IntiCom-DB: A Manually Curated Database of Inter-Tissue Communication Molecules and Their Communication Routes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature Data Curation and Annotation
2.2. Proteomic and Transcriptomic Profiles and Tissue Specificity Score Calculation
2.3. Cross-Dataset Mapping
2.4. Database Web Interface Construction
3. Results
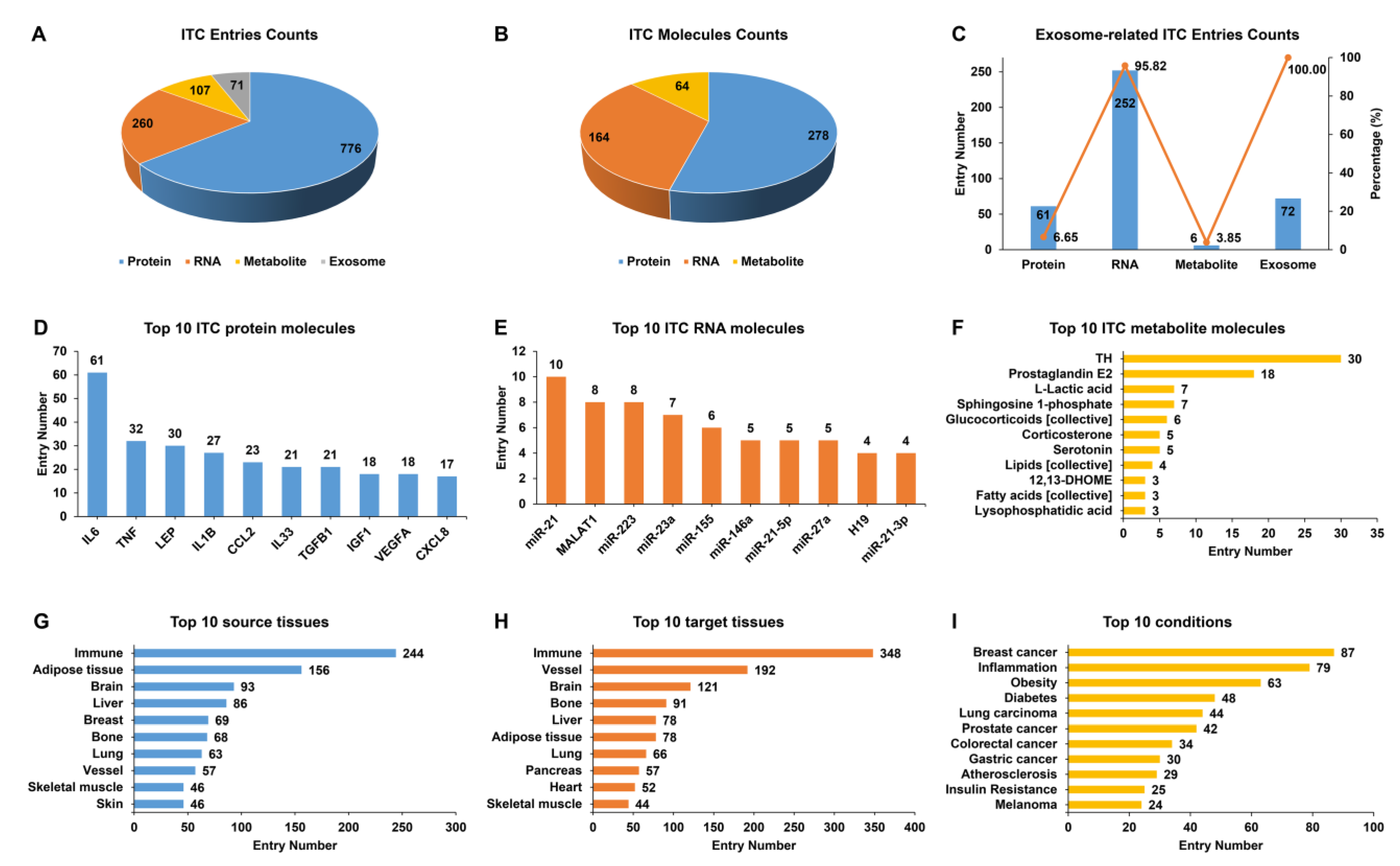
3.1. IntiCom-DB Dataset Overview
3.2. IntiCom-DB Web Interface
3.3. Common Biological Characteristics of Communication Proteins Revealed by the Analysis of IntiCom-DB Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Long, Q.; Argmann, C.; Houten, S.M.; Huang, T.; Peng, S.; Zhao, Y.; Tu, Z.; Consortium, G.T.; Zhu, J. Inter-tissue coexpression network analysis reveals DPP4 as an important gene in heart to blood communication. Genome Med. 2016, 8, 15. [Google Scholar] [CrossRef]
- Chevrier, N. Decoding the Body Language of Immunity: Tackling the Immune System at the Organism Level. Curr. Opin. Syst. Biol. 2019, 18, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Droujinine, I.A.; Perrimon, N. Defining the interorgan communication network: Systemic coordination of organismal cellular processes under homeostasis and localized stress. Front. Cell. Infect. Microbiol. 2013, 3, 82. [Google Scholar] [CrossRef]
- Castillo-Armengol, J.; Fajas, L.; Lopez-Mejia, I.C. Inter-organ communication: A gatekeeper for metabolic health. EMBO Rep. 2019, 20, e47903. [Google Scholar] [CrossRef]
- Droujinine, I.A.; Perrimon, N. Interorgan Communication Pathways in Physiology: Focus on Drosophila. Annu. Rev. Genet. 2016, 50, 539–570. [Google Scholar] [CrossRef]
- Seldin, M.M.; Koplev, S.; Rajbhandari, P.; Vergnes, L.; Rosenberg, G.M.; Meng, Y.; Pan, C.; Phuong, T.M.N.; Gharakhanian, R.; Che, N.; et al. A Strategy for Discovery of Endocrine Interactions with Application to Whole-Body Metabolism. Cell Metab. 2018, 27, 1138–1155.e6. [Google Scholar] [CrossRef]
- Seldin, M.M.; Lusis, A.J. Systems-based approaches for investigation of inter-tissue communication[S]. J. Lipid Res. 2019, 60, 450–455. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Kim, K.E.; Park, I.; Kim, J.; Kang, M.G.; Choi, W.G.; Shin, H.; Kim, J.S.; Rhee, H.W.; Suh, J.M. Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice. Nat. Commun. 2021, 12, 5204. [Google Scholar] [CrossRef]
- Williams, M.J. JJR Macleod: The co-discoverer of insulin. Proc. R. Coll. Physicians Edinb. 1993, 23, 1–125. [Google Scholar]
- Fasshauer, M.; Bluher, M. Adipokines in health and disease. Trends Pharmacol. Sci. 2015, 36, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Giudice, J.; Taylor, J.M. Muscle as a paracrine and endocrine organ. Curr. Opin. Pharmacol. 2017, 34, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Meex, R.C.R.; Watt, M.J. Hepatokines: Linking nonalcoholic fatty liver disease and insulin resistance. Nat. Rev. Endocrinol. 2017, 13, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Ahima, R.S.; Prabakaran, D.; Mantzoros, C.; Qu, D.; Lowell, B.; Maratos-Flier, E.; Flier, J.S. Role of leptin in the neuroendocrine response to fasting. Nature 1996, 382, 250–252. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, L.A.; Dembski, M.; Weng, X.; Deng, N.; Culpepper, J.; Devos, R.; Richards, G.J.; Campfield, L.A.; Clark, F.T.; Deeds, J.; et al. Identification and expression cloning of a leptin receptor, OB-R. Cell 1995, 83, 1263–1271. [Google Scholar] [CrossRef] [PubMed]
- Bostrom, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Bostrom, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef]
- Lu, K.; Shi, T.S.; Shen, S.Y.; Shi, Y.; Gao, H.L.; Wu, J.; Lu, X.; Gao, X.; Ju, H.X.; Wang, W.; et al. Defects in a liver-bone axis contribute to hepatic osteodystrophy disease progression. Cell Metab. 2022, 34, 441–457.e7. [Google Scholar] [CrossRef]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Yu, Y.; Du, H.; Wei, S.; Feng, L.; Li, J.; Yao, F.; Zhang, M.; Hatch, G.M.; Chen, L. Adipocyte-Derived Exosomal MiR-27a Induces Insulin Resistance in Skeletal Muscle Through Repression of PPARgamma. Theranostics 2018, 8, 2171–2188. [Google Scholar] [CrossRef]
- Sun, L.L.; Duan, M.J.; Ma, J.C.; Xu, L.; Mao, M.; Biddyut, D.; Wang, Q.; Yang, C.; Zhang, S.; Xu, Y.; et al. Myocardial infarction-induced hippocampal microtubule damage by cardiac originating microRNA-1 in mice. J. Mol. Cell. Cardiol. 2018, 120, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Duan, M.J.; Yan, M.L.; Wang, Q.; Mao, M.; Su, D.; Sun, L.L.; Li, K.X.; Qu, Y.; Sun, Q.; Zhang, X.Y.; et al. Overexpression of miR-1 in the heart attenuates hippocampal synaptic vesicle exocytosis by the posttranscriptional regulation of SNAP-25 through the transportation of exosomes. Cell Commun. Signal. 2018, 16, 91. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Vijayakumar, A.; Kahn, B.B. Metabolites as regulators of insulin sensitivity and metabolism. Nat. Rev. Mol. Cell Biol. 2018, 19, 654–672. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.; Hui, S.; Zeng, X.; Cowan, A.J.; Wang, L.; Chen, L.; Morscher, R.J.; Reyes, J.; Frezza, C.; Hwang, H.Y.; et al. Metabolite Exchange between Mammalian Organs Quantified in Pigs. Cell Metab. 2019, 30, 594–606.e3. [Google Scholar] [CrossRef] [PubMed]
- Priest, C.; Tontonoz, P. Inter-organ cross-talk in metabolic syndrome. Nat. Metab. 2019, 1, 1177–1188. [Google Scholar] [CrossRef]
- Cori, C.F.; Cori, G.T. Carbohydrate metabolism. Annu. Rev. Biochem. 1946, 15, 193–218. [Google Scholar] [CrossRef]
- Inagaki, T.; Choi, M.; Moschetta, A.; Peng, L.; Cummins, C.L.; McDonald, J.G.; Luo, G.; Jones, S.A.; Goodwin, B.; Richardson, J.A.; et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005, 2, 217–225. [Google Scholar] [CrossRef]
- Simcox, J.; Geoghegan, G.; Maschek, J.A.; Bensard, C.L.; Pasquali, M.; Miao, R.; Lee, S.; Jiang, L.; Huck, I.; Kershaw, E.E.; et al. Global Analysis of Plasma Lipids Identifies Liver-Derived Acylcarnitines as a Fuel Source for Brown Fat Thermogenesis. Cell Metab. 2017, 26, 509–522.e6. [Google Scholar] [CrossRef]
- Gyorgy, B.; Szabo, T.G.; Pasztoi, M.; Pal, Z.; Misjak, P.; Aradi, B.; Laszlo, V.; Pallinger, E.; Pap, E.; Kittel, A.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef]
- Gao, H.; Zhang, L.; Wang, Z.; Yan, K.; Zhao, L.; Xiao, W. Research Progress on Transorgan Regulation of the Cardiovascular and Motor System through Cardiogenic Exosomes. Int. J. Mol. Sci. 2022, 23, 5765. [Google Scholar] [CrossRef]
- Fevrier, B.; Raposo, G. Exosomes: Endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004, 16, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Bucan, V.; Vaslaitis, D.; Peck, C.T.; Strauss, S.; Vogt, P.M.; Radtke, C. Effect of Exosomes from Rat Adipose-Derived Mesenchymal Stem Cells on Neurite Outgrowth and Sciatic Nerve Regeneration After Crush Injury. Mol. Neurobiol. 2019, 56, 1812–1824. [Google Scholar] [CrossRef] [PubMed]
- Kozawa, S.; Ueda, R.; Urayama, K.; Sagawa, F.; Endo, S.; Shiizaki, K.; Kurosu, H.; Maria de Almeida, G.; Hasan, S.M.; Nakazato, K.; et al. The Body-wide Transcriptome Landscape of Disease Models. iScience 2018, 2, 238–268. [Google Scholar] [CrossRef] [PubMed]
- Argiles, J.M.; Stemmler, B.; Lopez-Soriano, F.J.; Busquets, S. Inter-tissue communication in cancer cachexia. Nat. Rev. Endocrinol. 2018, 15, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Ochoa, E.; Froldi, F.; Cheng, L.Y. Interorgan communication in development and cancer. Wiley Interdiscip. Rev. Dev. Biol. 2021, 10, e394. [Google Scholar] [CrossRef] [PubMed]
- Kir, S.; White, J.P.; Kleiner, S.; Kazak, L.; Cohen, P.; Baracos, V.E.; Spiegelman, B.M. Tumour-derived PTH-related protein triggers adipose tissue browning and cancer cachexia. Nature 2014, 513, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.M.; Li, Y.F.; Luo, J.; Wang, J.Q.; Wei, J.; Wang, J.Q.; Xiao, T.; Xie, C.; Hong, J.; Ning, G.; et al. Gpnmb secreted from liver promotes lipogenesis in white adipose tissue and aggravates obesity and insulin resistance. Nat. Metab. 2019, 1, 570–583. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Li, Y.; Wang, X.Y.; Zhang, D.; Zhang, H.; Wu, Q.; He, Y.Q.; Wang, J.Y.; Zhang, L.; Xia, H.; et al. Circulating miR-130b mediates metabolic crosstalk between fat and muscle in overweight/obesity. Diabetologia 2013, 56, 2275–2285. [Google Scholar] [CrossRef]
- Liu, T.; Sun, Y.C.; Cheng, P.; Shao, H.G. Adipose tissue macrophage-derived exosomal miR-29a regulates obesity-associated insulin resistance. Biochem. Biophys. Res. Commun. 2019, 515, 352–358. [Google Scholar] [CrossRef]
- Xu, H.; Du, X.; Xu, J.; Zhang, Y.; Tian, Y.; Liu, G.; Wang, X.; Ma, M.; Du, W.; Liu, Y.; et al. Pancreatic beta cell microRNA-26a alleviates type 2 diabetes by improving peripheral insulin sensitivity and preserving beta cell function. PLoS Biol. 2020, 18, e3000603. [Google Scholar] [CrossRef]
- Okada-Iwabu, M.; Yamauchi, T.; Iwabu, M.; Honma, T.; Hamagami, K.; Matsuda, K.; Yamaguchi, M.; Tanabe, H.; Kimura-Someya, T.; Shirouzu, M.; et al. A small-molecule AdipoR agonist for type 2 diabetes and short life in obesity. Nature 2013, 503, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Pan, X.; Wu, F.; Ye, D.; Zhang, Y.; Wang, Y.; Jin, L.; Lian, Q.; Huang, Y.; Ding, H.; et al. Fibroblast growth factor 21 prevents atherosclerosis by suppression of hepatic sterol regulatory element-binding protein-2 and induction of adiponectin in mice. Circulation 2015, 131, 1861–1871. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Dong, F.; Shi, G.; Lao, X.; Zheng, H. HORDB a comprehensive database of peptide hormones. Sci. Data 2022, 9, 187. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, M.; Lin, S.; Jian, R.; Li, X.; Chan, J.; Dong, G.; Fang, H.; Robinson, A.E.; Consortium, G.T.; et al. A Quantitative Proteome Map of the Human Body. Cell 2020, 183, 269–283.e19. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jiang, L.; Snyder, M.P. AdaTiSS: A Novel Data-Adaptive Robust Method for Identifying Tissue Specificity Scores. Bioinformatics 2021, 37, 4469–4476. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Xiao, L.; He, Y.; Peng, F.; Yang, J.; Yuan, C. Endometrial Cancer Cells Promote M2-Like Macrophage Polarization by Delivering Exosomal miRNA-21 under Hypoxia Condition. J. Immunol. Res. 2020, 2020, 9731049. [Google Scholar] [CrossRef]
- Brent, G.A. Mechanisms of thyroid hormone action. J. Clin. Investig. 2012, 122, 3035–3043. [Google Scholar] [CrossRef]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Abugessaisa, I., Kasukawa, T., Eds.; Springer Nature Singapore: Singapore, 2021; pp. 27–56. [Google Scholar]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Li, B.; Hong, J.; Hong, M.; Wang, Y.; Yu, T.; Zang, S.; Wu, Q. piRNA-823 delivered by multiple myeloma-derived extracellular vesicles promoted tumorigenesis through re-educating endothelial cells in the tumor environment. Oncogene 2019, 38, 5227–5238. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, J.; Li, Y.; Song, T.; Wu, Y.; Fang, S.; Bu, D.; Li, H.; Sun, L.; Pei, D.; et al. NONCODEV6: An updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 2021, 49, D165–D171. [Google Scholar] [CrossRef] [PubMed]
- Glazar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shi, Y.; Zhou, H.; Zhang, P.; Song, T.; Ying, Z.; Yu, H.; Li, Y.; Zhao, Y.; Zeng, X.; et al. piRBase: Integrating piRNA annotation in all aspects. Nucleic Acids Res. 2022, 50, D265–D272. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yang, L.; Wang, X.; Xing, H.; Zhao, H.; Xing, Y.; Zhou, F.; Wang, C.; Song, G.; Ma, H. Exploring the Multi-Tissue Crosstalk Relevant to Insulin Resistance Through Network-Based Analysis. Front. Endocrinol. 2021, 12, 756785. [Google Scholar] [CrossRef]
- Efremova, M.; Vento-Tormo, M.; Teichmann, S.A.; Vento-Tormo, R. CellPhoneDB: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020, 15, 1484–1506. [Google Scholar] [CrossRef]
- Browaeys, R.; Saelens, W.; Saeys, Y. NicheNet: Modeling intercellular communication by linking ligands to target genes. Nat. Methods 2020, 17, 159–162. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Uhlen, M.; Karlsson, M.J.; Hober, A.; Svensson, A.S.; Scheffel, J.; Kotol, D.; Zhong, W.; Tebani, A.; Strandberg, L.; Edfors, F.; et al. The human secretome. Sci. Signal. 2019, 12, eaaz0274. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borras, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Xie, G.Y.; Miao, Y.R.; Xia, M.; Wang, Y.; Lei, Q.; Zhang, Q.; Guo, A.Y. EVAtlas: A comprehensive database for ncRNA expression in human extracellular vesicles. Nucleic Acids Res. 2022, 50, D111–D117. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.K.; Lee, J.; Kim, S.R.; Choi, D.S.; Yoon, Y.J.; Kim, J.H.; Go, G.; Nhung, D.; Hong, K.; Jang, S.C.; et al. EVpedia: A community web portal for extracellular vesicles research. Bioinformatics 2015, 31, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Khor, E.C.; Baldock, P. The NPY system and its neural and neuroendocrine regulation of bone. Curr. Osteoporos. Rep. 2012, 10, 160–168. [Google Scholar] [CrossRef]


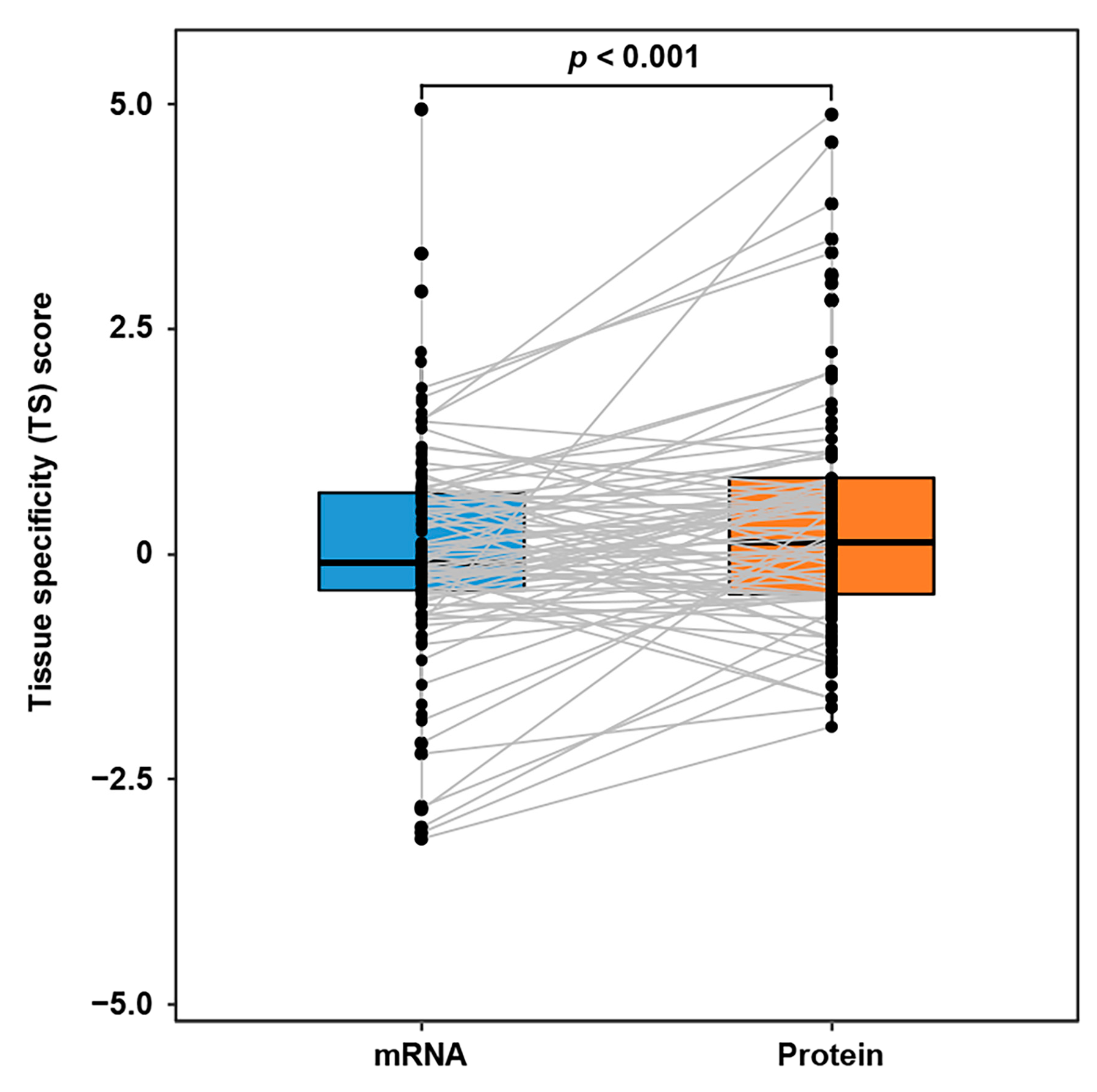
| Molecule * | Type | Tissue * | Statistic | p-Value * |
|---|---|---|---|---|
| ITC molecules | mRNA | Target tissues | 8179.5 | 1.01 × 10−2 |
| ITC molecules | Protein | Target tissues | 6290.5 | 6.70 × 10−3 |
| ITC molecules | mRNA | Source tissues | 7257.5 | 2.15 × 10−4 |
| ITC molecules | Protein | Source tissues | 5417.0 | 5.97 × 10−3 |
| ITC molecular partners | mRNA | Target tissues | 25,978.0 | 2.06 × 10−2 |
| ITC molecular partners | Protein | Target tissues | 29,329.0 | 3.54 × 10−6 |
| ITC molecular partners | mRNA | Source tissues | 21,192.5 | 9.50 × 10−4 |
| ITC molecular partners | Protein | Source tissues | 21,352.5 | 1.50 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, C.; Zhou, Y.; Han, Y.; Yi, J.; Pang, H.; Zheng, R.; Zhou, Y. IntiCom-DB: A Manually Curated Database of Inter-Tissue Communication Molecules and Their Communication Routes. Biology 2023, 12, 833. https://doi.org/10.3390/biology12060833
Xiong C, Zhou Y, Han Y, Yi J, Pang H, Zheng R, Zhou Y. IntiCom-DB: A Manually Curated Database of Inter-Tissue Communication Molecules and Their Communication Routes. Biology. 2023; 12(6):833. https://doi.org/10.3390/biology12060833
Chicago/Turabian StyleXiong, Changxian, Yiran Zhou, Yu Han, Jingkun Yi, Huai Pang, Ruimao Zheng, and Yuan Zhou. 2023. "IntiCom-DB: A Manually Curated Database of Inter-Tissue Communication Molecules and Their Communication Routes" Biology 12, no. 6: 833. https://doi.org/10.3390/biology12060833
APA StyleXiong, C., Zhou, Y., Han, Y., Yi, J., Pang, H., Zheng, R., & Zhou, Y. (2023). IntiCom-DB: A Manually Curated Database of Inter-Tissue Communication Molecules and Their Communication Routes. Biology, 12(6), 833. https://doi.org/10.3390/biology12060833








