Short-Term Alternate Feeding between Terrestrially Sourced Oil- and Fish Oil-Based Diets Modulates the Intestinal Microecology of Juvenile Turbot
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
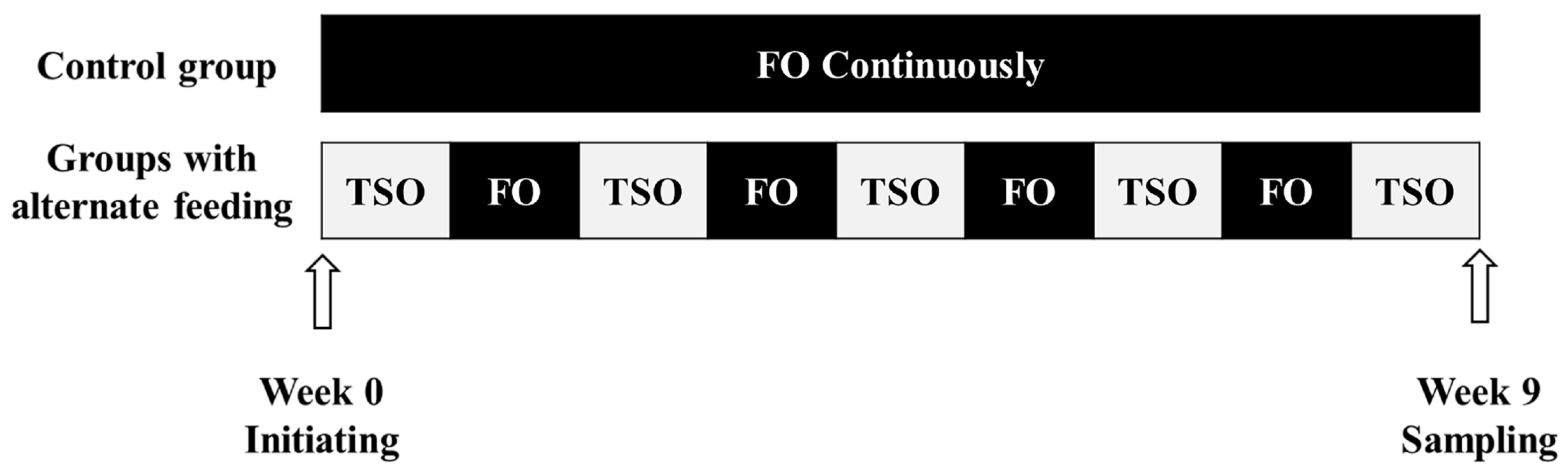
2.1. Experimental Diets
2.2. Fish Husbandry
2.3. Sample Collection
2.4. Intestinal Microbiota DNA Extraction, 16S rRNA Sequencing, and Data Analysis
2.5. Functional Predictions of Intestinal Microbiota
2.6. Calculation and Statistical Methods
3. Results
3.1. Growth Performance and Somatic Parameters
3.2. OTU Taxonomic Statistics and Venn Diagrams
3.3. Effects of Feed Alteration Strategies on Intestinal Microbiome Structure
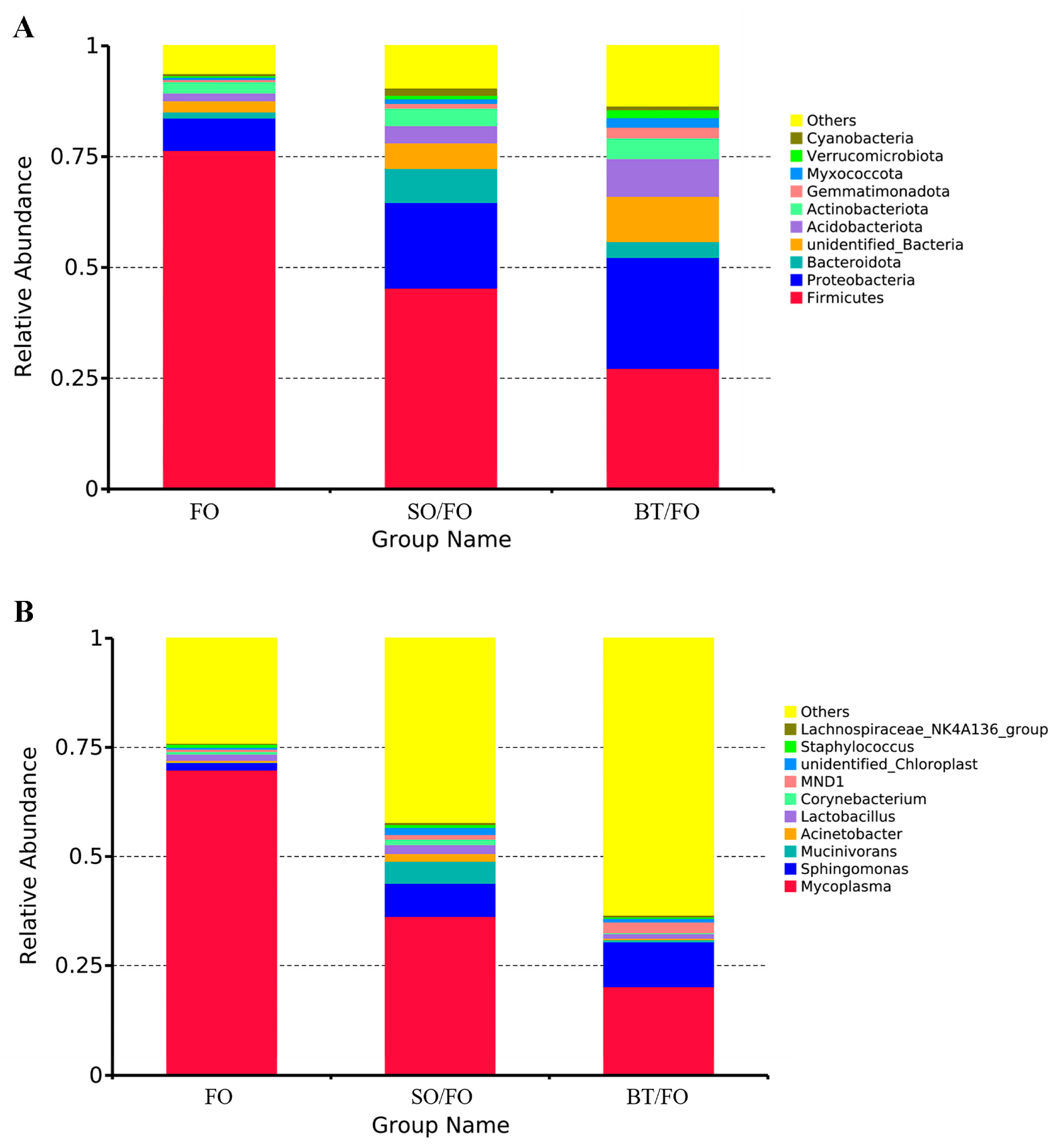
3.4. Effects of Alterative Feeding on Intestinal Microbiome Composition
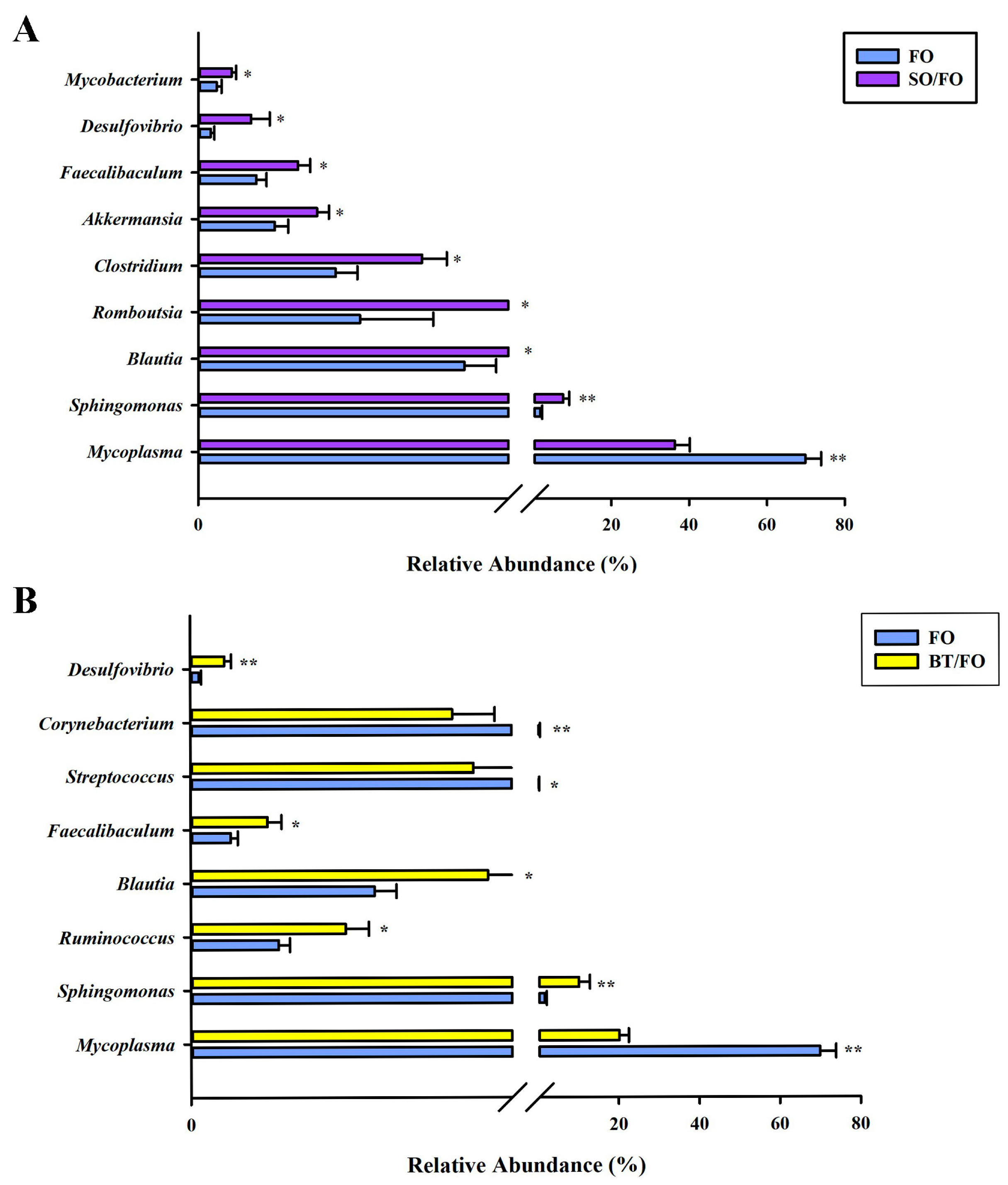
3.5. Analysis of Differential Bacteria among Groups
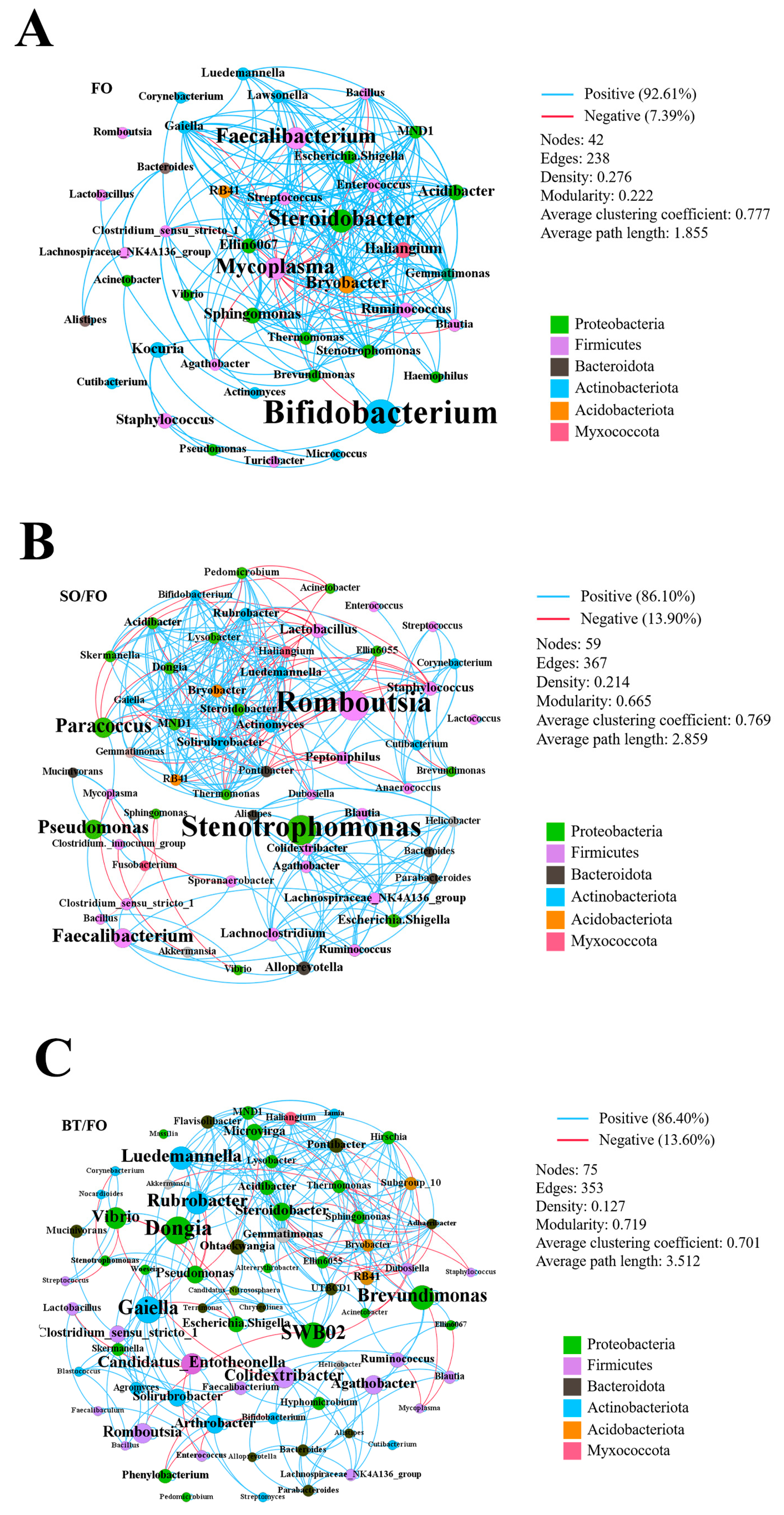
3.6. Microbial Co-Occurrence Network
3.7. Functional Predictions of Intestinal Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hodar, A.; Vasava, R.; Mahavadiya, D.; Joshi, N. Fish meal and fish oil replacement for aqua feed formulation by using alternative sources: A review. J. Exp. Zool. India 2020, 23, 13–21. [Google Scholar]
- Turchini, G.M.; Torstensen, B.E.; Ng, W.K. Fish oil replacement in finfish nutrition. Rev. Aquac. 2010, 1, 10–57. [Google Scholar] [CrossRef]
- Peng, M.; Xu, W.; Mai, K.; Zhou, H.; Zhang, Y.; Liufu, Z.; Zhang, K.; Ai, Q. Growth performance, lipid deposition and hepatic lipid metabolism related gene expression in juvenile turbot (Scophthalmus maximus L.) fed diets with various fish oil substitution levels by soybean oil. Aquaculture 2014, 433, 442–449. [Google Scholar] [CrossRef]
- Mu, H.; Shen, H.; Liu, J.; Xie, F.; Zhang, W.; Mai, K. High level of dietary soybean oil depresses the growth and anti-oxidative capacity and induces inflammatory response in large yellow croaker Larimichthys crocea. Fish Shellfish Immunol. 2018, 77, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Rombenso, A.N.; Trushenski, J.T.; Schwarz, M.H. Beef tallow is suitable as a primary lipid source in juvenile Florida pompano feeds. Aquacult. Nutr. 2017, 23, 1274–1286. [Google Scholar] [CrossRef]
- Lee, Y.; Lee, H.-Y. Revisiting the Bacterial Phylum Composition in Metabolic Diseases Focused on Host Energy Metabolism. Diabetes Metab. J. 2020, 44, 658–667. [Google Scholar] [CrossRef]
- Francis, D.S.; Turchini, G.M.; Smith, B.K.; Ryan, S.G.; De Silva, S.S. Effects of alternate phases of fish oil and vegetable oil-based diets in Murray cod. Aquacult. Res. 2009, 40, 1123–1134. [Google Scholar] [CrossRef]
- Eroldogan, O.T.; Elsabagh, M.; Emre, Y.; Turchini, G.M.; Yilmaz, H.A.; Eraslan, D.; Emre, N.; Evliyaoglu, E. Circadian feeding schedules in gilthead sea bream (Sparus aurata) and European sea bass (Dicentrarchus labrax): A comparative approach towards improving dietary fish oil utilization and n-3 LC-PUFA metabolism. Aquaculture 2018, 495, 806–814. [Google Scholar] [CrossRef]
- Xu, H.; Bi, Q.; Liao, Z.; Sun, B.; Jia, L.; Wei, Y.; Liang, M. Long-term alternate feeding between fish oil- and terrestrially sourced oil-based diets mitigated the adverse effects of terrestrially sourced oils on turbot fillet quality. Aquaculture 2021, 531, 735974. [Google Scholar] [CrossRef]
- Bondia-Pons, I.; Hyötyläinen, T.; Orešič, M. Role of Microbiota in Regulating Host Lipid Metabolism and Disease Risk. In Metabonomics and Gut Microbiota in Nutrition and Disease; Kochhar, S., Martin, F.-P., Eds.; Springer: London, UK, 2015; pp. 235–260. [Google Scholar]
- Koch, B.E.V.; Yang, S.; Lamers, G.; Stougaard, J.; Spaink, H.P. Intestinal microbiome adjusts the innate immune setpoint during colonization through negative regulation of MyD88. Nat. Commun. 2018, 9, 4099. [Google Scholar] [CrossRef]
- Semova, I.; Carten, J.D.; Stombaugh, J.; Mackey, L.C.; Knight, R.; Farber, S.A.; Rawls, J.F. Microbiota Regulate Intestinal Absorption and Metabolism of Fatty Acids in the Zebrafish. Cell Host Microbe 2012, 12, 277–288. [Google Scholar] [CrossRef]
- Hooper, L.V.; Gordon, J.I. Commensal host-bacterial relationships in the gut. Science 2001, 292, 1115–1118. [Google Scholar] [CrossRef] [PubMed]
- Pérez, T.; Balcázar, J.L.; Ruiz-Zarzuela, I.; Halaihel, N.; Vendrell, D.; De Blas, I.; Múzquiz, J.L. Host–microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol. 2010, 3, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.R.; Ran, C.; Ringo, E.; Zhou, Z.G. Progress in fish gastrointestinal microbiota research. Rev. Aquac. 2018, 10, 626–640. [Google Scholar] [CrossRef]
- Ou, W.; Yu, G.; Zhang, Y.; Mai, K. Recent progress in the understanding of the gut microbiota of marine fishes. Mar. Life Sci. Technol. 2021, 3, 434–448. [Google Scholar] [CrossRef]
- Egerton, S.; Culloty, S.; Whooley, J.; Stanton, C.; Ross, R.P. The Gut Microbiota of Marine Fish. Front. Microbiol. 2018, 9, 873. [Google Scholar] [CrossRef]
- Ghanbari, M.; Kneifel, W.; Domig, K.J. A new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture 2015, 448, 464–475. [Google Scholar] [CrossRef]
- You, C.; Chen, B.; Wang, M.; Wang, S.; Zhang, M.; Sun, Z.; Juventus, A.J.; Ma, H.; Li, Y. Effects of dietary lipid sources on the intestinal microbiome and health of golden pompano (Trachinotus ovatus). Fish Shellfish Immunol. 2019, 89, 187–197. [Google Scholar] [CrossRef]
- Castro, C.; Couto, A.; Diógenes, A.F.; Corraze, G.; Panserat, S.; Serra, C.R.; Oliva-Teles, A. Vegetable oil and carbohydrate-rich diets marginally affected intestine histomorphology, digestive enzymes activities, and gut microbiota of gilthead sea bream juveniles. Fish Physiol. Biochem. 2019, 45, 681–695. [Google Scholar] [CrossRef]
- Xu, H.; Mu, Y.; Zhang, Y.; Li, J.; Liang, M.; Zheng, K.; Wei, Y. Graded levels of fish protein hydrolysate in high plant diets for turbot (Scophthalmus maximus): Effects on growth performance and lipid accumulation. Aquaculture 2016, 454, 140–147. [Google Scholar] [CrossRef]
- Bo-Ra, K.; Jiwon, S.; Robin, G.; Hyung, L.J.; Wan, K.D.; Kuk-Hwan, S.; Ju-Hoon, L.; Bum, K.H.; Richard, I. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- De Roy, K.; Marzorati, M.; Negroni, A.; Thas, O.; Balloi, A.; Fava, F.; Verstraete, W.; Daffonchio, D.; Boon, N. Environmental conditions and community evenness determine the outcome of biological invasion. Nat. Commun. 2013, 4, 1383. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- Louis, P.; Young, P.; Holtrop, G.; Flint, H.J. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 2010, 12, 304–314. [Google Scholar] [CrossRef]
- Mu, C.; Yang, Y.; Luo, Z.; Guan, L.; Zhu, W. The Colonic Microbiome and Epithelial Transcriptome Are Altered in Rats Fed a High-Protein Diet Compared with a Normal-Protein Diet. J. Nutr. 2016, 146, 474–483. [Google Scholar] [CrossRef]
- Kong, C.; Gao, R.; Yan, X.; Huang, L.; Qin, H. Probiotics improve gut microbiota dysbiosis in obese mice fed a high-fat or high-sucrose diet. Nutrition 2018, 60, 175–184. [Google Scholar] [CrossRef]
- Kato, S.; Haruta, S.; Cui, Z.J.; Ishii, M.; Yokota, A.; Igarashi, Y. Clostridium straminisolvens sp. nov., A moderately thermophilic, aerotolerant and cellulolytic bacterium isolated from a cellulose-degrading bacterial community. Int. J. Syst. Evol. Microbiol. 2004, 54, 2043–2047. [Google Scholar] [CrossRef]
- Golubeva, A.V.; Joyce, S.A.; Moloney, G.; Burokas, A.; Sherwin, E.; Arboleya, S.; Flynn, I.; Khochanskiy, D.; Moya-Pérez, A.; Peterson, V.; et al. Microbiota-related Changes in Bile Acid & Tryptophan Metabolism are Associated with Gastrointestinal Dysfunction in a Mouse Model of Autism. EBioMedicine 2017, 24, 166–178. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef]
- Naito, Y.; Uchiyama, K.; Takagi, T. A next-generation beneficial microbe: Akkermansia muciniphila. J. Clin. Biochem. Nutr. 2018, 63, 33–35. [Google Scholar] [CrossRef]
- Chelakkot, C.; Choi, Y.; Kim, D.-K.; Park, H.T.; Ghim, J.; Kwon, Y.; Jeon, J.; Kim, M.-S.; Jee, Y.-K.; Gho, Y.S.; et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 2018, 50, e450. [Google Scholar] [CrossRef]
- Shen, J.; Tong, X.; Sud, N.; Khound, R.; Song, Y.; Maldonado-Gomez, M.X.; Walter, J.; Su, Q. Low-Density Lipoprotein Receptor Signaling Mediates the Triglyceride-Lowering Action of Akkermansia muciniphila in Genetic-Induced Hyperlipidemia. Atertio. Thromb. Vasc. Biol. 2016, 36, 1448–1456. [Google Scholar] [CrossRef] [PubMed]
- Ali, T.E.S.; El-Sayed, A.M.; Eissa, M.A.R.; Hanafi, H.M. Effect of dietary supplementation of sodium butyrate on growth performance and feed utilization of Nile tilapia (Oreochromis niloticus) fries. Indian J. Geo.-Mar. Sci. 2018, 47, 2071–2076. [Google Scholar]
- Robles, R.; Lozano, A.B.; Sevilla, A.; Marquez, L.; Nuez-Ortin, W.; Moyano, F.J. Effect of partially protected butyrate used as feed additive on growth and intestinal metabolism in sea bream (Sparus aurata). Fish Physiol. Biochem. 2013, 39, 1567–1580. [Google Scholar] [CrossRef]
- Wu, X.; Wang, L.G.; Xie, Q.P.; Tan, P. Effects of dietary sodium butyrate on growth, diet conversion, body chemical compositions and distal intestinal health in yellow drum (Nibea albiflora, Richardson). Aquacult. Res. 2020, 51, 69–79. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Z.; Dai, J.; Yang, P.; Xu, W.; Ai, Q.; Zhang, W.; Zhang, Y.; Zhang, Y.; Mai, K. Sodium butyrate supplementation in high-soybean meal diets for turbot (Scophthalmus maximus L.): Effects on inflammatory status, mucosal barriers and microbiota in the intestine. Fish Shellfish Immunol. 2019, 88, 65–75. [Google Scholar] [CrossRef]
- Ray, A.K.; Ghosh, K.; Ringo, E. Enzyme-producing bacteria isolated from fish gut: A review. Aquacult. Nutr. 2012, 18, 465–492. [Google Scholar] [CrossRef]
- Banerjee, G.; Ray, A.K. Bacterial symbiosis in the fish gut and its role in health and metabolism. Symbiosis 2017, 72, 1–11. [Google Scholar] [CrossRef]
- Ranjani, A.; Dhanasekaran, D.; Gopinath, P.M. An Introduction to Actinobacteria. In Actinobacteria–Basics and Biotechnological Applications; IntechOpen: London, UK, 2016; pp. 3–38. [Google Scholar]
- Zhou, Y.; Yuan, X.; Liang, X.F.; Fang, L.; Li, J.; Guo, X.; Bai, X.; He, S. Enhancement of growth and intestinal flora in grass carp: The effect of exogenous cellulase. Aquaculture 2013, 416–417, 1–7. [Google Scholar] [CrossRef]
- Cândido, F.G.; Valente, F.X.; Grześkowiak, Ł.M.; Moreira, A.P.B.; Rocha, D.M.U.P.; Alfenas, R.D.C.G. Impact of dietary fat on gut microbiota and low-grade systemic inflammation: Mechanisms and clinical implications on obesity. Int. J. Food Sci. Nutr. 2018, 69, 125–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yi, C.; Han, J.; Ming, T.; Zhou, J.; Lu, C.; Li, Y.; Su, X. Novel high-docosahexaenoic-acid tuna oil supplementation modulates gut microbiota and alleviates obesity in high-fat diet mice. Food Sci. Nutr. 2020, 8, 6513–6527. [Google Scholar] [CrossRef]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Chang, E.B.J.G. 43 Dietary Fat-Induced Taurocholic Acid Production Promotes Pathobiont and Colitis in IL-10-/- Mice. Gastroenterology 2012, 142, S-12. [Google Scholar] [CrossRef]
- Zhang-Sun, W.; Augusto, L.A.; Zhao, L.; Caroff, M. Desulfovibrio desulfuricans isolates from the gut of a single individual: Structural and biological lipid A characterization. FEBS Lett. 2015, 589, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Bozzetta, E.; Varello, K.; Giorgi, I.; Fioravanti, M.L.; Prearo, M.J.J.o.F.D. Mycobacterium marinum infection in a striped bass farm in Italy. J. Fish Dis. 2010, 33, 781–785. [Google Scholar] [CrossRef]
- Anderson, E.T.; Jr., S.F.; Asakawa, M.G.; Fatzinger, M.H.; Johnson, J.; Marchetere, K.; Goodale, L.; Risatti, G.R.; Harms, C.A.J.J.o.F.D. Splenic mycobacteriosis in an Atlantic guitarfish, Rhinobatos lentiginosus Garman. J. Fish Dis. 2012, 35, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Jeronimo, D.; Barbosa, A.; Ramos, M.; Marques, J.; Saraiva, A. Granulomas caused by Mycobacterium sp. in farmed Turbot Scopthalmus maximus (Linnaeus, 1758). Mediterr. Mar. Sci. 2013, 14, 424. [Google Scholar] [CrossRef]
- Donova, M.V.; Dovbnya, D.V.; Sukhodolskaya, G.V.; Khomutov, S.M.; Nikolayeva, V.M.; Kwon, I.; Han, K. Microbial conversion of sterol-containing soybean oil production waste. J. Chem. Technol. Biotechnol. 2005, 80, 55–60. [Google Scholar] [CrossRef]
- Rosengarten, R.; Citti, C.; Glew, M.; Lischewski, A.; Droeße, M.; Much, P.; Winner, F.; Brank, M.; Spergser, J. Host-pathogen interactions in mycoplasma pathogenesis: Virulence and survival strategies of minimalist prokaryotes. Int. J. Med. Microbiol. 2000, 290, 15–25. [Google Scholar] [CrossRef]
- Stadtländer, C.T.K.H.; Lotz, W.; Körting, W.; Kirchhoff, H. Piscine gill epithelial cell necrosis due to Mycoplasma mobile strain 163 K: Comparison of in-vivo and in-vitro infection. J. Comp. Pathol. 1995, 112, 351–359. [Google Scholar] [CrossRef]
- Brown, D.R. Mycoplasmosis and immunity of fish and reptiles. Front. Biosci.-Landmark 2002, 7, 1338–1346. [Google Scholar] [CrossRef]
- Wang, J.; Jaramillo-Torres, A.; Li, Y.; Kortner, T.M.; Gajardo, K.; Brevik, Ø.J.; Jakobsen, J.V.; Krogdahl, Å. Microbiota in intestinal digesta of Atlantic salmon (Salmo salar), observed from late freshwater stage until one year in seawater, and effects of functional ingredients: A case study from a commercial sized research site in the Arctic region. Anim. Microbiome 2021, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Holben, W.E.; Williams, P.; Gilbert, M.A.; Saarinen, M.; Sarkilahti, L.K.; Apajalahti, J.H.A. Phylogenetic analysis of intestinal microflora indicates a novel mycoplasma phylotype in farmed and wild salmon. Microb. Ecol. 2003, 46, 289. [Google Scholar] [CrossRef]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Deng, Y.; Luo, F.; He, Z.; Yang, Y. Phylogenetic Molecular Ecological Network of Soil Microbial Communities in Response to Elevated CO2. Mbio 2011, 2, e00122-11. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef]
- Olesen, J.M.; Bascompte, J.; Dupont, Y.L.; Jordano, P. The modularity of pollination networks. Proc. Natl. Acad. Sci. USA 2007, 104, 19891–19896. [Google Scholar] [CrossRef]
- Huyben, D.; Roehe, B.K.; Bekaert, M.; Ruyter, B.; Glencross, B. Dietary Lipid:Protein Ratio and n-3 Long-Chain Polyunsaturated Fatty Acids Alters the Gut Microbiome of Atlantic Salmon Under Hypoxic and Normoxic Conditions. Front. Microbiol. 2020, 11, 589898. [Google Scholar] [CrossRef]
- Huyben, D.; Rimoldi, S.; Ceccotti, C.; Montero, D.; Betancor, M.; Iannini, F.; Terova, G. Effect of dietary oil from Camelina sativa on the growth performance, fillet fatty acid profile and gut microbiome of gilthead Sea bream (Sparus aurata). PeerJ 2020, 8, e10430. [Google Scholar] [CrossRef]
- Yildirimer, C.C.; Brown, K.H. Intestinal microbiota lipid metabolism varies across rainbow trout (Oncorhynchus mykiss) phylogeographic divide. J. Appl. Microbiol. 2018, 125, 1614–1625. [Google Scholar] [CrossRef]
- Cario, E. Bacterial interactions with cells of the intestinal mucosa: Toll-like receptors and NOD2. Gut 2005, 54, 1182–1193. [Google Scholar] [CrossRef] [PubMed]


| Ingredients | Fish Oil | Soybean Oil | Beef Tallow |
|---|---|---|---|
| Fish meal | 40 | 40 | 40 |
| Soy protein concentrate | 5 | 5 | 5 |
| Soybean meal | 10 | 10 | 10 |
| Wheat meal | 21.98 | 21.98 | 21.98 |
| Casein | 4 | 4 | 4 |
| Brewer’s yeast | 8 | 8 | 8 |
| Mineral premix 1 | 0.5 | 0.5 | 0.5 |
| Vitamin premix 2 | 0.2 | 0.2 | 0.2 |
| Monocalcium phosphate | 1 | 1 | 1 |
| L-ascorbyl-2-polyphosphate | 0.2 | 0.2 | 0.2 |
| Choline chloride | 0.2 | 0.2 | 0.2 |
| Betaine | 0.3 | 0.3 | 0.3 |
| Ethoxyquin | 0.02 | 0.02 | 0.02 |
| Mold inhibitor 3 | 0.1 | 0.1 | 0.1 |
| Soya lecithin | 1 | 1 | 1 |
| Fish oil | 7.5 | ||
| Soybean oil | 7.5 | ||
| Beef tallow | 7.5 | ||
| Proximate composition | |||
| Moisture | 8.08 | 7.55 | 7.94 |
| Crude protein | 51.81 | 51.49 | 51.57 |
| Crude lipid | 11.56 | 11.87 | 11.70 |
| Ash | 9.50 | 9.76 | 9.46 |
| Fatty Acid | Oil | Diet | ||||
|---|---|---|---|---|---|---|
| Fish Oil | Soybean Oil | Beef Tallow | Fish Oil | Soybean Oil | Beef Tallow | |
| C14:0 | 6.88 | 0.09 | 2.67 | 4.82 | 1.51 | 2.65 |
| C16:0 | 20.80 | 11.00 | 36.50 | 18.52 | 13.06 | 24.60 |
| C18:0 | 4.37 | 4.02 | 20.30 | 4.27 | 3.76 | 11.69 |
| ∑SFA | 34.98 | 16.35 | 61.61 | 27.62 | 18.33 | 38.93 |
| C16:1n-7 | 6.50 | 0.09 | 1.38 | 4.71 | 1.54 | 2.05 |
| C18:1n-9 | 15.44 | 26.85 | 32.37 | 12.34 | 17.50 | 19.57 |
| C20:1n-9 | 4.67 | 0.56 | 0.12 | 2.64 | 0.46 | 0.26 |
| C22:1n-9 | 0.65 | ND | ND | 0.41 | 0.05 | 0.04 |
| C24:1n-9 | 0.64 | ND | ND | 0.65 | 0.21 | 0.23 |
| ∑MUFA | 27.90 | 27.49 | 33.87 | 20.75 | 19.76 | 22.15 |
| C18:2n-6 | 8.04 | 50.50 | 3.86 | 11.84 | 31.37 | 8.37 |
| C20:4n-6 | 1.06 | ND | 0.05 | 0.76 | 0.22 | 0.25 |
| n-6∑PUFA | 9.50 | 50.86 | 3.94 | 12.89 | 31.73 | 8.76 |
| C18:3n-3 | 2.50 | 5.24 | 0.26 | 2.15 | 3.44 | 0.95 |
| C20:3n-3 | 0.28 | ND | ND | 0.20 | 0.05 | 0.05 |
| C20:5n-3 | 9.52 | ND | ND | 7.96 | 2.90 | 2.92 |
| C22:6n-3 | 14.8 | ND | ND | 13.26 | 4.49 | 4.58 |
| n-3∑PUFA | 27.10 | 5.24 | 0.26 | 23.56 | 10.89 | 8.50 |
| n-3/n-6 | 2.85 | 0.10 | 0.06 | 1.83 | 0.34 | 0.97 |
| Parameters | FO | SO/FO | BT/FO |
|---|---|---|---|
| Growth performance | |||
| Initial weight g | 25.99 ± 0.01 | 25.98 ± 0.03 | 26.01 ± 0.01 |
| Final weight g | 56.28 ± 4.31 | 58.92 ± 1.52 | 55.31 ± 9.44 |
| Survival % | 63.86 ± 5.95 | 56.19 ± 3.30 | 61.43 ± 10.10 |
| Weight gain % | 116.50 ± 16.70 | 126.80 ± 5.80 | 132.9 ± 14.10 |
| Feed efficiency ratio | 0.94 ± 0.16 | 0.88 ± 0.08 | 0.80 ± 0.08 |
| Somatic parameters | |||
| VSI % | 5.70 ± 0.42 | 5.00 ± 0.08 | 5.72 ± 0.19 |
| HSI % | 1.35 ± 0.30 | 1.21 ± 0.17 | 1.71 ± 0.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Kong, Y.; Xu, H.; Bi, Q.; Liang, M.; Mai, K.; Zhang, Y. Short-Term Alternate Feeding between Terrestrially Sourced Oil- and Fish Oil-Based Diets Modulates the Intestinal Microecology of Juvenile Turbot. Biology 2023, 12, 650. https://doi.org/10.3390/biology12050650
Ma X, Kong Y, Xu H, Bi Q, Liang M, Mai K, Zhang Y. Short-Term Alternate Feeding between Terrestrially Sourced Oil- and Fish Oil-Based Diets Modulates the Intestinal Microecology of Juvenile Turbot. Biology. 2023; 12(5):650. https://doi.org/10.3390/biology12050650
Chicago/Turabian StyleMa, Xiuhua, Yaoyao Kong, Houguo Xu, Qingzhu Bi, Mengqing Liang, Kangsen Mai, and Yanjiao Zhang. 2023. "Short-Term Alternate Feeding between Terrestrially Sourced Oil- and Fish Oil-Based Diets Modulates the Intestinal Microecology of Juvenile Turbot" Biology 12, no. 5: 650. https://doi.org/10.3390/biology12050650
APA StyleMa, X., Kong, Y., Xu, H., Bi, Q., Liang, M., Mai, K., & Zhang, Y. (2023). Short-Term Alternate Feeding between Terrestrially Sourced Oil- and Fish Oil-Based Diets Modulates the Intestinal Microecology of Juvenile Turbot. Biology, 12(5), 650. https://doi.org/10.3390/biology12050650








