Prediction Model for Pre-Eclampsia Using Gestational-Age-Specific Serum Creatinine Distribution
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Selection of Predictors Related to PE Status
2.3. Establishment of the PE Prediction Model and Statistical Analysis
- (Step 1) We randomly divided the dataset using ratios of 0.7 and 0.3 and categorized them into training and testing sets, respectively.
- (Step 2) Using the candidate feature set (one of four feature sets), the training dataset, and logistic regression as the input variable, dataset, and classifier, respectively, we established the prediction model and measured the classification performance of adverse pregnancy outcomes in the testing dataset.
- (Step 3) For the same feature set, dataset, and classifier, we simultaneously measured the prediction performance of adverse pregnancy outcomes in the training dataset.
- (Step 4) We iterated steps 1 to 3 100 times, resulting in 100 pairs of predictive performance for the testing and training datasets.
3. Results
3.1. General Characteristics of Participants Included in the PE Prediction Model
3.2. Prediction Performance for PE
3.3. Predictive Performance for PE According to GW
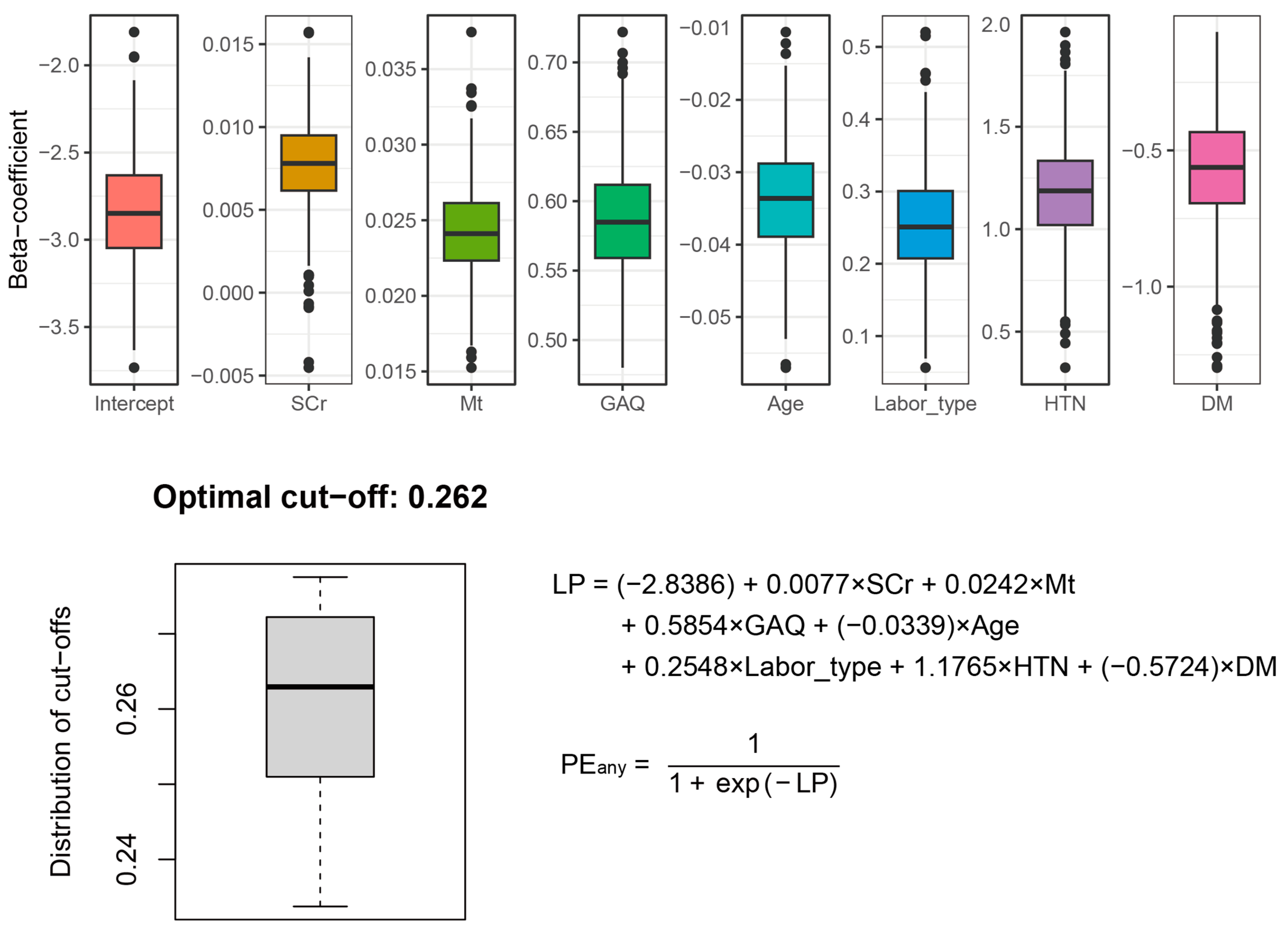
3.4. Final PE Prediction Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- ACOG Practice Bulletin No. 202: Gestational Hypertension and Preeclampsia. Obs. Gynecol 2019, 133, 1. [CrossRef]
- Ghulmiyyah, L.; Sibai, B. Maternal mortality from preeclampsia/eclampsia. Semin. Perinatol. 2012, 36, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Ma’ayeh, M.; Costantine, M.M. Prevention of preeclampsia. Semin. Fetal Neonatal Med. 2020, 25, 101123. [Google Scholar] [CrossRef]
- Ives, C.W.; Sinkey, R.; Rajapreyar, I.; Tita, A.T.N.; Oparil, S. Preeclampsia-Pathophysiology and Clinical Presentations: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2020, 76, 1690–1702. [Google Scholar] [CrossRef]
- Filipek, A.; Jurewicz, E. Preeclampsia—A disease of pregnant women. Postep. Biochem. 2018, 64, 229–232. [Google Scholar] [CrossRef]
- Duley, L. The global impact of pre-eclampsia and eclampsia. Semin. Perinatol. 2009, 33, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Rolnik, D.L.; Wright, D.; Poon, L.C.; O’Gorman, N.; Syngelaki, A.; de Paco Matallana, C.; Akolekar, R.; Cicero, S.; Janga, D.; Singh, M.; et al. Aspirin versus Placebo in Pregnancies at High Risk for Preterm Preeclampsia. N. Engl. J. Med. 2017, 377, 613–622. [Google Scholar] [CrossRef]
- MacDonald, T.M.; Walker, S.P.; Hannan, N.J.; Tong, S.; Kaitu’u-Lino, T.J. Clinical tools and biomarkers to predict preeclampsia. EBioMedicine 2022, 75, 103780. [Google Scholar] [CrossRef]
- Mikat, B.; Gellhaus, A.; Wagner, N.; Birdir, C.; Kimmig, R.; Köninger, A. Early detection of maternal risk for preeclampsia. ISRN Obs. Gynecol. 2012, 2012, 172808. [Google Scholar] [CrossRef]
- Bokslag, A.; van Weissenbruch, M.; Mol, B.W.; de Groot, C.J. Preeclampsia; short and long-term consequences for mother and neonate. Early Hum. Dev. 2016, 102, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, T.; Odutayo, A.; Keunen, J.; Hladunewich, M. The kidney in normal pregnancy and preeclampsia. Semin. Nephrol. 2011, 31, 4–14. [Google Scholar] [CrossRef]
- Lafayette, R. The kidney in preeclampsia. Kidney Int. 2005, 67, 1194–1203. [Google Scholar] [CrossRef]
- Beheiry, H.M.; Ali, I.A.; Rayis, D.A.; Saeed, A.M. Effect of pre-eclampsia on glomerular filtration rate in Sudanese women. Int. J. Res. Med. Sci. 2017, 5, 1053–1057. [Google Scholar] [CrossRef]
- Park, S.; Lee, S.M.; Park, J.S.; Hong, J.S.; Chin, H.J.; Na, K.Y.; Kim, D.K.; Oh, K.H.; Joo, K.W.; Kim, Y.S.; et al. Midterm eGFR and Adverse Pregnancy Outcomes: The Clinical Significance of Gestational Hyperfiltration. Clin. J. Am. Soc. Nephrol. 2017, 12, 1048–1056. [Google Scholar] [CrossRef] [PubMed]
- Kasraeian, M.; Asadi, N.; Vafaei, H.; Zamanpour, T.; Shahraki, H.R.; Bazrafshan, K. Evaluation of serum biomarkers for detection of preeclampsia severity in pregnant women. Pak. J. Med. Sci. 2018, 34, 869–873. [Google Scholar] [CrossRef] [PubMed]
- Harel, Z.; McArthur, E.; Hladunewich, M.; Dirk, J.S.; Wald, R.; Garg, A.X.; Ray, J.G. Serum Creatinine Levels Before, During, and After Pregnancy. JAMA 2019, 321, 205–207. [Google Scholar] [CrossRef]
- Harel, Z.; Park, A.L.; Ray, J.G. Blunted Glomerular Hyperfiltration in Pregnancy and Risk of Adverse Perinatal Outcomes. Am. J. Kidney Dis. 2020, 76, 297–299. [Google Scholar] [CrossRef]
- Kang, J.; Hwang, S.; Lee, T.S.; Cho, J.; Seo, D.M.; Choi, S.J.; Uh, Y. Gestational age-specific serum creatinine can predict adverse pregnancy outcomes. Sci. Rep. 2022, 12, 11224. [Google Scholar] [CrossRef]
- Hwang, S.; Gwon, C.; Seo, D.M.; Cho, J.; Kim, J.Y.; Uh, Y. A Deep Neural Network for Estimating Low-Density Lipoprotein Cholesterol from Electronic Health Records: Real-Time Routine Clinical Application. JMIR Med. Inform. 2021, 9, e29331. [Google Scholar] [CrossRef]
- Lee, J.; Jang, H.; Kim, J.; Min, S. Development of a suicide index model in general adolescents using the South Korea 2012–2016 national representative survey data. Sci. Rep. 2019, 9, 1846. [Google Scholar] [CrossRef]
- Moon, S.; Jang, J.-Y.; Kim, Y.; Oh, C.-M. Development and validation of a new diabetes index for the risk classification of present and new-onset diabetes: Multicohort study. Sci. Rep. 2021, 11, 15748. [Google Scholar] [CrossRef]
- Seo, M.J.; Ahn, S.G.; Lee, Y.J.; Kim, J.K. Development of the Hypertension Index Model in General Adult Using the Korea National Health and Nutritional Examination Survey and the Korean Genome and Epidemiology Study. J. Pers. Med. 2021, 11, 968. [Google Scholar] [CrossRef]
- Jacob, L.; Speed, T.P. The healthy ageing gene expression signature for Alzheimer’s disease diagnosis: A random sampling perspective. Genome Biol. 2018, 19, 97. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Lee, H.; The Alzheimer’s Disease Neuroimaging Initiative. Identification of Disease-Related Genes That Are Common between Alzheimer’s and Cardiovascular Disease Using Blood Genome-Wide Transcriptome Analysis. Biomedicines 2021, 9, 1525. [Google Scholar] [CrossRef]
- Jung, D.-H.; Hong, K.W.; Park, B.; Lee, Y.-J. Dietary iron to total energy intake ratio and type 2 diabetes incidence in a longitudinal 12-year analysis of the Korean Genome and Epidemiology Cohort Study. Eur. J. Nutr. 2021, 60, 4453–4461. [Google Scholar] [CrossRef]
- Michiels, S.; Koscielny, S.; Hill, C. Prediction of cancer outcome with microarrays: A multiple random validation strategy. Lancet 2005, 365, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Efron, B.; Tibshirani, R. On testing the significance of sets of genes. Ann. Appl. Stat. 2007, 1, 107–129. [Google Scholar] [CrossRef]
- Zander, T.; Hofmann, A.; Staratschek-Jox, A.; Classen, S.; Debey-Pascher, S.; Maisel, D.; Ansén, S.; Hahn, M.; Beyer, M.; Thomas, R.K.; et al. Blood-based gene expression signatures in non-small cell lung cancer. Clin. Cancer Res. 2011, 17, 3360–3367. [Google Scholar] [CrossRef]
- Goldenberg, R.L.; Culhane, J.F.; Iams, J.D.; Romero, R. Epidemiology and causes of preterm birth. Lancet 2008, 371, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Chappell, L.C.; Cluver, C.A.; Kingdom, J.; Tong, S. Pre-eclampsia. Lancet 2021, 398, 341–354. [Google Scholar] [CrossRef]
- Byerley, B.M.; Haas, D.M. A systematic overview of the literature regarding group prenatal care for high-risk pregnant women. BMC Pregnancy Childbirth 2017, 17, 329. [Google Scholar] [CrossRef] [PubMed]
- Power, M.L.; Wilson, E.K.; Hogan, S.O.; Loft, J.D.; Williams, J.L.; Mersereau, P.W.; Schulkin, J. Patterns of preconception, prenatal and postnatal care for diabetic women by obstetrician-gynecologists. J. Reprod. Med. 2013, 58, 7–14. [Google Scholar] [PubMed]
- Luke, B.; Brown, M.B.; Misiunas, R.; Anderson, E.; Nugent, C.; van de Ven, C.; Burpee, B.; Gogliotti, S. Specialized prenatal care and maternal and infant outcomes in twin pregnancy. Am. J. Obstet. Gynecol. 2003, 189, 934–938. [Google Scholar] [CrossRef] [PubMed]
- ACOG Committee Opinion No. 743: Low-Dose Aspirin Use During Pregnancy. Obs. Gynecol 2018, 132, e44–e52. [CrossRef]
- Davidson, K.W.; Barry, M.J.; Mangione, C.M.; Cabana, M.; Caughey, A.B.; Davis, E.M.; Donahue, K.E.; Doubeni, C.A.; Kubik, M.; Li, L.; et al. Aspirin Use to Prevent Preeclampsia and Related Morbidity and Mortality: US Preventive Services Task Force Recommendation Statement. JAMA 2021, 326, 1186–1191. [Google Scholar] [CrossRef]
- Taşkömür, A.T.; Erten, Ö. The role of cystatin C, neutrophil-lymphocyte ratio and platelet-lymphocyte ratio in the evaluation of kidney function in women with preeclampsia. Taiwan. J. Obstet. Gynecol. 2021, 60, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Lee, H. Shared Blood Transcriptomic Signatures between Alzheimer’s Disease and Diabetes Mellitus. Biomedicines 2021, 9, 34. [Google Scholar] [CrossRef]
- Chau, K.; Hennessy, A.; Makris, A. Placental growth factor and pre-eclampsia. J. Hum. Hypertens. 2017, 31, 782–786. [Google Scholar] [CrossRef]
- Roberts, J.M.; Rajakumar, A. Preeclampsia and soluble fms-like tyrosine kinase 1. J. Clin. Endocrinol. Metab. 2009, 94, 2252–2254. [Google Scholar] [CrossRef]
- Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [CrossRef]
- Gadde, R.; Cd, D.; Sheela, S.R. Placental protein 13: An important biological protein in preeclampsia. J. Circ. Biomark. 2018, 7, 1849454418786159. [Google Scholar] [CrossRef] [PubMed]
- Obrador, G.T.; Schultheiss, U.T.; Kretzler, M.; Langham, R.G.; Nangaku, M.; Pecoits-Filho, R.; Pollock, C.; Rossert, J.; Correa-Rotter, R.; Stenvinkel, P.; et al. Genetic and environmental risk factors for chronic kidney disease. Kidney Int. Suppl. 2017, 7, 88–106. [Google Scholar] [CrossRef]
- Hussein, W.; Lafayette, R.A. Renal function in normal and disordered pregnancy. Curr. Opin. Nephrol. Hypertens. 2014, 23, 46–53. [Google Scholar] [CrossRef]
- Cheung, K.L.; Lafayette, R.A. Renal physiology of pregnancy. Adv. Chronic Kidney Dis. 2013, 20, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Strevens, H.; Wide-Swensson, D.; Grubb, A. Serum cystatin C is a better marker for preeclampsia than serum creatinine or serum urate. Scand. J. Clin. Lab. Investig. 2001, 61, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Delude, C.M. Deep phenotyping: The details of disease. Nature 2015, 527, S14–S15. [Google Scholar] [CrossRef]
- Teasdale, S.; Morton, A. Changes in biochemical tests in pregnancy and their clinical significance. Obstet. Med. 2018, 11, 160–170. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
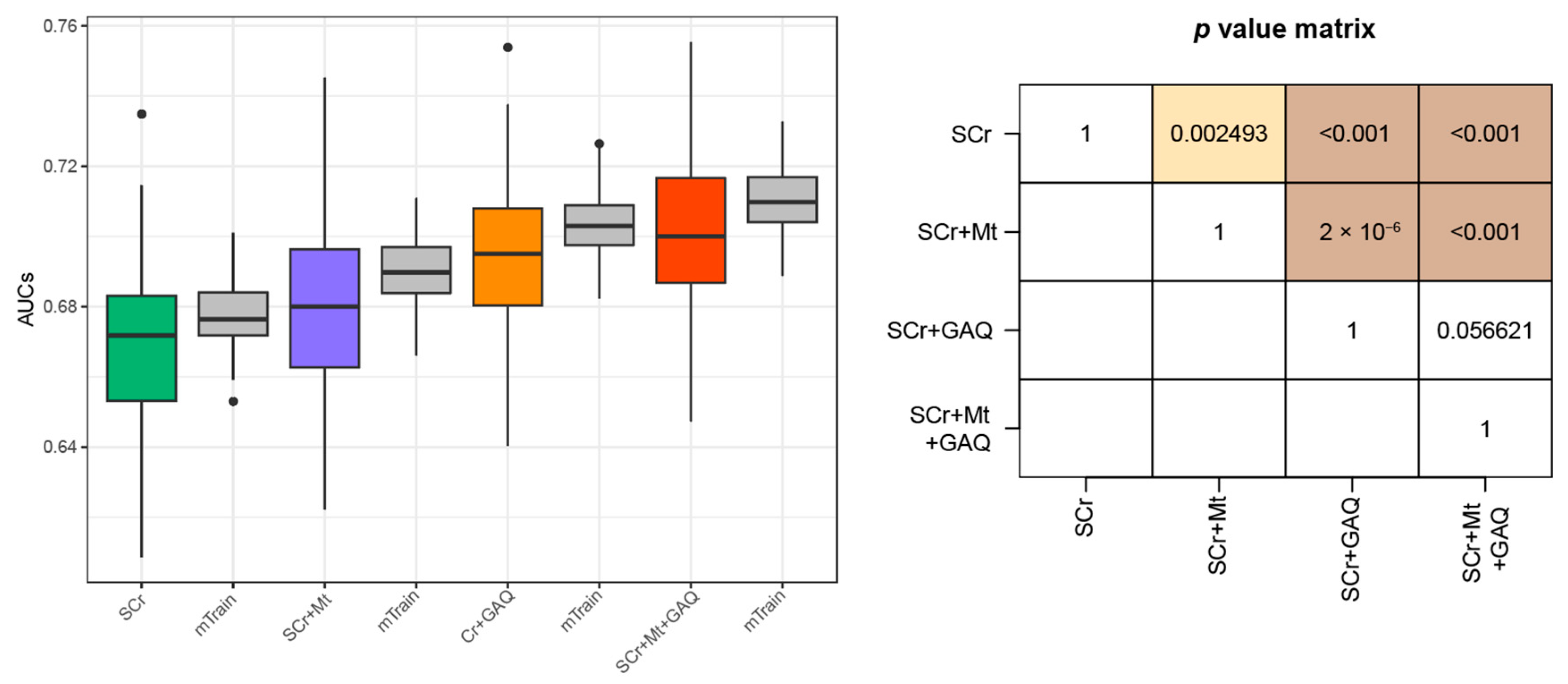
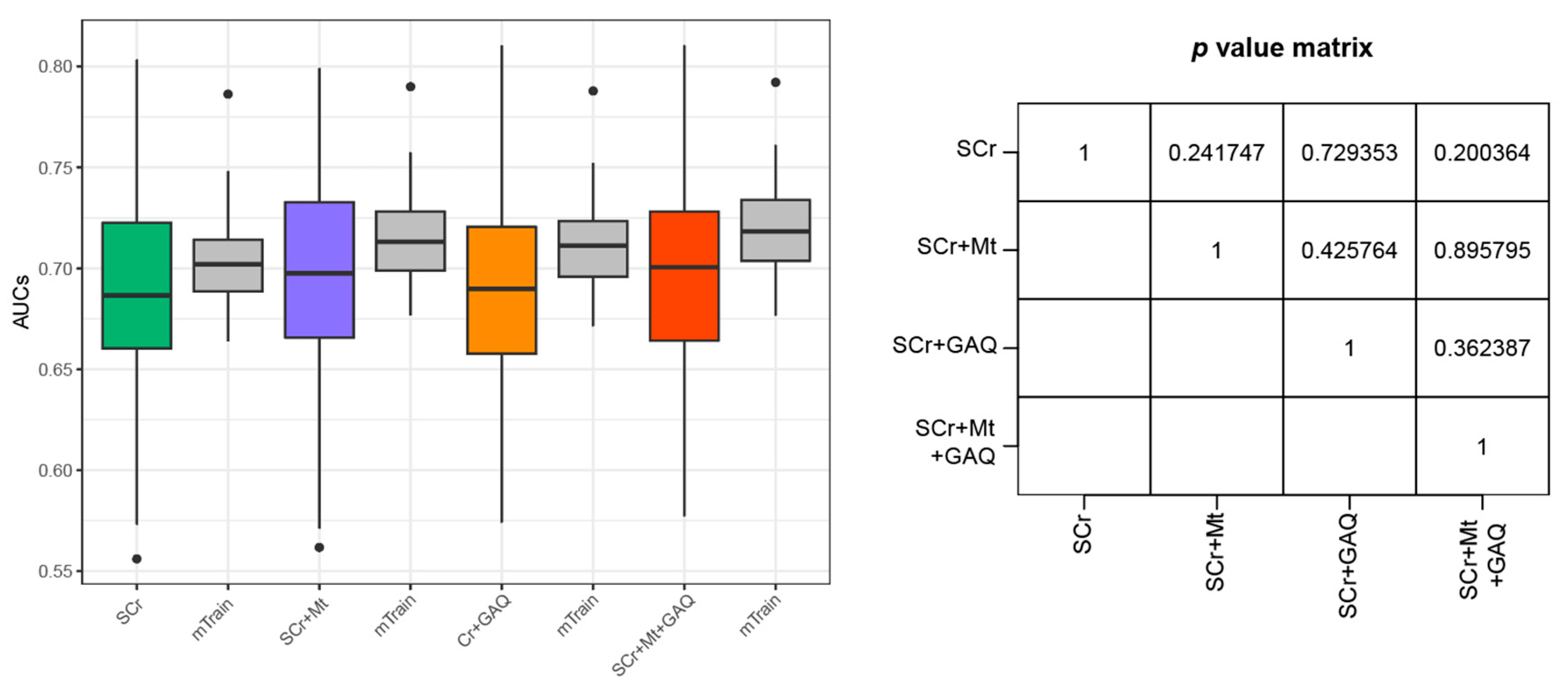
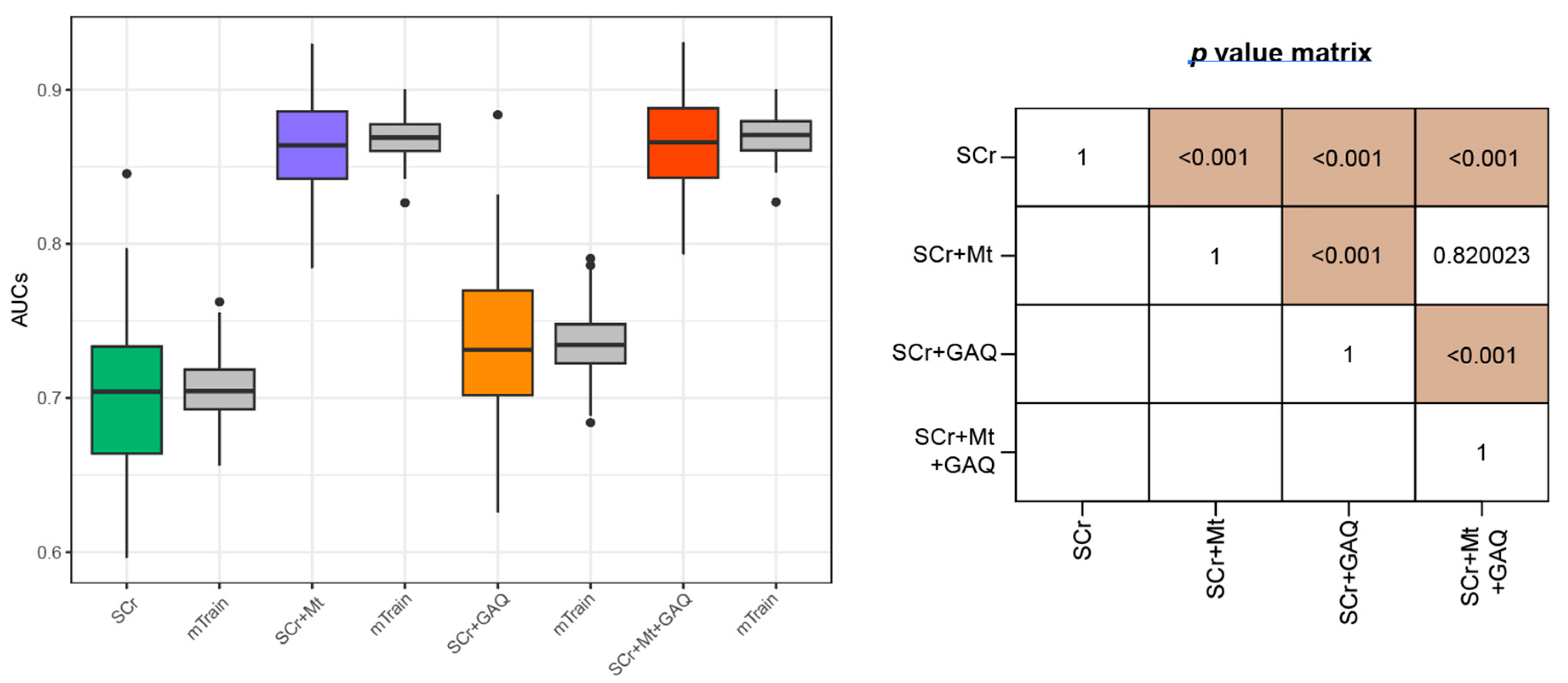

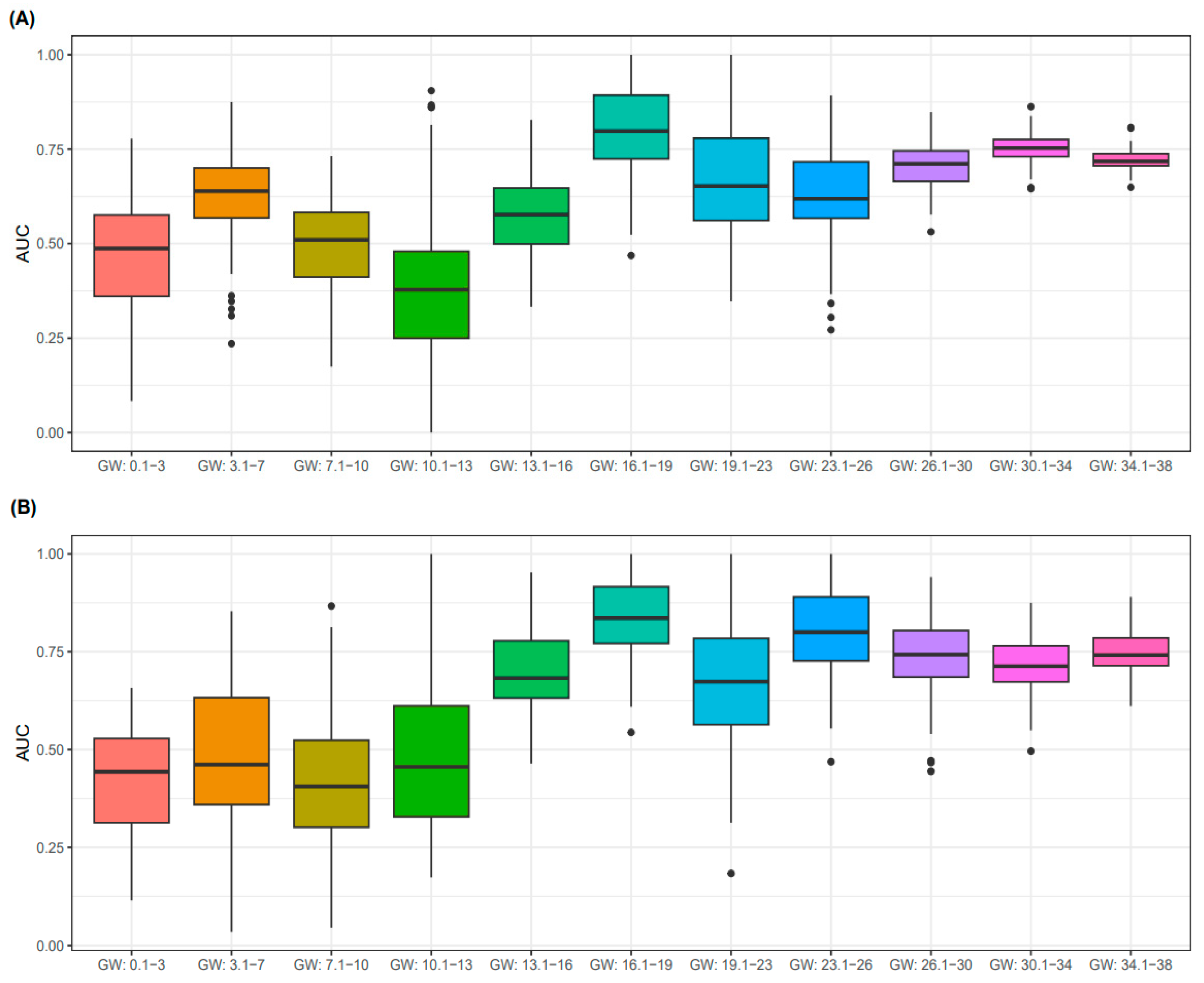
| All | First Trimester GW < 14 | Second Trimester 14 ≤ GW < 28 | Third Trimester GW ≥ 28 | p-for Trend | |
|---|---|---|---|---|---|
| Number of SCr measurements | 10,126 | 1302 | 1255 | 7569 | |
| GW of SCr sampling, weeks (mean ± SE) | 32.2 ± 0.12 | 7.3 ± 0.11 | 21.4 ± 0.12 | 38.2 ± 0.07 | <0.001 |
| GW of SCr sampling, weeks (median/IQR) | 35.6 (27.7–40.1) | 7.3 (4.1–10.6) | 21.4 (17.7–25.3) | 37.3 (34.7–40.3) | |
| Age, years (mean ± SE) | 33.36 ± 0.05 | 33.6 ± 0.12 | 33.3 ± 0.13 | 33.3 ± 0.06 | 0.081 |
| Labor types, n | |||||
| Nullipara | 4205 (41.5) | 430 (33) | 506 (40.3) | 3269 (43.2) | <0.001 |
| Multipara | 5921 (58.5) | 872 (67) | 749 (59.7) | 4300 (56.8) | <0.001 |
| Essential hypertension, n | 153 (1.5) | 12 (0.9) | 28 (2.2) | 113 (1.5) | 0.024 |
| Diabetes, n | 407 (4.0) | 70 (5.4) | 71 (5.7) | 266 (3.5) | <0.001 |
| PE, n | 1216 (12.0) | 53 (4.1) | 94 (7.5) | 1069 (14.1) | <0.001 |
| PE alone, n | 169 (1.7) | 17 (1.3) | 9 (0.7) | 143 (1.9) | 0.006 |
| PE + FGR, n | 114 (1.1) | 3 (0.2) | 3 (0.2) | 108 (1.4) | <0.001 a |
| PE + PTB, n | 432 (4.3) | 15 (1.2) | 27 (2.2) | 390 (5.2) | <0.001 |
| PE + FGR + PTB, n | 501 (4.9) | 18 (1.4) | 55 (4.4) | 428 (5.7) | <0.001 |
| BMI, kg/m2 (mean ± SE) a | 26.6 ± 0.13 | 23.2 ± 0.32 | 24.1 ± 0.38 | 27.1 ± 0.14 | <0.001 |
| SCr, μmol/L (mean ± SE) | 52.8 ± 0.38 | 55.2 ± 0.77 | 48.9 ± 1.22 | 53 ± 0.45 | 0.605 |
| BUN, mg/dL (mean ± SE) b | 9.2 ± 0.05 | 9.8 ± 0.15 | 8.3 ± 0.17 | 9.2 ± 0.06 | 0.333 |
| AST, U/L (mean ± SE) b | 28.1 ± 0.51 | 26.5 ± 1.99 | 26.3 ± 1.29 | 28.8 ± 0.55 | 0.06 |
| ALT, U/L (mean ± SE) b | 22.4 ± 0.57 | 26.8 ± 2.94 | 21.5 ± 1.11 | 21.8 ± 0.52 | 0.009 |
| ALP, U/L (mean ± SE) b | 126.1 ± 0.87 | 62.8 ± 0.97 | 73.9 ± 1.17 | 142.8 ± 1 | <0.001 |
| GGT, U/L (mean ± SE) b | 19.2 ± 0.41 | 24.9 ± 1.66 | 15.6 ± 0.59 | 18.9 ± 0.47 | <0.001 |
| LDH, U/L (mean ± SE) b | 229.2 ± 3.36 | 180.2 ± 2.28 | 206.2 ± 5.81 | 265.6 ± 5.69 | <0.001 |
| Group | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|
| All | 0.79 | 0.51 | 0.881 | 0.346 |
| GW (16.1–19) | 0.745 | 0.619 | 0.837 | 0.481 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, J.; Hwang, S.; Lee, T.; Ahn, K.; Seo, D.M.; Choi, S.J.; Uh, Y. Prediction Model for Pre-Eclampsia Using Gestational-Age-Specific Serum Creatinine Distribution. Biology 2023, 12, 816. https://doi.org/10.3390/biology12060816
Kang J, Hwang S, Lee T, Ahn K, Seo DM, Choi SJ, Uh Y. Prediction Model for Pre-Eclampsia Using Gestational-Age-Specific Serum Creatinine Distribution. Biology. 2023; 12(6):816. https://doi.org/10.3390/biology12060816
Chicago/Turabian StyleKang, Jieun, Sangwon Hwang, Taesic Lee, Kwangjin Ahn, Dong Min Seo, Seong Jin Choi, and Young Uh. 2023. "Prediction Model for Pre-Eclampsia Using Gestational-Age-Specific Serum Creatinine Distribution" Biology 12, no. 6: 816. https://doi.org/10.3390/biology12060816
APA StyleKang, J., Hwang, S., Lee, T., Ahn, K., Seo, D. M., Choi, S. J., & Uh, Y. (2023). Prediction Model for Pre-Eclampsia Using Gestational-Age-Specific Serum Creatinine Distribution. Biology, 12(6), 816. https://doi.org/10.3390/biology12060816






