Functional Characterization of the Putative POT from Clostridium perfringens
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Expression
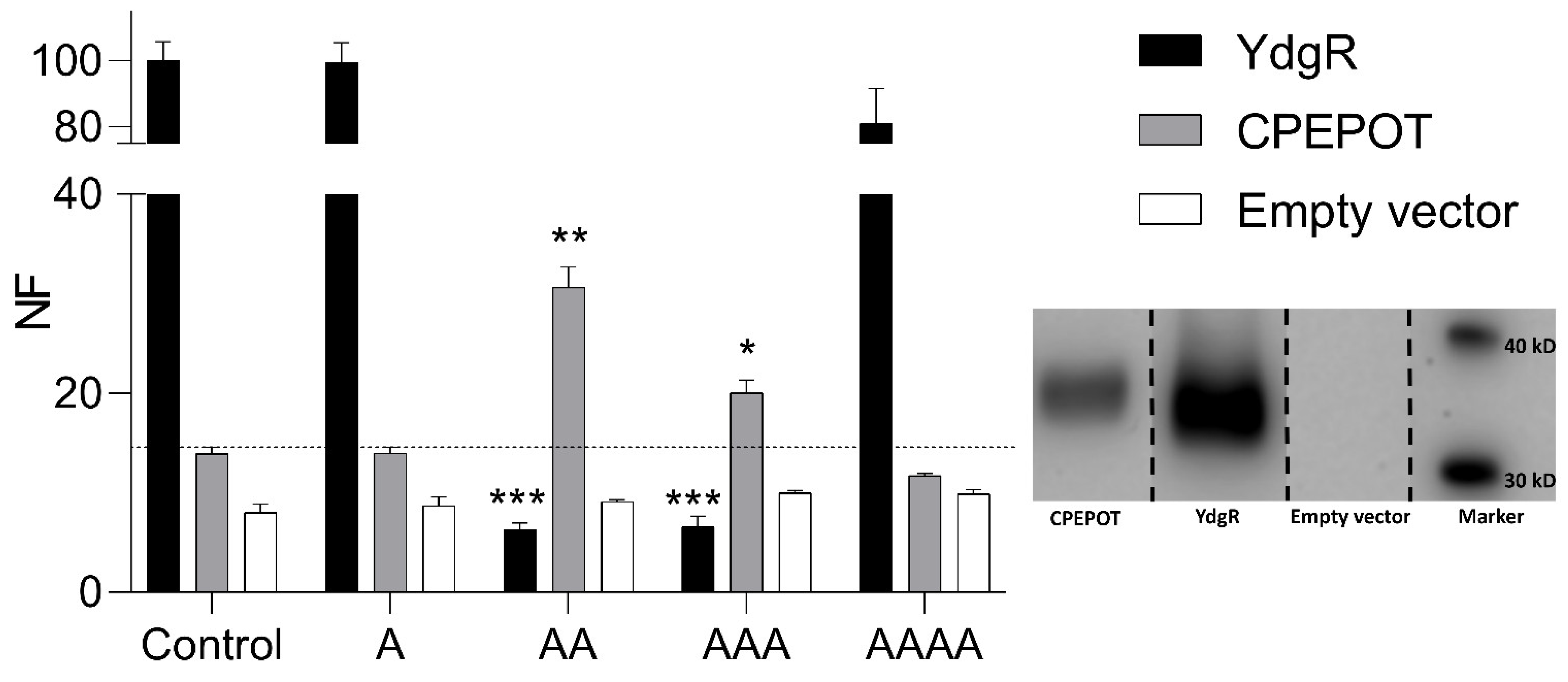
2.2. Western Blot
2.3. Uptake Assay
2.4. Peptide Specificity and pH Dependence Assay
2.5. Time-Dependent Uptake Assay
2.6. Uptake Assays in the Absence of a Proton Electrochemical Gradient
2.7. Bioinformatic Analysis
3. Results
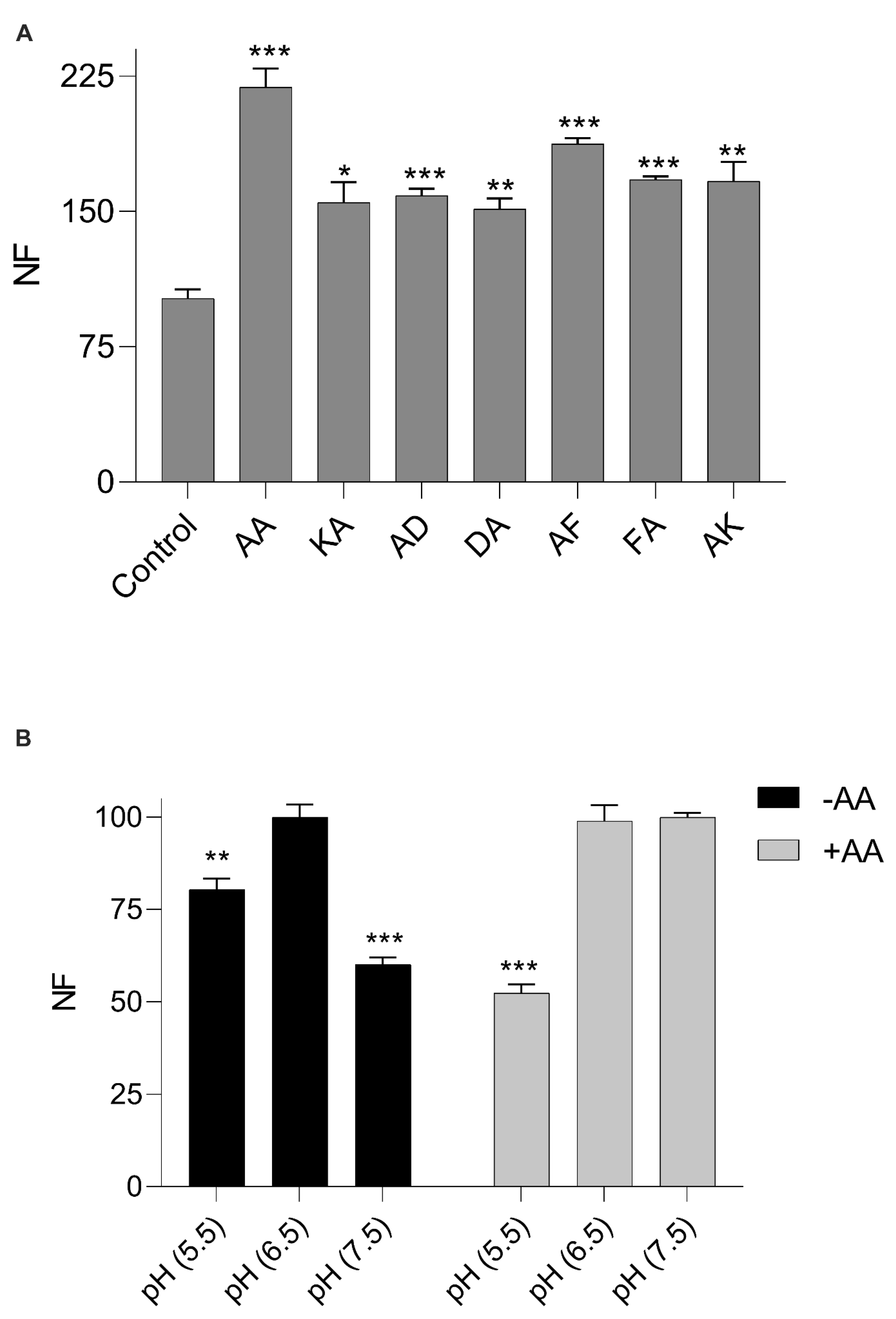
3.1. Increased β-Ala-Lys-AMCA Uptake in the Presence of Di- and Tripeptides
3.2. Increased β-Ala-Lys-AMCA Uptake in the Presence of Dipeptides
3.3. pH Dependency of β-Ala-Lys-AMCA Uptake
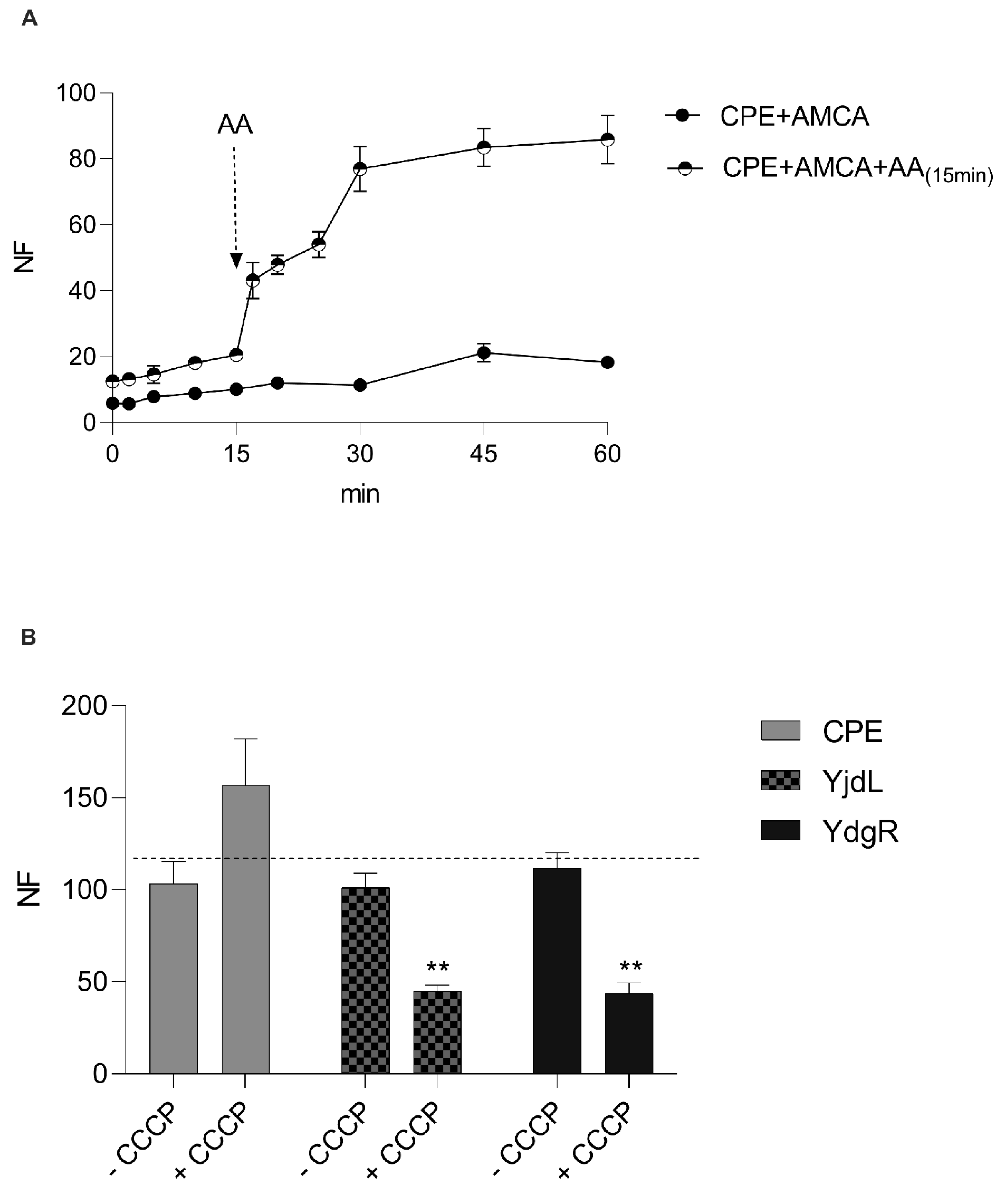
3.4. Addition of a “Stimulator” Peptide Results in a Higher Activity of CPEPOT
3.5. Removing the Proton Gradient Does Not Alter the Function of CPEPOT
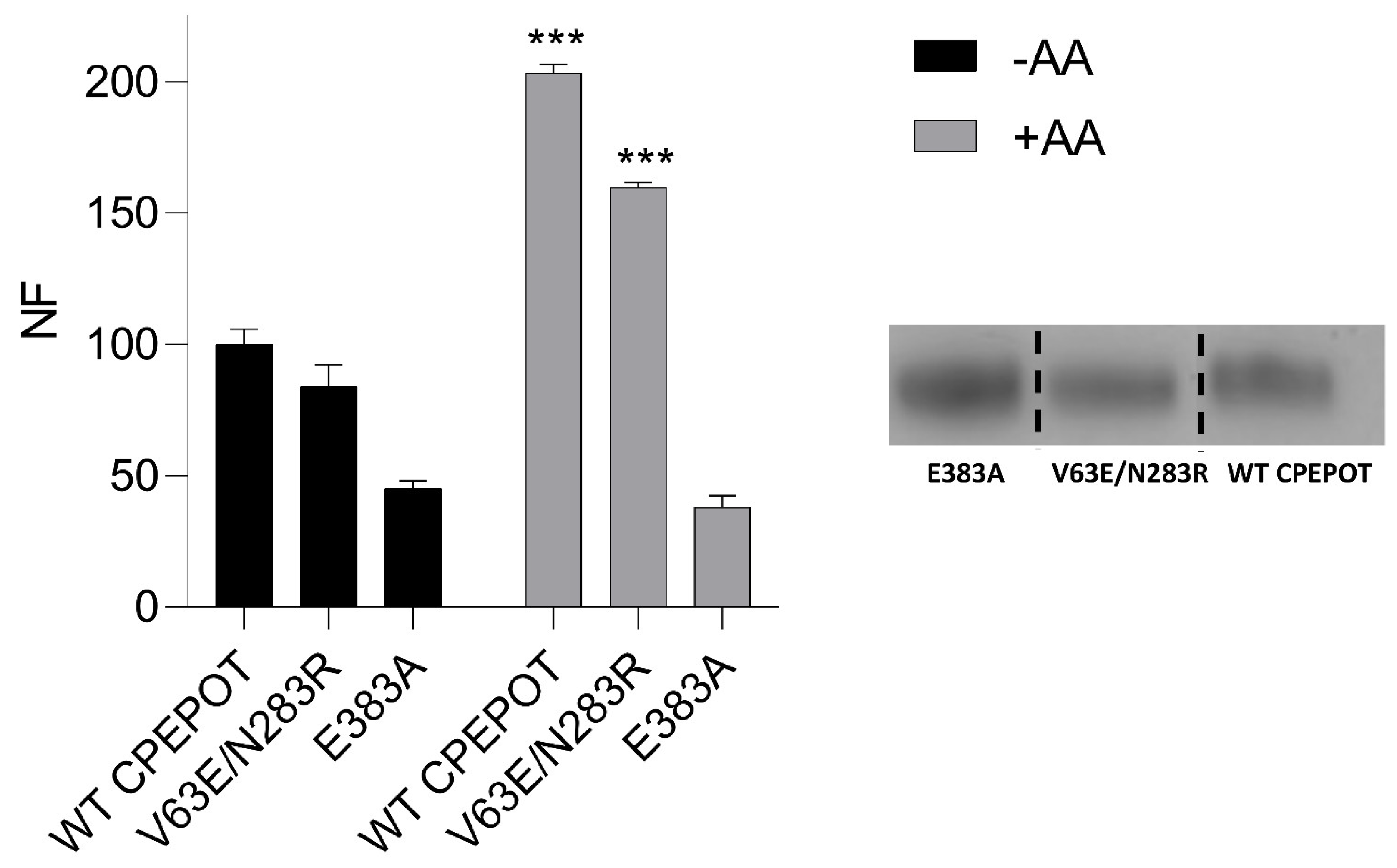
3.6. CPEPOT Lacks the Extra Helices and Gating Salt Bridges Observed in Prototypical POTs
3.7. Addition of a Periplasmic Salt Bridge Does Not Alter the Function of CPEPOT
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aduri, N.G.; Prabhala, B.K.; Ernst, H.A.; Jørgensen, F.S.; Olsen, L.; Mirza, O. Salt Bridge Swapping in the EXXERFXYY Motif of Proton-coupled Oligopeptide Transporters. J. Biol. Chem. 2015, 290, 29931–29940. [Google Scholar] [CrossRef]
- Boggavarapu, R.; Jeckelmann, J.-M.; Harder, D.; Ucurum, Z.; Fotiadis, D. Role of electrostatic interactions for ligand recognition and specificity of peptide transporters. BMC Biol. 2015, 13, 58. [Google Scholar] [CrossRef]
- Boll, M.; Herget, M.; Wagener, M.; Weber, W.M.; Markovich, D.; Biber, J.; Clauss, W.; Murer, H.; Daniel, H. Expression cloning and functional characterization of the kidney cortex high-affinity proton-coupled peptide transporter. Proc. Natl. Acad. Sci. USA 1996, 93, 284–289. [Google Scholar] [CrossRef]
- Boll, M.; Markovich, D.; Weber, W.M.; Korte, H.; Daniel, H.; Murer, H. Expression cloning of a cDNA from rabbit small intestine related to proton-coupled transport of peptides, ßlactam antibiotics and ACE-inhibitors. Pflügers Arch. 1994, 429, 146–149. [Google Scholar] [CrossRef]
- Daniel, H.; Morse, E.; Adibi, S. Determinants of substrate affinity for the oligopeptide/H+ symporter in the renal brush border membrane. J. Biol. Chem. 1992, 267, 9565–9573. [Google Scholar] [CrossRef]
- Doki, S.; Kato, H.E.; Solcan, N.; Iwaki, M.; Koyama, M.; Hattori, M.; Iwase, N.; Tsukazaki, T.; Sugita, Y.; Kandori, H.; et al. Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT. Proc. Natl. Acad. Sci. USA 2013, 110, 11343–11348. [Google Scholar] [CrossRef]
- Ernst, H.A.; Pham, A.; Hald, H.; Kastrup, J.S.; Rahman, M.; Mirza, O. Ligand binding analyses of the putative peptide transporter YjdL from E. coli display a significant selectivity towards dipeptides. Biochem. Biophys. Res. Commun. 2009, 389, 112–116. [Google Scholar] [CrossRef]
- Fei, Y.-J.; Kanai, Y.; Nussberger, S.; Ganapathy, V.; Leibach, F.H.; Romero, M.F.; Singh, S.K.; Boron, W.F.; Hediger, M.A. Expression cloning of a mammalian proton-coupled oligopeptide transporter. Nature 1994, 368, 563–566. [Google Scholar] [CrossRef]
- Fowler, P.W.; Orwick-Rydmark, M.; Radestock, S.; Solcan, N.; Dijkman, P.M.; Lyons, J.A.; Kwok, J.; Caffrey, M.; Watts, A.; Forrest, L.R.; et al. Gating Topology of the Proton-Coupled Oligopeptide Symporters. Structure 2015, 23, 290–301. [Google Scholar] [CrossRef]
- Guan, L.; Kaback, H.R. Properties of a LacY Efflux Mutant. Biochemistry 2009, 48, 9250–9255. [Google Scholar] [CrossRef]
- Guettou, F.; Quistgaard, E.M.; Trésaugues, L.; Moberg, P.; Jegerschöld, C.; Zhu, L.; Jong, A.J.O.; Nordlund, P.; Löw, C. Structural insights into substrate recognition in proton-dependent oligopeptide transporters. EMBO Rep. 2013, 14, 804–810. [Google Scholar] [CrossRef]
- Jensen, J.M.; Ernst, H.A.; Wang, X.; Hald, H.; Ditta, A.C.; Ismat, F.; Rahman, M.; Mirza, O.A. Functional investigation of conserved membrane-embedded glutamate residues in the proton-coupled peptide transporter YjdL. Protein Pept. Lett. 2012, 19, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.M.; Ismat, F.; Szakonyi, G.; Rahman, M.; Mirza, O. Probing the Putative Active Site of YjdL: An Unusual Proton-Coupled Oligopeptide Transporter from E. coli. PLoS ONE 2012, 7, e47780. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Kaczorowski, G.J.; Robertson, D.E.; Kaback, H.R. Mechanism of lactose translocation in membrane vesicles from Escherichia coli. 2. Effect of imposed.DELTA..PSI.,.DELTA.pH, and.DELTA..lovin..mu.H+. Biochemistry 1979, 18, 3697–3704. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Killer, M.; Finocchio, G.; Mertens, H.D.T.; Svergun, D.I.; Pardon, E.; Steyaert, J.; Löw, C. Cryo-EM Structure of an Atypical Proton-Coupled Peptide Transporter: Di- and Tripeptide Permease C. Front. Mol. Biosci. 2022, 9, 917725. [Google Scholar] [CrossRef]
- Killer, M.; Wald, J.; Pieprzyk, J.; Marlovits, T.C.; Löw, C. Structural snapshots of human PepT1 and PepT2 reveal mechanistic insights into substrate and drug transport across epithelial membranes. Sci. Adv. 2021, 7, eabk3259. [Google Scholar] [CrossRef]
- Kiu, R.; Hall, L.J. An update on the human and animal enteric pathogen Clostridium perfringens. Emerg. Microbes Infect. 2018, 7, 1–15. [Google Scholar] [CrossRef]
- Kottra, G.; Daniel, H. Bidirectional electrogenic transport of peptides by the proton-coupled carrier PEPT1 in Xenopus laevis oocytes: Its asymmetry and symmetry. J. Physiol. 2001, 536 Pt 2, 495–503. [Google Scholar] [CrossRef]
- Liu, W.; Liang, R.; Ramamoorthy, S.; Fei, Y.-J.; Ganapathy, M.E.; Hediger, M.A.; Ganapathy, V.; Leibach, F.H. Molecular cloning of PEPT 2, a new member of the H+/peptide cotransporter family, from human kidney. Biochim. Biophys. Acta Biomembr. 1995, 1235, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Malle, E.; Zhou, H.; Neuhold, J.; Spitzenberger, B.; Klepsch, F.; Pollak, T.; Bergner, O.; Ecker, G.; Stolt-Bergner, P.C. Random Mutagenesis of the Prokaryotic Peptide Transporter YdgR Identifies Potential Periplasmic Gating Residues. J. Biol. Chem. 2011, 286, 23121–23131. [Google Scholar] [CrossRef]
- Gohari, I.M.; Navarro, M.A.; Li, J.; Shrestha, A.; Uzal, F.; McClane, B.A. Pathogenicity and virulence of Clostridium perfringens. Virulence 2021, 12, 723–753. [Google Scholar] [CrossRef] [PubMed]
- Newstead, S. Recent advances in understanding proton coupled peptide transport via the POT family. Curr. Opin. Struct. Biol. 2017, 45, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Newstead, S.; Drew, D.; Cameron, A.D.; Postis, V.L.G.; Xia, X.; Fowler, P.W.; Ingram, J.C.; Carpenter, E.P.; Sansom, M.S.P.; McPherson, M.J.; et al. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J. 2011, 30, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.L.; Deme, J.C.; Wu, Z.; Kuteyi, G.; Huo, J.; Owens, R.J.; Biggin, P.C.; Lea, S.M.; Newstead, S. Cryo-EM structure of PepT2 reveals structural basis for proton-coupled peptide and prodrug transport in mammals. Sci. Adv. 2021, 7, eabh3355. [Google Scholar] [CrossRef] [PubMed]
- Prabhala, B.K.; Aduri, N.G.; Sharma, N.; Shaheen, A.; Sharma, A.; Iqbal, M.; Hansen, P.R.; Brasen, C.; Gajhede, M.; Rahman, M.; et al. The prototypical proton-coupled oligopeptide transporter YdgR from Escherichia coli facilitates chloramphenicol uptake into bacterial cells. J. Biol. Chem. 2018, 293, 1007–1017. [Google Scholar] [CrossRef]
- Prabhala, B.K.; Rahman, M.; Nour-Eldin, H.H.; Jørgensen, F.S.; Mirza, O. PTR2/POT/NPF transporters: What makes them tick? Adv. Protein Chem. Struct. Biol. 2021, 123, 219–240. [Google Scholar] [CrossRef]
- Sakata, K.; Yamashita, T.; Maeda, M.; Moriyama, Y.; Shimada, S.; Tohyama, M. Cloning of a lymphatic peptide/histidine transporter. Biochem. J. 2001, 356 Pt 1, 53–60. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Sharma, N.; Aduri, N.G.; Iqbal, A.; Prabhala, B.K.; Mirza, O. Peptide Selectivity of the Proton-Coupled Oligopeptide Transporter from Neisseria meningitidis. Microb. Physiol. 2016, 26, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Smith, D.E.; Brosius, F.C. Developmental Expression of PEPT1 and PEPT2 in Rat Small Intestine, Colon, and Kidney. Pediatr. Res. 2001, 49, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Solcan, N.; Kwok, J.; Fowler, P.W.; Cameron, A.D.; Drew, D.; Iwata, S.; Newstead, S. Alternating access mechanism in the POT family of oligopeptide transporters. EMBO J. 2012, 31, 3411–3421. [Google Scholar] [CrossRef]
- Sonna, L.A.; Ambudkar, S.V.; Maloney, P.C. The mechanism of glucose 6-phosphate transport by Escherichia coli. J. Biol. Chem. 1988, 263, 6625–6630. [Google Scholar] [CrossRef] [PubMed]
- Ural-Blimke, Y.; Flayhan, A.; Strauss, J.; Rantos, V.; Bartels, K.; Nielsen, R.; Pardon, E.; Steyaert, J.; Kosinski, J.; Quistgaard, E.M.; et al. Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1. J. Am. Chem. Soc. 2019, 141, 2404–2412. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Weitz, D.; Harder, D.; Casagrande, F.; Fotiadis, D.; Obrdlik, P.; Kelety, B.; Daniel, H. Functional and Structural Characterization of a Prokaryotic Peptide Transporter with Features Similar to Mammalian PEPT1. J. Biol. Chem. 2007, 282, 2832–2839. [Google Scholar] [CrossRef]
- Yamashita, T.; Shimada, S.; Guo, W.; Sato, K.; Kohmura, E.; Hayakawa, T.; Takagi, T.; Tohyama, M. Cloning and Functional Expression of a Brain Peptide/Histidine Transporter. J. Biol. Chem. 1997, 272, 10205–10211. [Google Scholar] [CrossRef]
- Zhao, Y.; Mao, G.; Liu, M.; Zhang, L.; Wang, X.; Zhang, X.C. Crystal Structure of the E. coli Peptide Transporter YbgH. Structure 2014, 22, 1152–1160. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gharabli, H.; Rafiq, M.; Iqbal, A.; Yan, R.; Aduri, N.G.; Sharma, N.; Prabhala, B.K.; Mirza, O. Functional Characterization of the Putative POT from Clostridium perfringens. Biology 2023, 12, 651. https://doi.org/10.3390/biology12050651
Gharabli H, Rafiq M, Iqbal A, Yan R, Aduri NG, Sharma N, Prabhala BK, Mirza O. Functional Characterization of the Putative POT from Clostridium perfringens. Biology. 2023; 12(5):651. https://doi.org/10.3390/biology12050651
Chicago/Turabian StyleGharabli, Hani, Maria Rafiq, Anna Iqbal, Ruyu Yan, Nanda G. Aduri, Neha Sharma, Bala K. Prabhala, and Osman Mirza. 2023. "Functional Characterization of the Putative POT from Clostridium perfringens" Biology 12, no. 5: 651. https://doi.org/10.3390/biology12050651
APA StyleGharabli, H., Rafiq, M., Iqbal, A., Yan, R., Aduri, N. G., Sharma, N., Prabhala, B. K., & Mirza, O. (2023). Functional Characterization of the Putative POT from Clostridium perfringens. Biology, 12(5), 651. https://doi.org/10.3390/biology12050651





