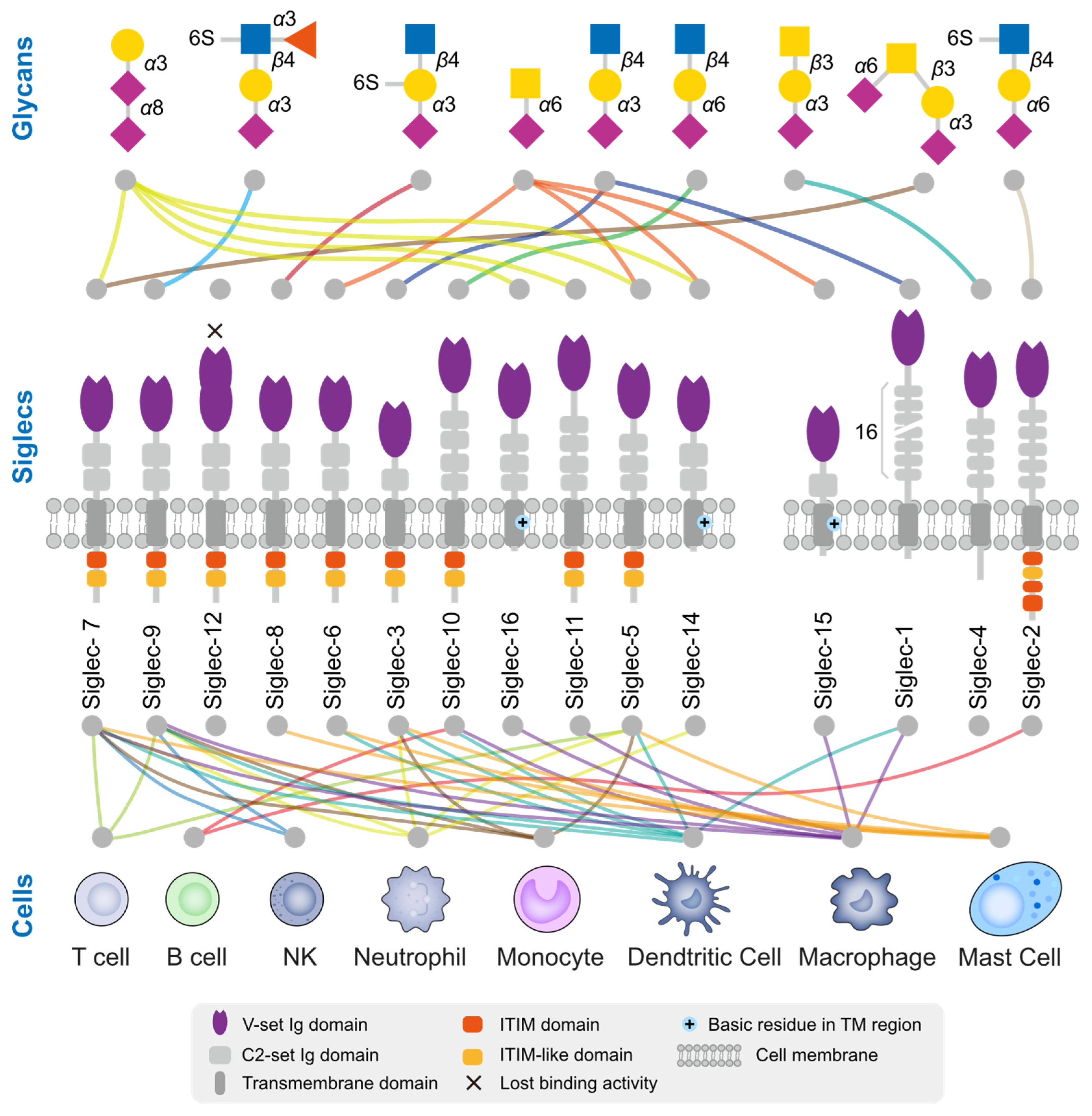
3.2. Tumor Cells Deceive Immune Cells through the Binding of Sialylated Ligands to Siglecs
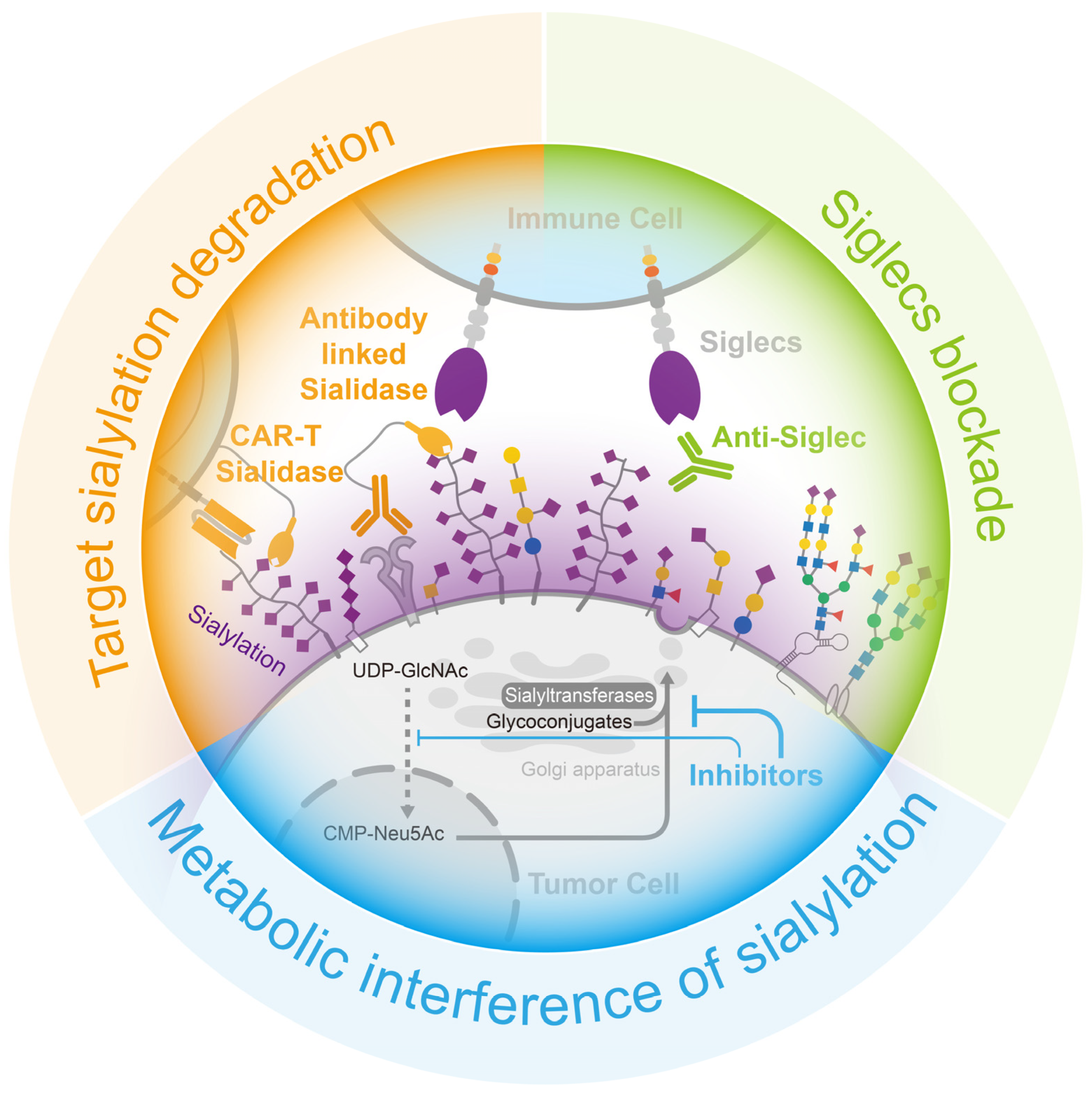
4. Opportunities for Cancer Treatment via the Removal of Sialylation-Caused Immune Suppression
4.1. Metabolic Interference with Sialylation
4.2. Target Sialylation Degradation
4.3. Siglecs Blockade
5. Discussion and Future Perspectives
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, X.; Varki, A. Advances in the biology and chemistry of sialic acids. ACS Chem. Biol. 2010, 5, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Schauer, R.; Kamerling, J.P. Exploration of the Sialic Acid World. Adv. Carbohydr. Chem. Biochem. 2018, 75, 1–213. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yang, G.; Guan, F. Biological Functions and Analytical Strategies of Sialic Acids in Tumor. Cells 2020, 9, 273. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Kim, G.; Shafer, S.; Chen, Z.; Kubo, S.; Ji, Y.; Luo, J.; Yang, W.; Perner, S.P.; Kanellopoulou, C.; et al. Mucus sialylation determines intestinal host-commensal homeostasis. Cell 2022, 185, 1172–1188 e1128. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X. Sialic acid metabolism and sialyltransferases: Natural functions and applications. Appl. Microbiol. Biotechnol. 2012, 94, 887–905. [Google Scholar] [CrossRef]
- Teoh, S.T.; Ogrodzinski, M.P.; Ross, C.; Hunter, K.W.; Lunt, S.Y. Sialic Acid Metabolism: A Key Player in Breast Cancer Metastasis Revealed by Metabolomics. Front. Oncol. 2018, 8, 174. [Google Scholar] [CrossRef]
- Harduin-Lepers, A.; Vallejo-Ruiz, V.; Krzewinski-Recchi, M.A.; Samyn-Petit, B.; Julien, S.; Delannoy, P. The human sialyltransferase family. Biochimie 2001, 83, 727–737. [Google Scholar] [CrossRef]
- Seyrantepe, V.; Landry, K.; Trudel, S.; Hassan, J.A.; Morales, C.R.; Pshezhetsky, A.V. Neu4, a novel human lysosomal lumen sialidase, confers normal phenotype to sialidosis and galactosialidosis cells. J. Biol. Chem. 2004, 279, 37021–37029. [Google Scholar] [CrossRef]
- Seyrantepe, V.; Poupetova, H.; Froissart, R.; Zabot, M.T.; Maire, I.; Pshezhetsky, A.V. Molecular pathology of NEU1 gene in sialidosis. Hum. Mutat. 2003, 22, 343–352. [Google Scholar] [CrossRef]
- Zhou, X.; Zhai, Y.; Liu, C.; Yang, G.; Guo, J.; Li, G.; Sun, C.; Qi, X.; Li, X.; Guan, F. Sialidase NEU1 suppresses progression of human bladder cancer cells by inhibiting fibronectin-integrin alpha5beta1 interaction and Akt signaling pathway. Cell Commun. Signal. 2020, 18, 44. [Google Scholar] [CrossRef]
- Hyun, S.W.; Feng, C.; Liu, A.; Lillehoj, E.P.; Trotta, R.; Kingsbury, T.J.; Passaniti, A.; Lugkey, K.N.; Chauhan, S.; Cipollo, J.F.; et al. Altered sialidase expression in human myeloid cells undergoing apoptosis and differentiation. Sci. Rep. 2022, 12, 14173. [Google Scholar] [CrossRef]
- Miyagi, T.; Takahashi, K.; Hata, K.; Shiozaki, K.; Yamaguchi, K. Sialidase significance for cancer progression. Glycoconj. J. 2012, 29, 567–577. [Google Scholar] [CrossRef]
- Elhanani, O.; Ben-Uri, R.; Keren, L. Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell 2023, 41, 404–420. [Google Scholar] [CrossRef]
- Carlino, M.S.; Larkin, J.; Long, G.V. Immune checkpoint inhibitors in melanoma. Lancet 2021, 398, 1002–1014. [Google Scholar] [CrossRef]
- Nagasaki, J.; Ishino, T.; Togashi, Y. Mechanisms of resistance to immune checkpoint inhibitors. Cancer Sci. 2022, 113, 3303–3312. [Google Scholar] [CrossRef]
- Pietrobono, S.; Stecca, B. Aberrant Sialylation in Cancer: Biomarker and Potential Target for Therapeutic Intervention? Cancers 2021, 13, 2014. [Google Scholar] [CrossRef]
- Chang, X.; Obianwuna, U.E.; Wang, J.; Zhang, H.; Qi, G.; Qiu, K.; Wu, S. Glycosylated proteins with abnormal glycosylation changes are potential biomarkers for early diagnosis of breast cancer. Int. J. Biol. Macromol. 2023, 236, 123855. [Google Scholar] [CrossRef]
- Hu, M.; Zhang, R.; Yang, J.; Zhao, C.; Liu, W.; Huang, Y.; Lyu, H.; Xiao, S.; Guo, D.; Zhou, C.; et al. The role of N-glycosylation modification in the pathogenesis of liver cancer. Cell Death Dis. 2023, 14, 222. [Google Scholar] [CrossRef]
- Zheng, L.; Yang, Q.; Li, F.; Zhu, M.; Yang, H.; Tan, T.; Wu, B.; Liu, M.; Xu, C.; Yin, J.; et al. The Glycosylation of Immune Checkpoints and Their Applications in Oncology. Pharmaceuticals 2022, 15, 1451. [Google Scholar] [CrossRef]
- Casalino, L.; Gaieb, Z.; Goldsmith, J.A.; Hjorth, C.K.; Dommer, A.C.; Harbison, A.M.; Fogarty, C.A.; Barros, E.P.; Taylor, B.C.; McLellan, J.S.; et al. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent. Sci. 2020, 6, 1722–1734. [Google Scholar] [CrossRef]
- Gonzalez-Gil, A.; Schnaar, R.L. Siglec Ligands. Cells 2021, 10, 1260. [Google Scholar] [CrossRef] [PubMed]
- Vajaria, B.N.; Patel, P.S. Glycosylation: A hallmark of cancer? Glycoconj. J. 2017, 34, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Gao, Y.; Diao, X.; Li, J.; Jiang, H.; Zhao, H. Diagnostic value of sialyl-Tn immunocytochemistry in breast cancer presenting with pathological nipple discharge. Cancer Med. 2021, 10, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, D.; Hongo, H.; Kosaka, T.; Aoki, N.; Oya, M.; Sato, T. The sialyl-Tn antigen synthase genes regulates migration-proliferation dichotomy in prostate cancer cells under hypoxia. Glycoconj. J. 2023, 40, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, S.; Abreu, C.M.; Ferreira, D.; Lima, L.; Ferreira, J.A.; Santos, L.L.; Ribeiro, R.; Grenha, V.; Martinez-Fernandez, M.; Duenas, M.; et al. Phenotypic Analysis of Urothelial Exfoliated Cells in Bladder Cancer via Microfluidic Immunoassays: Sialyl-Tn as a Novel Biomarker in Liquid Biopsies. Front. Oncol. 2020, 10, 1774. [Google Scholar] [CrossRef]
- Benbrook, D.M.; Deng, W.; Gold, M.A.; Rai, R.; Conrad, R.; van der Wel, H.; Husain, S.; Moore, K.; Spirtos, N.; Jackson, A.L.; et al. Association of Sialyl Tn antigen with cervical cancer lymph node status: An NRG oncology/GOG study. Gynecol. Oncol. 2023, 171, 67–75. [Google Scholar] [CrossRef]
- Munkley, J. The Role of Sialyl-Tn in Cancer. Int. J. Mol. Sci. 2016, 17, 275. [Google Scholar] [CrossRef]
- Bai, R.; Luan, X.; Zhang, Y.; Robbe-Masselot, C.; Brockhausen, I.; Gao, Y. The expression and functional analysis of the sialyl-T antigen in prostate cancer. Glycoconj. J. 2020, 37, 423–433. [Google Scholar] [CrossRef]
- Manfioletti, G.; Fedele, M. Epithelial-Mesenchymal Transition (EMT) 2021. Int. J. Mol. Sci. 2022, 23, 5848. [Google Scholar] [CrossRef]
- Ricardo, S.; Marcos-Silva, L.; Valente, C.; Coelho, R.; Gomes, R.; David, L. Mucins MUC16 and MUC1 are major carriers of SLe(a) and SLe(x) in borderline and malignant serous ovarian tumors. Virchows Arch. 2016, 468, 715–722. [Google Scholar] [CrossRef]
- Foley, D.A.; Swartzentruber, K.G.; Colley, K.J. Identification of sequences in the polysialyltransferases ST8Sia II and ST8Sia IV that are required for the protein-specific polysialylation of the neural cell adhesion molecule, NCAM. J. Biol. Chem. 2009, 284, 15505–15516. [Google Scholar] [CrossRef]
- Seki, T.; Arai, Y. Expression of highly polysialylated NCAM in the neocortex and piriform cortex of the developing and the adult rat. Anat. Embryol. 1991, 184, 395–401. [Google Scholar] [CrossRef]
- Li, J.; Yang, R.; Yang, H.; Chen, S.; Wang, L.; Li, M.; Yang, S.; Feng, Z.; Bi, J. NCAM regulates the proliferation, apoptosis, autophagy, EMT, and migration of human melanoma cells via the Src/Akt/mTOR/cofilin signaling pathway. J. Cell. Biochem. 2020, 121, 1192–1204. [Google Scholar] [CrossRef]
- Hauser, M.A.; Kindinger, I.; Laufer, J.M.; Spate, A.K.; Bucher, D.; Vanes, S.L.; Krueger, W.A.; Wittmann, V.; Legler, D.F. Distinct CCR7 glycosylation pattern shapes receptor signaling and endocytosis to modulate chemotactic responses. J. Leukoc. Biol. 2016, 99, 993–1007. [Google Scholar] [CrossRef]
- Verhallen, L.; Lackman, J.J.; Wendt, R.; Gustavsson, M.; Yang, Z.; Narimatsu, Y.; Sorensen, D.M.; Lafferty, K.M.; Gouwy, M.; Marques, P.E.; et al. “Glyco-sulfo barcodes” regulate chemokine receptor function. Cell. Mol. Life Sci. 2023, 80, 55. [Google Scholar] [CrossRef]
- Yabe, U.; Sato, C.; Matsuda, T.; Kitajima, K. Polysialic acid in human milk. CD36 is a new member of mammalian polysialic acid-containing glycoprotein. J. Biol. Chem. 2003, 278, 13875–13880. [Google Scholar] [CrossRef]
- Werneburg, S.; Buettner, F.F.; Erben, L.; Mathews, M.; Neumann, H.; Muhlenhoff, M.; Hildebrandt, H. Polysialylation and lipopolysaccharide-induced shedding of E-selectin ligand-1 and neuropilin-2 by microglia and THP-1 macrophages. Glia 2016, 64, 1314–1330. [Google Scholar] [CrossRef]
- Jarahian, M.; Marofi, F.; Maashi, M.S.; Ghaebi, M.; Khezri, A.; Berger, M.R. Re-Expression of Poly/Oligo-Sialylated Adhesion Molecules on the Surface of Tumor Cells Disrupts Their Interaction with Immune-Effector Cells and Contributes to Pathophysiological Immune Escape. Cancers 2021, 13, 5203. [Google Scholar] [CrossRef]
- Daniotti, J.L.; Lardone, R.D.; Vilcaes, A.A. Dysregulated Expression of Glycolipids in Tumor Cells: From Negative Modulator of Anti-tumor Immunity to Promising Targets for Developing Therapeutic Agents. Front. Oncol. 2015, 5, 300. [Google Scholar] [CrossRef]
- Kasprowicz, A.; Sophie, G.D.; Lagadec, C.; Delannoy, P. Role of GD3 Synthase ST8Sia I in Cancers. Cancers 2022, 14, 1299. [Google Scholar] [CrossRef]
- Rodrigues, E.; Macauley, M.S. Hypersialylation in Cancer: Modulation of Inflammation and Therapeutic Opportunities. Cancers 2018, 10, 207. [Google Scholar] [CrossRef] [PubMed]
- Dobie, C.; Skropeta, D. Insights into the role of sialylation in cancer progression and metastasis. Br. J. Cancer 2021, 124, 76–90. [Google Scholar] [CrossRef] [PubMed]
- Gc, S.; Bellis, S.L.; Hjelmeland, A.B. ST6Gal1: Oncogenic signaling pathways and targets. Front. Mol. Biosci. 2022, 9, 962908. [Google Scholar] [CrossRef] [PubMed]
- Gc, S.; Tuy, K.; Rickenbacker, L.; Jones, R.; Chakraborty, A.; Miller, C.R.; Beierle, E.A.; Hanumanthu, V.S.; Tran, A.N.; Mobley, J.A.; et al. alpha2,6 Sialylation mediated by ST6GAL1 promotes glioblastoma growth. JCI Insight 2022, 7, e158799. [Google Scholar] [CrossRef]
- Smithson, M.; Irwin, R.; Williams, G.; Alexander, K.L.; Smythies, L.E.; Nearing, M.; McLeod, M.C.; Al Diffalha, S.; Bellis, S.L.; Hardiman, K.M. Sialyltransferase ST6GAL-1 mediates resistance to chemoradiation in rectal cancer. J. Biol. Chem. 2022, 298, 101594. [Google Scholar] [CrossRef]
- Duarte, H.O.; Rodrigues, J.G.; Gomes, C.; Hensbergen, P.J.; Ederveen, A.L.H.; de Ru, A.H.; Mereiter, S.; Polonia, A.; Fernandes, E.; Ferreira, J.A.; et al. ST6Gal1 targets the ectodomain of ErbB2 in a site-specific manner and regulates gastric cancer cell sensitivity to trastuzumab. Oncogene 2021, 40, 3719–3733. [Google Scholar] [CrossRef]
- Wichert, B.; Milde-Langosch, K.; Galatenko, V.; Schmalfeldt, B.; Oliveira-Ferrer, L. Prognostic role of the sialyltransferase ST6GAL1 in ovarian cancer. Glycobiology 2018, 28, 898–903. [Google Scholar] [CrossRef]
- Wang, L.; Chen, X.; Wang, L.; Wang, S.; Li, W.; Liu, Y.; Zhang, J. Knockdown of ST6Gal-I expression in human hepatocellular carcinoma cells inhibits their exosome-mediated proliferation- and migration-promoting effects. IUBMB Life 2021, 73, 1378–1391. [Google Scholar] [CrossRef]
- Zhang, M.; Qi, T.; Yang, L.; Kolarich, D.; Heisterkamp, N. Multi-Faceted Effects of ST6Gal1 Expression on Precursor B-Lineage Acute Lymphoblastic Leukemia. Front. Oncol. 2022, 12, 828041. [Google Scholar] [CrossRef]
- Kurz, E.; Chen, S.; Vucic, E.; Baptiste, G.; Loomis, C.; Agrawal, P.; Hajdu, C.; Bar-Sagi, D.; Mahal, L.K. Integrated Systems Analysis of the Murine and Human Pancreatic Cancer Glycomes Reveals a Tumor-Promoting Role for ST6GAL1. Mol. Cell. Proteomics 2021, 20, 100160. [Google Scholar] [CrossRef]
- Britain, C.M.; Holdbrooks, A.T.; Anderson, J.C.; Willey, C.D.; Bellis, S.L. Sialylation of EGFR by the ST6Gal-I sialyltransferase promotes EGFR activation and resistance to gefitinib-mediated cell death. J. Ovarian Res. 2018, 11, 12. [Google Scholar] [CrossRef]
- Liu, Q.; Ma, H.; Sun, X.; Liu, B.; Xiao, Y.; Pan, S.; Zhou, H.; Dong, W.; Jia, L. The regulatory ZFAS1/miR-150/ST6GAL1 crosstalk modulates sialylation of EGFR via PI3K/Akt pathway in T-cell acute lymphoblastic leukemia. J. Exp. Clin. Cancer Res. 2019, 38, 199. [Google Scholar] [CrossRef]
- Kitazume, S.; Imamaki, R.; Ogawa, K.; Komi, Y.; Futakawa, S.; Kojima, S.; Hashimoto, Y.; Marth, J.D.; Paulson, J.C.; Taniguchi, N. Alpha2,6-sialic acid on platelet endothelial cell adhesion molecule (PECAM) regulates its homophilic interactions and downstream antiapoptotic signaling. J. Biol. Chem. 2010, 285, 6515–6521. [Google Scholar] [CrossRef]
- Wang, L.; Li, S.; Yu, X.; Han, Y.; Wu, Y.; Wang, S.; Chen, X.; Zhang, J.; Wang, S. alpha2,6-Sialylation promotes immune escape in hepatocarcinoma cells by regulating T cell functions and CD147/MMP signaling. J. Physiol. Biochem. 2019, 75, 199–207. [Google Scholar] [CrossRef]
- Hait, N.C.; Maiti, A.; Wu, R.; Andersen, V.L.; Hsu, C.C.; Wu, Y.; Chapla, D.G.; Takabe, K.; Rusiniak, M.E.; Bshara, W.; et al. Extracellular sialyltransferase st6gal1 in breast tumor cell growth and invasiveness. Cancer Gene Ther. 2022, 29, 1662–1675. [Google Scholar] [CrossRef]
- Takashima, S.; Tsuji, S.; Tsujimoto, M. Comparison of the enzymatic properties of mouse beta-galactoside alpha2,6-sialyltransferases, ST6Gal I and II. J. Biochem. 2003, 134, 287–296. [Google Scholar] [CrossRef]
- Cheng, J.; Wang, R.; Zhong, G.; Chen, X.; Cheng, Y.; Li, W.; Yang, Y. ST6GAL2 Downregulation Inhibits Cell Adhesion and Invasion and is Associated with Improved Patient Survival in Breast Cancer. Onco Targets Ther. 2020, 13, 903–914. [Google Scholar] [CrossRef]
- Xu, G.; Chen, J.; Wang, G.; Xiao, J.; Zhang, N.; Chen, Y.; Yu, H.; Wang, G.; Zhao, Y. Resveratrol Inhibits the Tumorigenesis of Follicular Thyroid Cancer via ST6GAL2-Regulated Activation of the Hippo Signaling Pathway. Mol. Ther. Oncolytics 2020, 16, 124–133. [Google Scholar] [CrossRef]
- Wu, X.; Zhao, J.; Ruan, Y.; Sun, L.; Xu, C.; Jiang, H. Sialyltransferase ST3GAL1 promotes cell migration, invasion, and TGF-beta1-induced EMT and confers paclitaxel resistance in ovarian cancer. Cell Death Dis. 2018, 9, 1102. [Google Scholar] [CrossRef]
- Chong, Y.K.; Sandanaraj, E.; Koh, L.W.; Thangaveloo, M.; Tan, M.S.; Koh, G.R.; Toh, T.B.; Lim, G.G.; Holbrook, J.D.; Kon, O.L.; et al. ST3GAL1-Associated Transcriptomic Program in Glioblastoma Tumor Growth, Invasion, and Prognosis. J. Natl. Cancer Inst. 2016, 108, djv326. [Google Scholar] [CrossRef]
- Pietrobono, S.; Anichini, G.; Sala, C.; Manetti, F.; Almada, L.L.; Pepe, S.; Carr, R.M.; Paradise, B.D.; Sarkaria, J.N.; Davila, J.I.; et al. ST3GAL1 is a target of the SOX2-GLI1 transcriptional complex and promotes melanoma metastasis through AXL. Nat. Commun. 2020, 11, 5865. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.D.; Fan, T.C.; Hung, J.T.; Yeo, H.L.; Wang, S.H.; Kuo, C.W.; Khoo, K.H.; Pai, L.M.; Yu, J.; Yu, A.L. Sialylation of CD55 by ST3GAL1 Facilitates Immune Evasion in Cancer. Cancer Immunol. Res. 2021, 9, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Sipione, S.; Monyror, J.; Galleguillos, D.; Steinberg, N.; Kadam, V. Gangliosides in the Brain: Physiology, Pathophysiology and Therapeutic Applications. Front. Neurosci. 2020, 14, 572965. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Lin, S.; Cui, W.; Newman, P.J. Overlapping and unique substrate specificities of ST3GAL1 and 2 during hematopoietic and megakaryocytic differentiation. Blood Adv. 2022, 6, 3945–3955. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Aoki, H.; Ito, A.; Ueno, S.; Wada, T.; Mitsuzuka, K.; Satoh, M.; Arai, Y.; Miyagi, T. Human alpha2,3-sialyltransferase (ST3Gal II) is a stage-specific embryonic antigen-4 synthase. J. Biol. Chem. 2003, 278, 26474–26479. [Google Scholar] [CrossRef] [PubMed]
- Kannagi, R.; Cochran, N.A.; Ishigami, F.; Hakomori, S.; Andrews, P.W.; Knowles, B.B.; Solter, D. Stage-specific embryonic antigens (SSEA-3 and -4) are epitopes of a unique globo-series ganglioside isolated from human teratocarcinoma cells. EMBO J. 1983, 2, 2355–2361. [Google Scholar] [CrossRef] [PubMed]
- Deschuyter, M.; Leger, D.Y.; Verboom, A.; Chaunavel, A.; Maftah, A.; Petit, J.M. ST3GAL2 knock-down decreases tumoral character of colorectal cancer cells in vitro and in vivo. Am. J. Cancer Res. 2022, 12, 280–302. [Google Scholar]
- Kono, M.; Ohyama, Y.; Lee, Y.C.; Hamamoto, T.; Kojima, N.; Tsuji, S. Mouse beta-galactoside alpha 2,3-sialyltransferases: Comparison of in vitro substrate specificities and tissue specific expression. Glycobiology 1997, 7, 469–479. [Google Scholar] [CrossRef]
- Okajima, T.; Fukumoto, S.; Miyazaki, H.; Ishida, H.; Kiso, M.; Furukawa, K.; Urano, T.; Furukawa, K. Molecular cloning of a novel alpha2,3-sialyltransferase (ST3Gal VI) that sialylates type II lactosamine structures on glycoproteins and glycolipids. J. Biol. Chem. 1999, 274, 11479–11486. [Google Scholar] [CrossRef]
- Quirino, M.W.L.; Albuquerque, A.P.B.; De Souza, M.F.D.; Da Silva Filho, A.F.; Martins, M.R.; Da Rocha Pitta, M.G.; Pereira, M.C.; De Melo Rego, M.J.B. alpha2,3 sialic acid processing enzymes expression in gastric cancer tissues reveals that ST3Gal3 but not Neu3 are associated with Lauren’s classification, angiolymphatic invasion and histological grade. Eur. J. Histochem. 2022, 66, 3330. [Google Scholar] [CrossRef]
- Rodriguez, E.; Boelaars, K.; Brown, K.; Eveline Li, R.J.; Kruijssen, L.; Bruijns, S.C.M.; van Ee, T.; Schetters, S.T.T.; Crommentuijn, M.H.W.; van der Horst, J.C.; et al. Sialic acids in pancreatic cancer cells drive tumour-associated macrophage differentiation via the Siglec receptors Siglec-7 and Siglec-9. Nat. Commun. 2021, 12, 1270. [Google Scholar] [CrossRef]
- Narimatsu, Y.; Joshi, H.J.; Nason, R.; Van Coillie, J.; Karlsson, R.; Sun, L.; Ye, Z.; Chen, Y.H.; Schjoldager, K.T.; Steentoft, C.; et al. An Atlas of Human Glycosylation Pathways Enables Display of the Human Glycome by Gene Engineered Cells. Mol. Cell 2019, 75, 394–407 e395. [Google Scholar] [CrossRef]
- Sun, M.; Zhao, X.; Liang, L.; Pan, X.; Lv, H.; Zhao, Y. Sialyltransferase ST3GAL6 mediates the effect of microRNA-26a on cell growth, migration, and invasion in hepatocellular carcinoma through the protein kinase B/mammalian target of rapamycin pathway. Cancer Sci. 2017, 108, 267–276. [Google Scholar] [CrossRef]
- Dalangood, S.; Zhu, Z.; Ma, Z.; Li, J.; Zeng, Q.; Yan, Y.; Shen, B.; Yan, J.; Huang, R. Identification of glycogene-type and validation of ST3GAL6 as a biomarker predicts clinical outcome and cancer cell invasion in urinary bladder cancer. Theranostics 2020, 10, 10078–10091. [Google Scholar] [CrossRef]
- Liu, J.; Li, M.; Wu, J.; Qi, Q.; Li, Y.; Wang, S.; Liang, S.; Zhang, Y.; Zhu, Z.; Huang, R.; et al. Identification of ST3GAL5 as a prognostic biomarker correlating with CD8(+) T cell exhaustion in clear cell renal cell carcinoma. Front. Immunol. 2022, 13, 979605. [Google Scholar] [CrossRef]
- Marcos, N.T.; Pinho, S.; Grandela, C.; Cruz, A.; Samyn-Petit, B.; Harduin-Lepers, A.; Almeida, R.; Silva, F.; Morais, V.; Costa, J.; et al. Role of the human ST6GalNAc-I and ST6GalNAc-II in the synthesis of the cancer-associated sialyl-Tn antigen. Cancer Res. 2004, 64, 7050–7057. [Google Scholar] [CrossRef]
- Rajesh, C.; Radhakrishnan, P. The (Sialyl) Tn antigen: Contributions to immunosuppression in gastrointestinal cancers. Front. Oncol. 2022, 12, 1093496. [Google Scholar] [CrossRef]
- Wang, W.Y.; Cao, Y.X.; Zhou, X.; Wei, B.; Zhan, L.; Sun, S.Y. Stimulative role of ST6GALNAC1 in proliferation, migration and invasion of ovarian cancer stem cells via the Akt signaling pathway. Cancer Cell Int. 2019, 19, 86. [Google Scholar] [CrossRef]
- Kvorjak, M.; Ahmed, Y.; Miller, M.L.; Sriram, R.; Coronnello, C.; Hashash, J.G.; Hartman, D.J.; Telmer, C.A.; Miskov-Zivanov, N.; Finn, O.J.; et al. Cross-talk between Colon Cells and Macrophages Increases ST6GALNAC1 and MUC1-sTn Expression in Ulcerative Colitis and Colitis-Associated Colon Cancer. Cancer Immunol. Res. 2020, 8, 167–178. [Google Scholar] [CrossRef]
- Murugesan, G.; Correia, V.G.; Palma, A.S.; Chai, W.; Li, C.; Feizi, T.; Martin, E.; Laux, B.; Franz, A.; Fuchs, K.; et al. Siglec-15 recognition of sialoglycans on tumor cell lines can occur independently of sialyl Tn antigen expression. Glycobiology 2021, 31, 44–54. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Liu, L.N.; Flies, D.B.; Nie, X.; Toki, M.; Zhang, J.; Song, C.; Zarr, M.; Zhou, X.; et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat. Med. 2019, 25, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Murugaesu, N.; Iravani, M.; van Weverwijk, A.; Ivetic, A.; Johnson, D.A.; Antonopoulos, A.; Fearns, A.; Jamal-Hanjani, M.; Sims, D.; Fenwick, K.; et al. An in vivo functional screen identifies ST6GalNAc2 sialyltransferase as a breast cancer metastasis suppressor. Cancer Discov. 2014, 4, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, C.M.; Reginato, M.J. Sticking to sugars at the metastatic site: Sialyltransferase ST6GalNAc2 acts as a breast cancer metastasis suppressor. Cancer Discov. 2014, 4, 275–277. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Zhao, Y. ST6GalNAcII mediates tumor invasion through PI3K/Akt/NF-kappaB signaling pathway in follicular thyroid carcinoma. Oncol. Rep. 2016, 35, 2131–2140. [Google Scholar] [CrossRef] [PubMed]
- Reticker-Flynn, N.E.; Bhatia, S.N. Aberrant glycosylation promotes lung cancer metastasis through adhesion to galectins in the metastatic niche. Cancer Discov. 2015, 5, 168–181. [Google Scholar] [CrossRef]
- Adams, O.J.; Stanczak, M.A.; von Gunten, S.; Laubli, H. Targeting sialic acid-Siglec interactions to reverse immune suppression in cancer. Glycobiology 2018, 28, 640–647. [Google Scholar] [CrossRef]
- Dai, T.; Li, J.; Liang, R.B.; Yu, H.; Lu, X.; Wang, G. Identification and Experimental Validation of the Prognostic Significance and Immunological Correlation of Glycosylation-Related Signature and ST6GALNAC4 in Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2023, 10, 531–551. [Google Scholar] [CrossRef]
- Ikehara, Y.; Shimizu, N.; Kono, M.; Nishihara, S.; Nakanishi, H.; Kitamura, T.; Narimatsu, H.; Tsuji, S.; Tatematsu, M. A novel glycosyltransferase with a polyglutamine repeat; a new candidate for GD1alpha synthase (ST6GalNAc V)(1). FEBS Lett. 1999, 463, 92–96. [Google Scholar] [CrossRef]
- Okajima, T.; Fukumoto, S.; Ito, H.; Kiso, M.; Hirabayashi, Y.; Urano, T.; Furukawa, K. Molecular cloning of brain-specific GD1alpha synthase (ST6GalNAc V) containing CAG/Glutamine repeats. J. Biol. Chem. 1999, 274, 30557–30562. [Google Scholar] [CrossRef]
- Dai, J.; Li, Q.; Quan, J.; Webb, G.; Liu, J.; Gao, K. Construction of a lipid metabolism-related and immune-associated prognostic score for gastric cancer. BMC Med. Genom. 2023, 16, 93. [Google Scholar] [CrossRef]
- Kroes, R.A.; He, H.; Emmett, M.R.; Nilsson, C.L.; Leach, F.E., 3rd; Amster, I.J.; Marshall, A.G.; Moskal, J.R. Overexpression of ST6GalNAcV, a ganglioside-specific alpha2,6-sialyltransferase, inhibits glioma growth in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 12646–12651. [Google Scholar] [CrossRef]
- Miyazaki, K.; Ohmori, K.; Izawa, M.; Koike, T.; Kumamoto, K.; Furukawa, K.; Ando, T.; Kiso, M.; Yamaji, T.; Hashimoto, Y.; et al. Loss of disialyl Lewis(a), the ligand for lymphocyte inhibitory receptor sialic acid-binding immunoglobulin-like lectin-7 (Siglec-7) associated with increased sialyl Lewis(a) expression on human colon cancers. Cancer Res. 2004, 64, 4498–4505. [Google Scholar] [CrossRef]
- Yeh, S.C.; Wang, P.Y.; Lou, Y.W.; Khoo, K.H.; Hsiao, M.; Hsu, T.L.; Wong, C.H. Glycolipid GD3 and GD3 synthase are key drivers for glioblastoma stem cells and tumorigenicity. Proc. Natl. Acad. Sci. USA 2016, 113, 5592–5597. [Google Scholar] [CrossRef]
- Hao, J.; Zeltz, C.; Pintilie, M.; Li, Q.; Sakashita, S.; Wang, T.; Cabanero, M.; Martins-Filho, S.N.; Wang, D.Y.; Pasko, E.; et al. Characterization of Distinct Populations of Carcinoma-Associated Fibroblasts from Non-Small Cell Lung Carcinoma Reveals a Role for ST8SIA2 in Cancer Cell Invasion. Neoplasia 2019, 21, 482–493. [Google Scholar] [CrossRef]
- Gong, L.; Zhou, X.; Yang, J.; Jiang, Y.; Yang, H. Effects of the regulation of polysialyltransferase ST8SiaII on the invasiveness and metastasis of small cell lung cancer cells. Oncol. Rep. 2017, 37, 131–138. [Google Scholar] [CrossRef]
- Ma, X.; Dong, W.; Su, Z.; Zhao, L.; Miao, Y.; Li, N.; Zhou, H.; Jia, L. Functional roles of sialylation in breast cancer progression through miR-26a/26b targeting ST8SIA4. Cell Death Dis. 2016, 7, e2561. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, Y.; Jiang, L.; Miao, X.; Zhou, H.; Jia, L. Glycomic alterations are associated with multidrug resistance in human leukemia. Int. J. Biochem. Cell Biol. 2012, 44, 1244–1253. [Google Scholar] [CrossRef]
- Baeza-Kallee, N.; Berges, R.; Souberan, A.; Colin, C.; Denicolai, E.; Appay, R.; Tchoghandjian, A.; Figarella-Branger, D. Glycolipids Recognized by A2B5 Antibody Promote Proliferation, Migration, and Clonogenicity in Glioblastoma Cells. Cancers 2019, 11, 1267. [Google Scholar] [CrossRef]
- Penrose, H.M.; Cable, C.; Heller, S.; Ungerleider, N.; Nakhoul, H.; Baddoo, M.; Hartono, A.B.; Lee, S.B.; Burow, M.E.; Flemington, E.F.; et al. Loss of Forkhead Box O3 Facilitates Inflammatory Colon Cancer: Transcriptome Profiling of the Immune Landscape and Novel Targets. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 391–408. [Google Scholar] [CrossRef]
- Huang, R.; Zheng, Z.; Xian, S.; Zhang, J.; Jia, J.; Song, D.; Yan, P.; Yin, H.; Hu, P.; Zhu, X.; et al. Identification of prognostic and bone metastatic alternative splicing signatures in bladder cancer. Bioengineered 2021, 12, 5289–5304. [Google Scholar] [CrossRef]
- Friedman, D.J.; Crotts, S.B.; Shapiro, M.J.; Rajcula, M.; McCue, S.; Liu, X.; Khazaie, K.; Dong, H.; Shapiro, V.S. ST8Sia6 Promotes Tumor Growth in Mice by Inhibiting Immune Responses. Cancer Immunol. Res. 2021, 9, 952–966. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; He, L.; Yang, L.; Fang, X.; Peng, L. Potential Role of NEU1 in Hepatocellular Carcinoma: A Study Based on Comprehensive Bioinformatical Analysis. Front. Mol. Biosci. 2021, 8, 651525. [Google Scholar] [CrossRef] [PubMed]
- Qorri, B.; Harless, W.; Szewczuk, M.R. Novel Molecular Mechanism of Aspirin and Celecoxib Targeting Mammalian Neuraminidase-1 Impedes Epidermal Growth Factor Receptor Signaling Axis and Induces Apoptosis in Pancreatic Cancer Cells. Drug Des. Dev. Ther. 2020, 14, 4149–4167. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.R.; Zhang, L.P.; Huang, S.Y.; Zhu, Y.F.; Li, W.J.; Fang, S.Y.; Shen, L.; Gao, Y.L. Effects of sialidase NEU1 siRNA on proliferation, apoptosis, and invasion in human ovarian cancer. Mol. Cell. Biochem. 2016, 411, 213–219. [Google Scholar] [CrossRef]
- Peng, Q.; Gao, L.; Cheng, H.B.; Wang, J.S.; Wang, J. Sialidase NEU1 May Serve as a Potential Biomarker of Proliferation, Migration and Prognosis in Melanoma. World J. Oncol. 2022, 13, 222–234. [Google Scholar] [CrossRef]
- Garcia-Dominguez, D.J.; Hajji, N.; Lopez-Alemany, R.; Sanchez-Molina, S.; Figuerola-Bou, E.; Moron Civanto, F.J.; Rello-Varona, S.; Andres-Leon, E.; Benito, A.; Keun, H.C.; et al. Selective histone methyltransferase G9a inhibition reduces metastatic development of Ewing sarcoma through the epigenetic regulation of NEU1. Oncogene 2022, 41, 2638–2650. [Google Scholar] [CrossRef]
- Nath, S.; Mondal, S.; Butti, R.; Prasanna Gunasekaran, V.; Chatterjee, U.; Halder, A.; Kundu, G.C.; Mandal, C. Desialylation of Sonic-Hedgehog by Neu2 Inhibits Its Association with Patched1 Reducing Stemness-Like Properties in Pancreatic Cancer Sphere-forming Cells. Cells 2020, 9, 1512. [Google Scholar] [CrossRef]
- Nath, S.; Mandal, C.; Chatterjee, U.; Mandal, C. Association of cytosolic sialidase Neu2 with plasma membrane enhances Fas-mediated apoptosis by impairing PI3K-Akt/mTOR-mediated pathway in pancreatic cancer cells. Cell Death Dis. 2018, 9, 210. [Google Scholar] [CrossRef]
- Satyavarapu, E.M.; Nath, S.; Mandal, C. Desialylation of Atg5 by sialidase (Neu2) enhances autophagosome formation to induce anchorage-dependent cell death in ovarian cancer cells. Cell Death Discov. 2021, 7, 26. [Google Scholar] [CrossRef]
- Iioka, H.; Saito, K.; Kondo, E. Crumbs3 regulates the expression of glycosphingolipids on the plasma membrane to promote colon cancer cell migration. Biochem. Biophys. Res. Commun. 2019, 519, 287–293. [Google Scholar] [CrossRef]
- Zhang, X.; Dou, P.; Akhtar, M.L.; Liu, F.; Hu, X.; Yang, L.; Yang, D.; Zhang, X.; Li, Y.; Qiao, S.; et al. NEU4 inhibits motility of HCC cells by cleaving sialic acids on CD44. Oncogene 2021, 40, 5427–5440. [Google Scholar] [CrossRef]
- Shiozaki, K.; Yamaguchi, K.; Takahashi, K.; Moriya, S.; Miyagi, T. Regulation of sialyl Lewis antigen expression in colon cancer cells by sialidase NEU4. J. Biol. Chem. 2011, 286, 21052–21061. [Google Scholar] [CrossRef]
- Grzeski, M.; Taube, E.T.; Braicu, E.I.; Sehouli, J.; Blanchard, V.; Klein, O. In Situ N-Glycosylation Signatures of Epithelial Ovarian Cancer Tissue as Defined by MALDI Mass Spectrometry Imaging. Cancers 2022, 14, 1021. [Google Scholar] [CrossRef]
- McDowell, C.T.; Klamer, Z.; Hall, J.; West, C.A.; Wisniewski, L.; Powers, T.W.; Angel, P.M.; Mehta, A.S.; Lewin, D.N.; Haab, B.B.; et al. Imaging Mass Spectrometry and Lectin Analysis of N-Linked Glycans in Carbohydrate Antigen-Defined Pancreatic Cancer Tissues. Mol. Cell. Proteom. 2021, 20, 100012. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, Q.; Xu, Y.; Guo, L.; Lai, Z.; Li, Z. Graphitic carbon nitride quantum dots as analytical probe for viewing sialic acid on the surface of cells and tissues. Anal. Chim. Acta 2020, 1095, 204–211. [Google Scholar] [CrossRef]
- Greco, B.; Malacarne, V.; De Girardi, F.; Scotti, G.M.; Manfredi, F.; Angelino, E.; Sirini, C.; Camisa, B.; Falcone, L.; Moresco, M.A.; et al. Disrupting N-glycan expression on tumor cells boosts chimeric antigen receptor T cell efficacy against solid malignancies. Sci. Transl. Med. 2022, 14, eabg3072. [Google Scholar] [CrossRef]
- Ghasempour, S.; Freeman, S.A. The glycocalyx and immune evasion in cancer. FEBS J. 2023, 290, 55–65. [Google Scholar] [CrossRef]
- Bull, C.; den Brok, M.H.; Adema, G.J. Sweet escape: Sialic acids in tumor immune evasion. Biochim. Biophys. Acta 2014, 1846, 238–246. [Google Scholar] [CrossRef]
- Banfer, S.; Jacob, R. Galectins. Curr. Biol. 2022, 32, R406–R408. [Google Scholar] [CrossRef]
- Gilson, R.C.; Gunasinghe, S.D.; Johannes, L.; Gaus, K. Galectin-3 modulation of T-cell activation: Mechanisms of membrane remodelling. Prog. Lipid Res. 2019, 76, 101010. [Google Scholar] [CrossRef]
- Rabinovich, G.A.; Toscano, M.A. Turning ‘sweet’ on immunity: Galectin-glycan interactions in immune tolerance and inflammation. Nat. Rev. Immunol. 2009, 9, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.T.; Stowell, S.R. The role of galectins in immunity and infection. Nat. Rev. Immunol. 2023, 1–16. [Google Scholar] [CrossRef]
- Zhuo, Y.; Bellis, S.L. Emerging role of alpha2,6-sialic acid as a negative regulator of galectin binding and function. J. Biol. Chem. 2011, 286, 5935–5941. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.; Azevedo, C.M.; Silva, M.C.; Faria, G.; Dantas, C.S.; Vicente, M.M.; Pinho, S.S. Glycans as shapers of tumour microenvironment: A sweet driver of T-cell-mediated anti-tumour immune response. Immunology 2023, 168, 217–232. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Guo, S.; Guan, X.; Kang, Y.; Liu, J.; Yang, X. Immunological Classification of Tumor Types and Advances in Precision Combination Immunotherapy. Front. Immunol. 2022, 13, 790113. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef]
- van de Wall, S.; Santegoets, K.C.M.; van Houtum, E.J.H.; Bull, C.; Adema, G.J. Sialoglycans and Siglecs Can Shape the Tumor Immune Microenvironment. Trends Immunol. 2020, 41, 274–285. [Google Scholar] [CrossRef]
- van Houtum, E.J.H.; Bull, C.; Cornelissen, L.A.M.; Adema, G.J. Siglec Signaling in the Tumor Microenvironment. Front. Immunol. 2021, 12, 790317. [Google Scholar] [CrossRef]
- Crocker, P.R.; Paulson, J.C.; Varki, A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007, 7, 255–266. [Google Scholar] [CrossRef]
- Siddiqui, S.S.; Vaill, M.; Do, R.; Khan, N.; Verhagen, A.L.; Zhang, W.; Lenz, H.J.; Johnson-Pais, T.L.; Leach, R.J.; Fraser, G.; et al. Human-specific polymorphic pseudogenization of SIGLEC12 protects against advanced cancer progression. FASEB Bioadv. 2021, 3, 69–82. [Google Scholar] [CrossRef]
- Bull, C.; Nason, R.; Sun, L.; Van Coillie, J.; Madriz Sorensen, D.; Moons, S.J.; Yang, Z.; Arbitman, S.; Fernandes, S.M.; Furukawa, S.; et al. Probing the binding specificities of human Siglecs by cell-based glycan arrays. Proc. Natl. Acad. Sci. USA 2021, 118, e2026102118. [Google Scholar] [CrossRef]
- Jandus, C.; Boligan, K.F.; Chijioke, O.; Liu, H.; Dahlhaus, M.; Demoulins, T.; Schneider, C.; Wehrli, M.; Hunger, R.E.; Baerlocher, G.M.; et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J. Clin. Investig. 2014, 124, 1810–1820. [Google Scholar] [CrossRef]
- Shao, J.Y.; Yin, W.W.; Zhang, Q.F.; Liu, Q.; Peng, M.L.; Hu, H.D.; Hu, P.; Ren, H.; Zhang, D.Z. Siglec-7 Defines a Highly Functional Natural Killer Cell Subset and Inhibits Cell-Mediated Activities. Scand. J. Immunol. 2016, 84, 182–190. [Google Scholar] [CrossRef]
- Wisnovsky, S.; Mockl, L.; Malaker, S.A.; Pedram, K.; Hess, G.T.; Riley, N.M.; Gray, M.A.; Smith, B.A.H.; Bassik, M.C.; Moerner, W.E.; et al. Genome-wide CRISPR screens reveal a specific ligand for the glycan-binding immune checkpoint receptor Siglec-7. Proc. Natl. Acad. Sci. USA 2021, 118, e2015024118. [Google Scholar] [CrossRef]
- Yoshimura, A.; Asahina, Y.; Chang, L.Y.; Angata, T.; Tanaka, H.; Kitajima, K.; Sato, C. Identification and functional characterization of a Siglec-7 counter-receptor on K562 cells. J. Biol. Chem. 2021, 296, 100477. [Google Scholar] [CrossRef]
- Alphey, M.S.; Attrill, H.; Crocker, P.R.; van Aalten, D.M. High resolution crystal structures of Siglec-7. Insights into ligand specificity in the Siglec family. J. Biol. Chem. 2003, 278, 3372–3377. [Google Scholar] [CrossRef]
- Zhang, J.Q.; Nicoll, G.; Jones, C.; Crocker, P.R. Siglec-9, a novel sialic acid binding member of the immunoglobulin superfamily expressed broadly on human blood leukocytes. J. Biol. Chem. 2000, 275, 22121–22126. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Ito, A.; Withers, D.A.; Taima, T.; Kakoi, N.; Saito, S.; Arai, Y. Ganglioside DSGb5, preferred ligand for Siglec-7, inhibits NK cell cytotoxicity against renal cell carcinoma cells. Glycobiology 2010, 20, 1373–1379. [Google Scholar] [CrossRef]
- Nicoll, G.; Avril, T.; Lock, K.; Furukawa, K.; Bovin, N.; Crocker, P.R. Ganglioside GD3 expression on target cells can modulate NK cell cytotoxicity via siglec-7-dependent and -independent mechanisms. Eur. J. Immunol. 2003, 33, 1642–1648. [Google Scholar] [CrossRef]
- Daly, J.; Sarkar, S.; Natoni, A.; Stark, J.C.; Riley, N.M.; Bertozzi, C.R.; Carlsten, M.; O’Dwyer, M.E. Targeting hypersialylation in multiple myeloma represents a novel approach to enhance NK cell-mediated tumor responses. Blood Adv. 2022, 6, 3352–3366. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.Y.; Liang, S.Y.; Lu, S.C.; Tseng, H.C.; Tsai, H.Y.; Tang, C.J.; Sugata, M.; Chen, Y.J.; Chen, Y.J.; Wu, S.J.; et al. Molecular Basis and Role of Siglec-7 Ligand Expression on Chronic Lymphocytic Leukemia B Cells. Front. Immunol. 2022, 13, 840388. [Google Scholar] [CrossRef] [PubMed]
- Belisle, J.A.; Horibata, S.; Jennifer, G.A.; Petrie, S.; Kapur, A.; Andre, S.; Gabius, H.J.; Rancourt, C.; Connor, J.; Paulson, J.C.; et al. Identification of Siglec-9 as the receptor for MUC16 on human NK cells, B cells, and monocytes. Mol. Cancer 2010, 9, 118. [Google Scholar] [CrossRef]
- Smith, B.A.H.; Deutzmann, A.; Correa, K.M.; Delaveris, C.S.; Dhanasekaran, R.; Dove, C.G.; Sullivan, D.K.; Wisnovsky, S.; Stark, J.C.; Pluvinage, J.V.; et al. MYC-driven synthesis of Siglec ligands is a glycoimmune checkpoint. Proc. Natl. Acad. Sci. USA 2023, 120, e2215376120. [Google Scholar] [CrossRef] [PubMed]
- Barkal, A.A.; Brewer, R.E.; Markovic, M.; Kowarsky, M.; Barkal, S.A.; Zaro, B.W.; Krishnan, V.; Hatakeyama, J.; Dorigo, O.; Barkal, L.J.; et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 2019, 572, 392–396. [Google Scholar] [CrossRef]
- Wang, J.; Manni, M.; Barenwaldt, A.; Wieboldt, R.; Kirchhammer, N.; Ivanek, R.; Stanczak, M.; Zippelius, A.; Konig, D.; Rodrigues Manutano, N.; et al. Siglec Receptors Modulate Dendritic Cell Activation and Antigen Presentation to T Cells in Cancer. Front. Cell Dev. Biol. 2022, 10, 828916. [Google Scholar] [CrossRef]
- Haas, Q.; Boligan, K.F.; Jandus, C.; Schneider, C.; Simillion, C.; Stanczak, M.A.; Haubitz, M.; Seyed Jafari, S.M.; Zippelius, A.; Baerlocher, G.M.; et al. Siglec-9 Regulates an Effector Memory CD8(+) T-cell Subset That Congregates in the Melanoma Tumor Microenvironment. Cancer Immunol. Res. 2019, 7, 707–718. [Google Scholar] [CrossRef]
- Vuchkovska, A.; Glanville, D.G.; Scurti, G.M.; Nishimura, M.I.; White, P.; Ulijasz, A.T.; Iwashima, M. Siglec-5 is an inhibitory immune checkpoint molecule for human T cells. Immunology 2022, 166, 238–248. [Google Scholar] [CrossRef]
- Jiang, K.Y.; Qi, L.L.; Kang, F.B.; Wang, L. The intriguing roles of Siglec family members in the tumor microenvironment. Biomark. Res. 2022, 10, 22. [Google Scholar] [CrossRef]
- Yamaguchi, S.I.; Xie, Q.; Ito, F.; Terao, K.; Kato, Y.; Kuroiwa, M.; Omori, S.; Taniura, H.; Kinoshita, K.; Takahashi, T.; et al. Carbon nanotube recognition by human Siglec-14 provokes inflammation. Nat. Nanotechnol. 2023. [Google Scholar] [CrossRef]
- Thiesler, H.; Gretenkort, L.; Hoffmeister, L.; Albers, I.; Ohlmeier, L.; Rockle, I.; Verhagen, A.; Banan, R.; Kopcke, N.; Kronke, N.; et al. Proinflammatory macrophage activation by the polysialic acid-Siglec-16 axis is linked to increased survival of glioblastoma patients. Clin. Cancer Res. 2023, OF1–OF14. [Google Scholar] [CrossRef]
- Bull, C.; Boltje, T.J.; Wassink, M.; de Graaf, A.M.; van Delft, F.L.; den Brok, M.H.; Adema, G.J. Targeting aberrant sialylation in cancer cells using a fluorinated sialic acid analog impairs adhesion, migration, and in vivo tumor growth. Mol. Cancer Ther. 2013, 12, 1935–1946. [Google Scholar] [CrossRef] [PubMed]
- Hinderlich, S.; Stasche, R.; Zeitler, R.; Reutter, W. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver. Purification and characterization of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. J. Biol. Chem. 1997, 272, 24313–24318. [Google Scholar] [CrossRef] [PubMed]
- Miro, L.; Lopez, J.; Guerrero, P.E.; Martinez-Bosch, N.; Manero-Ruperez, N.; Moreno, M.; Ortiz, M.R.; Llop, E.; Navarro, P.; Peracaula, R. Sialyltransferase Inhibitor Ac(5)3F(ax)Neu5Ac Reverts the Malignant Phenotype of Pancreatic Cancer Cells, and Reduces Tumor Volume and Favors T-Cell Infiltrates in Mice. Cancers 2022, 14, 6133. [Google Scholar] [CrossRef] [PubMed]
- Macauley, M.S.; Arlian, B.M.; Rillahan, C.D.; Pang, P.C.; Bortell, N.; Marcondes, M.C.; Haslam, S.M.; Dell, A.; Paulson, J.C. Systemic blockade of sialylation in mice with a global inhibitor of sialyltransferases. J. Biol. Chem. 2014, 289, 35149–35158. [Google Scholar] [CrossRef]
- Bull, C.; Boltje, T.J.; van Dinther, E.A.; Peters, T.; de Graaf, A.M.; Leusen, J.H.; Kreutz, M.; Figdor, C.G.; den Brok, M.H.; Adema, G.J. Targeted delivery of a sialic acid-blocking glycomimetic to cancer cells inhibits metastatic spread. ACS Nano 2015, 9, 733–745. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, C.H.; Mo, J.; Zheng, X.J.; Chen, Y.F.; Yang, A.Q.; Zhang, Y.H.; Wang, P.Y.; Yuan, X.; Ye, X.S. Self-Assembled Core-Shell Nanoscale Coordination Polymer Nanoparticles Carrying a Sialyltransferase Inhibitor for Cancer Metastasis Inhibition. ACS Appl. Mater. Interfaces 2023, 15, 7713–7724. [Google Scholar] [CrossRef]
- Gray, M.A.; Stanczak, M.A.; Mantuano, N.R.; Xiao, H.; Pijnenborg, J.F.A.; Malaker, S.A.; Miller, C.L.; Weidenbacher, P.A.; Tanzo, J.T.; Ahn, G.; et al. Targeted glycan degradation potentiates the anticancer immune response in vivo. Nat. Chem. Biol. 2020, 16, 1376–1384. [Google Scholar] [CrossRef]
- King, T.; Posey, A.D. Co-Expression of an Engineered Cell-Surface Sialidase by Cart Cells Improves Anti-Cancer Activity of Nk Cells in Solid Tumors. Cytotherapy 2019, 21, S27. [Google Scholar] [CrossRef]
- Gutierrez, M.; Hamid, O.; Shum, E.; Wise, D.R.; Balar, A.V.; Weber, J.S.; LoRusso, P.; Shafi, S.; Rimm, D.L.; Tolcher, A.W.; et al. Trial in progress: A phase I/II, open-label, dose-escalation, safety and tolerability study of NC318 in subjects with advanced or metastatic solid tumors. J. Clin. Oncol. 2020, 38, TPS3166. [Google Scholar] [CrossRef]
- Shum, E.; Myint, H.; Shaik, J.; Zhou, Q.; Barbu, E.; Morawski, A.; Abukharma, H.; Liu, L.; Nelson, M.; Zeidan, S.; et al. 490 Clinical benefit through Siglec-15 targeting with NC318 antibody in subjects with Siglec-15 positive advanced solid tumors. J. ImmunoTherapy Cancer 2021, 9, A520–A521. [Google Scholar] [CrossRef]
- Xiao, X.; Peng, Y.; Wang, Z.; Zhang, L.; Yang, T.; Sun, Y.; Chen, Y.; Zhang, W.; Chang, X.; Huang, W.; et al. A novel immune checkpoint siglec-15 antibody inhibits LUAD by modulating mphi polarization in TME. Pharmacol. Res. 2022, 181, 106269. [Google Scholar] [CrossRef]
- Choi, H.; Ho, M.; Adeniji, O.S.; Giron, L.; Bordoloi, D.; Kulkarni, A.J.; Puchalt, A.P.; Abdel-Mohsen, M.; Muthumani, K. Development of Siglec-9 Blocking Antibody to Enhance Anti-Tumor Immunity. Front. Oncol. 2021, 11, 778989. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Chi, K.; Zhang, C.; Liu, Q.; Yang, G. Sialylation: A Cloak for Tumors to Trick the Immune System in the Microenvironment. Biology 2023, 12, 832. https://doi.org/10.3390/biology12060832
Zhou X, Chi K, Zhang C, Liu Q, Yang G. Sialylation: A Cloak for Tumors to Trick the Immune System in the Microenvironment. Biology. 2023; 12(6):832. https://doi.org/10.3390/biology12060832
Chicago/Turabian StyleZhou, Xiaoman, Kaijun Chi, Chairui Zhang, Quan Liu, and Ganglong Yang. 2023. "Sialylation: A Cloak for Tumors to Trick the Immune System in the Microenvironment" Biology 12, no. 6: 832. https://doi.org/10.3390/biology12060832
APA StyleZhou, X., Chi, K., Zhang, C., Liu, Q., & Yang, G. (2023). Sialylation: A Cloak for Tumors to Trick the Immune System in the Microenvironment. Biology, 12(6), 832. https://doi.org/10.3390/biology12060832





