Lipid Droplets from Plants and Microalgae: Characteristics, Extractions, and Applications
Abstract
Simple Summary
Abstract
1. Introduction
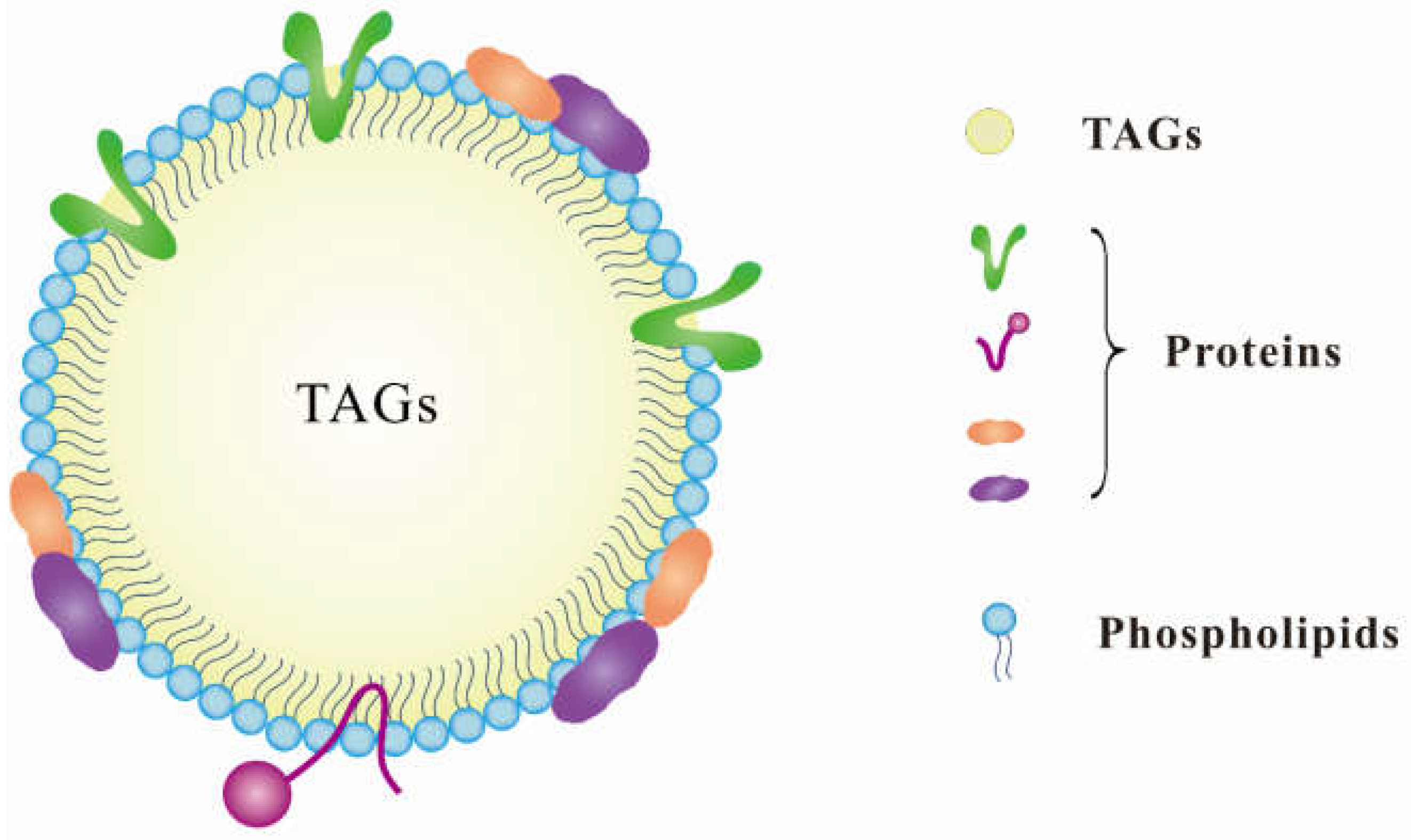
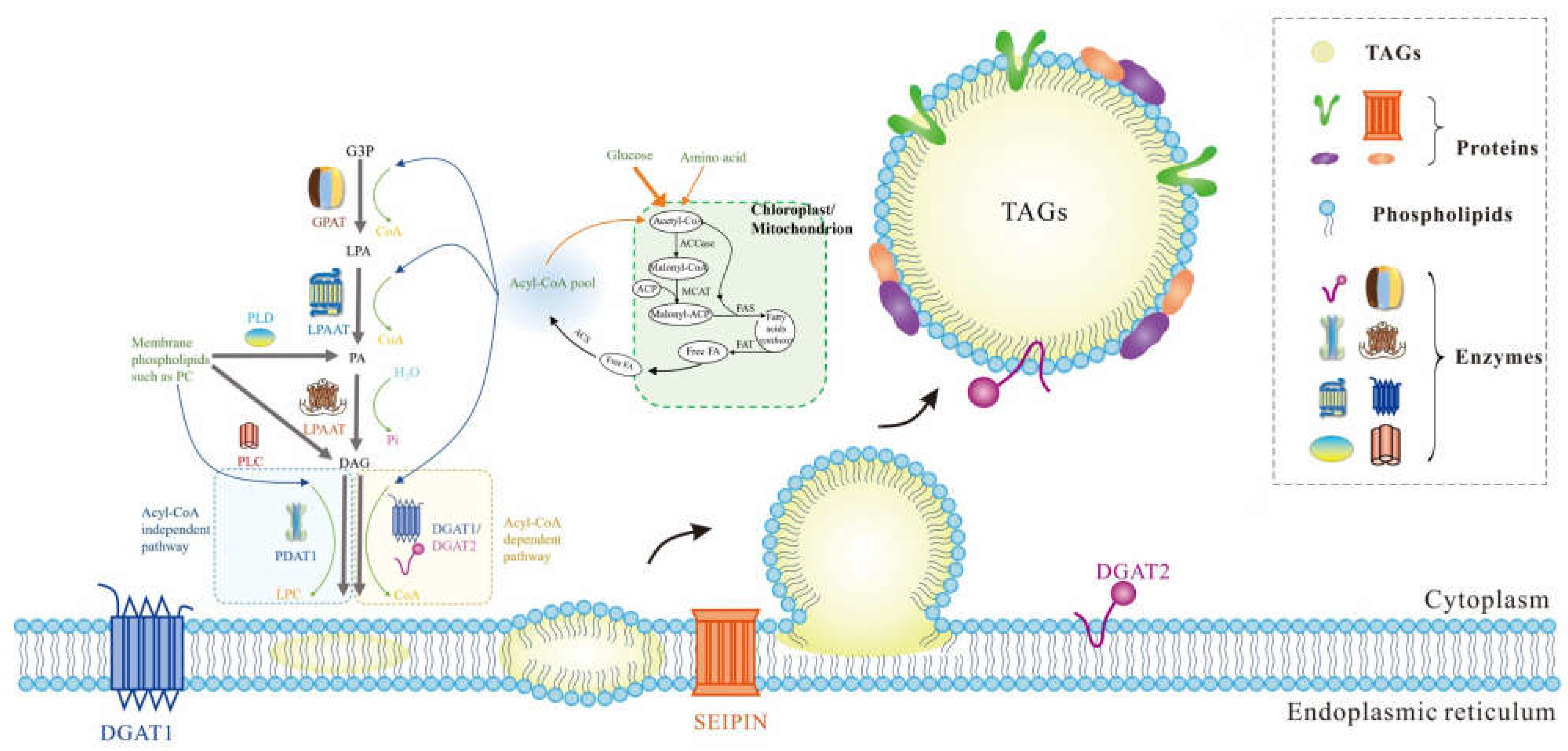
2. Characteristics of Lipid Droplets
| Organisms | TAGs | Protein | Phospholipid | Others (Sterols, Wax Esters, Steryl Esters, Carotenoids) | References | |
|---|---|---|---|---|---|---|
| Plants | Soybean | <40.1% | 8.8% | Na* | Na* | [28] |
| Peanuts | <98.1% | 1.27% | 0.77% | Na* | [29] | |
| Sunflower seed | <92.6% | 7.3% | Na* | Na* | [30] | |
| Maize | 97.66% | 1.43% | 0.91% | Na* | [20] | |
| Rapeseed | 94.21% | 3.46% | 1.97% | 0.36% | [27] | |
| Coconut | <38.2% | 4.1% | Na* | 0.15% | [31] | |
| Safflower | 97% | 2.5% | 0.7% | Na* | [32] | |
| Cotton | 96.99% | 1.70% | 1.18% | 0.13% | [27] | |
| Flax | 97.65% | 1.34% | 0.90% | 0.11% | [27] | |
| Sesame | 97.37% | 0.59% | 0.57% | 0.13% | [27] | |
| Mustard | 94.64% | 3.25% | 1.60% | 0.17% | [27] | |
| Algae | Chlamydomonas reinhardtii | 85–95% | Na* | <5% | 10% | [2] |
| Dunaliella salina | >90% | Na* | <10% | Na* | [33] | |
| Thraustochytrid | 81% | 0.5% | Na* | Na* | [14] | |
| Diatom | 58% | 2.3% | Na* | Na* | [34] |
3. Features of Proteins Associated with Lipid Droplets
| Proteins | Function | Molecular Mass | References |
|---|---|---|---|
| Oleosin | Involved in the structure; influence the size and stability of LDs | 15–25 kD | [70,71] |
| Caleosin | Involved in peroxygenase activity | 20–30 kD | [72,73] |
| Steroleosin | Involved in brassinosteroid metabolism | 40–41 kD | [74,75] |
| α-Dioxygenase | Phytoalexin synthesis | ~73 kD | [76] |
| LD-associated protein (LDAP) | Involved in the formation and turnover of LDs; related to stress | ~25 kD | [16] |
| Oil-body-associated protein (OBAP1) | Regulating the size of LDs | ~27 kD | [77] |
| CGI-58 | Involved in LDs’ homeostasis | Na* | [78] |
| PXA1 | Involved in LDs’ metabolism and signaling | Na* | [79] |
| SEIPIN | Modulating the accumulation of TAG and influencing the proliferation of LDs | Na | [80] |
| Sugar-dependent 1 (SDP1) | Involved in the degradation of LDs | Na* | [81] |
4. Lipid Droplet Extraction Strategies
4.1. Physical Method
4.1.1. Grinding
4.1.2. Pressing
4.1.3. High-Pressure Homogenization
4.2. Chemical Methods
4.3. Biological Methods (Enzymatic Lysis)
5. Factors Affecting the Stability of Lipid Droplets after Extraction
5.1. pH
5.2. Ionic Strength
5.3. Thermal Treatment
6. Potential Applications of Lipid Droplets
6.1. Potential Food Applications
6.2. Biotechnological Applications
6.2.1. Growth Factors
6.2.2. Insulin
6.2.3. Vaccines
6.2.4. Astaxanthin
6.3. Biofuel Applications
6.4. Bioplastic Applications
7. Limitations of This Review
8. Conclusions and Suggestions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ischebeck, T.; Krawczyk, H.E.; Mullen, R.T.; Dyer, J.M.; Chapman, K.D. In Lipid droplets in plants and algae: Distribution, formation, turnover and function, Semin. Cell Dev. Biol. 2020, 108, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Goold, H.; Beisson, F.; Peltier, G.; Li-Beisson, Y. Microalgal lipid droplets: Composition, diversity, biogenesis and functions. Plant Cell Rep. 2015, 34, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.J. The dynamic roles of intracellular lipid droplets: From archaea to mammals. Protoplasma 2012, 249, 541–585. [Google Scholar] [CrossRef] [PubMed]
- Pyc, M.; Cai, Y.; Greer, M.S.; Yurchenko, O.; Chapman, K.D.; Dyer, J.M.; Mullen, R.T. Turning over a new leaf in lipid droplet biology. Trends Plant Sci. 2017, 22, 596–609. [Google Scholar] [CrossRef]
- Barbosa, A.D.; Siniossoglou, S. Function of lipid droplet-organelle interactions in lipid homeostasis. Biochim. Biophys. Acta (Bba)-Mol. Cell Res. 2017, 1864, 1459–1468. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Huang, X.; Song, B.-L.; Yang, H. The biogenesis of lipid droplets: Lipids take center stage. Prog. Lipid Res. 2019, 75, 100989. [Google Scholar] [CrossRef] [PubMed]
- Olzmann, J.A.; Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef]
- Jayaram, S.; Bal, A. Oleosomes (lipid bodies) in nitrogen-fixing peanut nodules. Plantcell Environ. 1991, 14, 195–203. [Google Scholar] [CrossRef]
- Shimada, T.L.; Hara-Nishimura, I. Leaf oil bodies are subcellular factories producing antifungal oxylipins. Curr. Opin. Plant Biol. 2015, 25, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-Y.; Chung, C.-I.; Lin, Y.-C.; Hsing, Y.-I.C.; Huang, A.H. Oil bodies and oleosins in Physcomitrella possess characteristics representative of early trends in evolution. Plant Physiol. 2009, 150, 1192–1203. [Google Scholar] [CrossRef] [PubMed]
- Huang, A. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 1996, 110, 1055. [Google Scholar] [CrossRef] [PubMed]
- Vieler, A.; Brubaker, S.B.; Vick, B.; Benning, C. A lipid droplet protein of Nannochloropsis with functions partially analogous to plant oleosins. Plant Physiol. 2012, 158, 1562–1569. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.-L.; Huang, M.-D.; Chen, T.-L.L.; Huang, A.H. Oleosin of subcellular lipid droplets evolved in green algae. Plant Physiol. 2013, 161, 1862–1874. [Google Scholar] [CrossRef] [PubMed]
- Nham Tran, T.L.; Miranda, A.F.; Gupta, A.; Puri, M.; Ball, A.S.; Adhikari, B.; Mouradov, A. The Nutritional and Pharmacological Potential of New Australian Thraustochytrids Isolated from Mangrove Sediments. Mar. Drugs 2020, 18, 151. [Google Scholar] [CrossRef]
- Ischebeck, T. Lipids in pollen—They are different. Biochim. Biophys. Acta (Bba)-Mol. Cell Biol. Lipids 2016, 1861, 1315–1328. [Google Scholar] [CrossRef] [PubMed]
- Gidda, S.K.; Park, S.; Pyc, M.; Yurchenko, O.; Cai, Y.; Wu, P.; Andrews, D.W.; Chapman, K.D.; Dyer, J.M.; Mullen, R.T. Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol. 2016, 170, 2052–2071. [Google Scholar] [CrossRef]
- Bhatla, S.; Kaushik, V.; Yadav, M. Use of oil bodies and oleosins in recombinant protein production and other biotechnological applications. Biotechnol. Adv. 2010, 28, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, E. Food emulsions and foams: Stabilization by particles. Curr. Opin. Colloid Interface Sci. 2010, 15, 40–49. [Google Scholar] [CrossRef]
- Menegazzo, M.L.; Fonseca, G.G. Biomass recovery and lipid extraction processes for microalgae biofuels production: A review. Renew. Sustain. Energy Rev. 2019, 107, 87–107. [Google Scholar] [CrossRef]
- Tzen, J.; Huang, A. Surface structure and properties of plant seed oil bodies. J. Cell Biol. 1992, 117, 327–335. [Google Scholar] [CrossRef]
- Laibach, N.; Post, J.; Twyman, R.M.; Gronover, C.S.; Prüfer, D. The characteristics and potential applications of structural lipid droplet proteins in plants. J. Biotechnol. 2015, 201, 15–27. [Google Scholar] [CrossRef]
- Bouvier-Navé, P.; Berna, A.; Noiriel, A.; Compagnon, V.; Carlsson, A.S.; Banas, A.; Stymne, S.; Schaller, H. Involvement of the phospholipid sterol acyltransferase1 in plant sterol homeostasis and leaf senescence. Plant Physiol. 2010, 152, 107–119. [Google Scholar] [CrossRef]
- Eizadora, T.Y.; Zendejas, F.J.; Lane, P.D.; Gaucher, S.; Simmons, B.A.; Lane, T.W. Triacylglycerol accumulation and profiling in the model diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum (Baccilariophyceae) during starvation. J. Appl. Phycol. 2009, 21, 669. [Google Scholar]
- Huang, A.H. Oil bodies and oleosins in seeds. Annu. Rev. Plant Biol. 1992, 43, 177–200. [Google Scholar] [CrossRef]
- Purkrtova, Z.; Jolivet, P.; Miquel, M.; Chardot, T. Structure and function of seed lipid body-associated proteins. Comptes Rendus Biol. 2008, 331, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Nikiforidis, C.V. Structure and functions of oleosomes (oil bodies). Adv. Colloid Interface Sci. 2019, 274, 102039. [Google Scholar] [CrossRef]
- Tzen, J.T.; Cao, Y.; Laurent, P.; Ratnayake, C.; Huang, A.H. Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiol. 1993, 101, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Iwanaga, D.; Gray, D.A.; Fisk, I.D.; Decker, E.A.; Weiss, J.; McClements, D.J. Extraction and characterization of oil bodies from soy beans: A natural source of pre-emulsified soybean oil. J. Agric. Food Chem. 2007, 55, 8711–8716. [Google Scholar] [CrossRef]
- Jacks, T.; Hensarling, T.; Neucere, J.; Yatsu, L.; Barker, R. Isolation and physicochemical characterization of the half-unit membranes of oilseed lipid bodies. J. Am. Oil Chem. Soc. 1990, 67, 353–361. [Google Scholar] [CrossRef]
- White, D.; Fisk, I.; Mitchell, J.; Wolf, B.; Hill, S.; Gray, D. Sunflower-seed oil body emulsions: Rheology and stability assessment of a natural emulsion. Food Hydrocoll. 2008, 22, 1224–1232. [Google Scholar] [CrossRef]
- Dave, A.C.; Ye, A.; Singh, H. Structural and interfacial characteristics of oil bodies in coconuts (Cocos nucifera L.). Food Chem. 2019, 276, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Slack, C.; Bertaud, W.; Shaw, B.; Holland, R.; Browse, J.; Wright, H. Some studies on the composition and surface properties of oil bodies from the seed cotyledons of safflower (Carthamus tinctorius) and linseed (Linum ustatissimum). Biochem. J. 1980, 190, 551–561. [Google Scholar] [CrossRef]
- Davidi, L.; Katz, A.; Pick, U. Characterization of major lipid droplet proteins from Dunaliella. Planta 2012, 236, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Nojima, D.; Yoshino, T.; Maeda, Y.; Tanaka, M.; Nemoto, M.; Tanaka, T. Proteomics analysis of oil body-associated proteins in the oleaginous diatom. J. Proteome Res. 2013, 12, 5293–5301. [Google Scholar] [CrossRef] [PubMed]
- Adrio, J.L. Oleaginous yeasts: Promising platforms for the production of oleochemicals and biofuels. Biotechnol. Bioeng. 2017, 114, 1915–1920. [Google Scholar] [CrossRef]
- Baud, S.; Boutin, J.-P.; Miquel, M.; Lepiniec, L.; Rochat, C. An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 2002, 40, 151–160. [Google Scholar] [CrossRef]
- Browse, J.; Warwick, N.; Somerville, C.; Slack, C. Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16: 3′plant Arabidopsis thaliana. Biochem. J. 1986, 235, 25–31. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, S.; Liu, H.; Shen, H.; Lin, X.; Yang, F.; Zhou, Y.J.; Jin, G.; Ye, M.; Zou, H. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat. Commun. 2012, 3, 1112. [Google Scholar] [CrossRef] [PubMed]
- Klug, L.; Daum, G. Yeast lipid metabolism at a glance. FEMS Yeast Res. 2014, 14, 369–388. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Beisson, F.; Riekhof, W. Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J. 2015, 82, 504–522. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Sommerfeld, M.; Jarvis, E.; Ghirardi, M.; Posewitz, M.; Seibert, M.; Darzins, A. Microalgal triacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J. 2008, 54, 621–639. [Google Scholar] [CrossRef] [PubMed]
- Ferrier, D.R. Biochemistry; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2014. [Google Scholar]
- Yao, L.; Shen, H.; Wang, N.; Tatlay, J.; Li, L.; Tan, T.W.; Lee, Y.K. Elevated acetyl-CoA by amino acid recycling fuels microalgal neutral lipid accumulation in exponential growth phase for biofuel production. Plant Biotechnol. J. 2017, 15, 497–509. [Google Scholar] [CrossRef]
- Oliver, D.J.; Nikolau, B.J.; Wurtele, E.S. Acetyl-CoA—Life at the metabolic nexus. Plant Sci. 2009, 176, 597–601. [Google Scholar] [CrossRef]
- Baud, S.; Lepiniec, L. Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 2010, 49, 235–249. [Google Scholar] [CrossRef]
- Weselake, R.J.; Taylor, D.C.; Rahman, M.H.; Shah, S.; Laroche, A.; McVetty, P.B.; Harwood, J.L. Increasing the flow of carbon into seed oil. Biotechnol. Adv. 2009, 27, 866–878. [Google Scholar] [CrossRef]
- Chang, L.; Lu, H.; Chen, H.; Tang, X.; Zhao, J.; Zhang, H.; Chen, Y.; Chen, W. Lipid metabolism research in oleaginous fungus Mortierella alpina: Current progress and future prospects. Biotechnol. Adv. 2021, 54, 107794. [Google Scholar] [CrossRef] [PubMed]
- Leyland, B.; Boussiba, S.; Khozin-Goldberg, I. A review of diatom lipid droplets. Biology 2020, 9, 38. [Google Scholar] [CrossRef]
- Wilfling, F.; Haas, J.T.; Walther, T.C.; Farese Jr, R.V. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 2014, 29, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Wilfling, F.; Wang, H.; Haas, J.T.; Krahmer, N.; Gould, T.J.; Uchida, A.; Cheng, J.-X.; Graham, M.; Christiano, R.; Fröhlich, F. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell 2013, 24, 384–399. [Google Scholar] [CrossRef]
- Huang, A.H. Plant lipid droplets and their associated proteins: Potential for rapid advances. Plant Physiol. 2018, 176, 1894–1918. [Google Scholar] [CrossRef] [PubMed]
- Nham Tran, T.L.; Miranda, A.F.; Mouradov, A.; Adhikari, B. Physicochemical characteristics of protein isolated from thraustochytrid oilcake. Foods 2020, 9, 779. [Google Scholar] [CrossRef]
- Kapchie, V.N.; Yao, L.; Hauck, C.C.; Wang, T.; Murphy, P.A. Oxidative stability of soybean oil in oleosomes as affected by pH and iron. Food Chem. 2013, 141, 2286–2293. [Google Scholar] [CrossRef]
- Bair, C.; Snyder, H. Electron microscopy of soybean lipid bodies. J. Am. Oil Chem. Soc. 1980, 57, 279–282. [Google Scholar] [CrossRef]
- Sukhotu, R.; Guo, S.; Xing, J.; Hu, Q.; Wang, R.; Shi, X.; Nishinari, K.; Fang, Y.; Guo, S. Changes in physiochemical properties and stability of peanut oil body emulsions by applying gum arabic. LWT-Food Sci. Technol. 2016, 68, 432–438. [Google Scholar] [CrossRef]
- Sukhotu, R.; Shi, X.; Hu, Q.; Nishinari, K.; Fang, Y.; Guo, S. Aggregation behaviour and stability of maize germ oil body suspension. Food Chem. 2014, 164, 1–6. [Google Scholar] [CrossRef]
- Zhao, D.; Li, T.; Li, Z.; Sun, J.; Tao, J. Characteristics of Paeonia ostii seed oil body and OLE17. 5 determining oil body morphology. Food Chem. 2020, 319, 126548. [Google Scholar] [CrossRef]
- De Chirico, S.; Di Bari, V.; Guzmán, M.J.R.; Nikiforidis, C.V.; Foster, T.; Gray, D. Assessment of rapeseed oil body (oleosome) lipolytic activity as an effective predictor of emulsion purity and stability. Food Chem. 2020, 316, 126355. [Google Scholar] [CrossRef] [PubMed]
- Vechtel, B.; Eichenberger, W.; Ruppel, G. Lipid bodies in Eremosphaera viridis De Bary (Chlorophyceae). Plant Cell Physiol. 1992, 33, 41–48. [Google Scholar]
- Lin, I.-P.; Jiang, P.-L.; Chen, C.-S.; Tzen, J.T. A unique caleosin serving as the major integral protein in oil bodies isolated from Chlorella sp. cells cultured with limited nitrogen. Plant Physiol. Biochem. 2012, 61, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Moellering, E.R.; Benning, C. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 2010, 9, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hao, T.-B.; Balamurugan, S.; Yang, W.-D.; Liu, J.-S.; Dong, H.-P.; Li, H.-Y. A lipid droplet-associated protein involved in lipid droplet biogenesis and triacylglycerol accumulation in the oleaginous microalga Phaeodactylum tricornutum. Algal Res. 2017, 26, 215–224. [Google Scholar] [CrossRef]
- Zienkiewicz, K.; Du, Z.-Y.; Ma, W.; Vollheyde, K.; Benning, C. Stress-induced neutral lipid biosynthesis in microalgae—Molecular, cellular and physiological insights. Biochim. Biophys. Acta (Bba)-Mol. Cell Biol. Lipids 2016, 1861, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-Y.; Huang, A.H. Unique motifs and length of hairpin in oleosin target the cytosolic side of endoplasmic reticulum and budding lipid droplet. Plant Physiol. 2017, 174, 2248–2260. [Google Scholar] [CrossRef]
- Le Lay, S.; Dugail, I. Connecting lipid droplet biology and the metabolic syndrome. Prog. Lipid Res. 2009, 48, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhao, L.; Ying, Y.; Kong, X.; Hua, Y.; Chen, Y. The characterization of soybean oil body integral oleosin isoforms and the effects of alkaline pH on them. Food Chem. 2015, 177, 288–294. [Google Scholar] [CrossRef]
- Gschloessl, B.; Guermeur, Y.; Cock, J.M. HECTAR: A method to predict subcellular targeting in heterokonts. BMC Bioinform. 2008, 9, 393. [Google Scholar] [CrossRef]
- HaileMariam, M.; Eguez, R.V.; Singh, H.; Bekele, S.; Ameni, G.; Pieper, R.; Yu, Y. S-Trap, an ultrafast sample-preparation approach for shotgun proteomics. J. Proteome Res. 2018, 17, 2917–2924. [Google Scholar] [CrossRef] [PubMed]
- Zougman, A.; Selby, P.J.; Banks, R.E. Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics 2014, 14, 1006–1000. [Google Scholar] [CrossRef]
- Qu, R.; Wang, S.; Lin, Y.; Vance, V.B.; Huang, A. Characteristics and biosynthesis of membrane proteins of lipid bodies in the scutella of maize (Zea mays L.). Biochem. J. 1986, 235, 57–65. [Google Scholar] [CrossRef]
- Huang, M.-D.; Huang, A.H. Bioinformatics reveal five lineages of oleosins and the mechanism of lineage evolution related to structure/function from green algae to seed plants. Plant Physiol. 2015, 169, 453–470. [Google Scholar] [CrossRef]
- Bourgeois, C.; Gomaa, A.I.; Lefèvre, T.; Cansell, M.; Subirade, M. Interaction of oil bodies proteins with phospholipid bilayers: A molecular level elucidation as revealed by infrared spectroscopy. Int. J. Biol. Macromol. 2019, 122, 873–881. [Google Scholar] [CrossRef]
- Song, W.; Qin, Y.; Zhu, Y.; Yin, G.; Wu, N.; Li, Y.; Hu, Y. Delineation of plant caleosin residues critical for functional divergence, positive selection and coevolution. BMC Evol. Biol. 2014, 14, 124. [Google Scholar] [CrossRef]
- Lin, L.-J.; Tai, S.S.; Peng, C.-C.; Tzen, J.T. Steroleosin, a sterol-binding dehydrogenase in seed oil bodies. Plant Physiol. 2002, 128, 1200–1211. [Google Scholar] [CrossRef]
- Pasaribu, B.; Chung, T.-y.; Chen, C.-S.; Jiang, P.-L.; Tzen, J.T. Identification of steroleosin in oil bodies of pine megagametophytes. Plant Physiol. Biochem. 2016, 101, 173–181. [Google Scholar] [CrossRef]
- Shimada, T.L.; Takano, Y.; Shimada, T.; Fujiwara, M.; Fukao, Y.; Mori, M.; Okazaki, Y.; Saito, K.; Sasaki, R.; Aoki, K. Leaf oil body functions as a subcellular factory for the production of a phytoalexin in Arabidopsis. Plant Physiol. 2014, 164, 105–118. [Google Scholar] [CrossRef] [PubMed]
- López-Ribera, I.; La Paz, J.L.; Repiso, C.; García, N.; Miquel, M.; Hernández, M.L.; Martínez-Rivas, J.M.; Vicient, C.M. The evolutionary conserved oil body associated protein OBAP1 participates in the regulation of oil body size. Plant Physiol. 2014, 164, 1237–1249. [Google Scholar] [CrossRef]
- James, C.N.; Horn, P.J.; Case, C.R.; Gidda, S.K.; Zhang, D.; Mullen, R.T.; Dyer, J.M.; Anderson, R.G.; Chapman, K.D. Disruption of the Arabidopsis CGI-58 homologue produces Chanarin–Dorfman-like lipid droplet accumulation in plants. Proc. Natl. Acad. Sci. USA 2010, 107, 17833–17838. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Gidda, S.K.; James, C.N.; Horn, P.J.; Khuu, N.; Seay, D.C.; Keereetaweep, J.; Chapman, K.D.; Mullen, R.T.; Dyer, J.M. The α/β hydrolase CGI-58 and peroxisomal transport protein PXA1 coregulate lipid homeostasis and signaling in Arabidopsis. Plant Cell 2013, 25, 1726–1739. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Goodman, J.M.; Pyc, M.; Mullen, R.T.; Dyer, J.M.; Chapman, K.D. Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell 2015, 27, 2616–2636. [Google Scholar] [CrossRef] [PubMed]
- Thazar-Poulot, N.; Miquel, M.; Fobis-Loisy, I.; Gaude, T. Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Proc. Natl. Acad. Sci. USA 2015, 112, 4158–4163. [Google Scholar] [CrossRef] [PubMed]
- Zaaboul, F.; Raza, H.; Cao, C.; Yuanfa, L. The impact of roasting, high pressure homogenization and sterilization on peanut milk and its oil bodies. Food Chem. 2019, 280, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Vishwanathan, K.; Singh, V.; Subramanian, R. Wet grinding characteristics of soybean for soymilk extraction. J. Food Eng. 2011, 106, 28–34. [Google Scholar] [CrossRef]
- Romero-Guzmán, M.; Jung, L.; Kyriakopoulou, K.; Boom, R.; Nikiforidis, C. Efficient single-step rapeseed oleosome extraction using twin-screw press. J. Food Eng. 2020, 276, 109890. [Google Scholar] [CrossRef]
- Geciova, J.; Bury, D.; Jelen, P. Methods for disruption of microbial cells for potential use in the dairy industry—A review. Int. Dairy J. 2002, 12, 541–553. [Google Scholar] [CrossRef]
- Kapchie, V.N.; Wei, D.; Hauck, C.; Murphy, P.A. Enzyme-assisted aqueous extraction of oleosomes from soybeans (Glycine max). J. Agric. Food Chem. 2008, 56, 1766–1771. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Yin, J.; Gao, Z.; Huang, H.; Ji, X.; Dou, C. Disruption of Chlorella vulgaris cells for the release of biodiesel-producing lipids: A comparison of grinding, ultrasonication, bead milling, enzymatic lysis, and microwaves. Appl. Biochem. Biotechnol. 2011, 164, 1215–1224. [Google Scholar] [CrossRef]
- Halim, R.; Danquah, M.K.; Webley, P.A. Extraction of oil from microalgae for biodiesel production: A review. Biotechnol. Adv. 2012, 30, 709–732. [Google Scholar] [CrossRef] [PubMed]
- Rawat, I.; Kumar, R.R.; Mutanda, T.; Bux, F. Biodiesel from microalgae: A critical evaluation from laboratory to large scale production. Appl. Energy 2013, 103, 444–467. [Google Scholar] [CrossRef]
- Chisti, Y.; Moo-Young, M. Disruption of microbial cells for intracellular products. Enzym. Microb. Technol. 1986, 8, 194–204. [Google Scholar] [CrossRef]
- Halim, R.; Rupasinghe, T.W.; Tull, D.L.; Webley, P.A. Mechanical cell disruption for lipid extraction from microalgal biomass. Bioresour. Technol. 2013, 140, 53–63. [Google Scholar] [CrossRef]
- Dong, T.; Knoshaug, E.P.; Pienkos, P.T.; Laurens, L.M. Lipid recovery from wet oleaginous microbial biomass for biofuel production: A critical review. Appl. Energy 2016, 177, 879–895. [Google Scholar] [CrossRef]
- Grimi, N.; Dubois, A.; Marchal, L.; Jubeau, S.; Lebovka, N.; Vorobiev, E. Selective extraction from microalgae Nannochloropsis sp. using different methods of cell disruption. Bioresour. Technol. 2014, 153, 254–259. [Google Scholar] [CrossRef]
- Drira, N.; Piras, A.; Rosa, A.; Porcedda, S.; Dhaouadi, H. Microalgae from domestic wastewater facility’s high rate algal pond: Lipids extraction, characterization and biodiesel production. Bioresour. Technol. 2016, 206, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.S.; Parameswaran, P.; Li, A.; Baez, M.; Rittmann, B.E. Effects of pulsed electric field treatment on enhancing lipid recovery from the microalga, Scenedesmus. Bioresour. Technol. 2014, 173, 457–461. [Google Scholar] [CrossRef]
- Roux, J.-M.; Lamotte, H.; Achard, J.-L. An overview of microalgae lipid extraction in a biorefinery framework. Energy Procedia 2017, 112, 680–688. [Google Scholar] [CrossRef]
- Miranda, J.; Passarinho, P.C.; Gouveia, L. Pre-treatment optimization of Scenedesmus obliquus microalga for bioethanol production. Bioresour. Technol. 2012, 104, 342–348. [Google Scholar] [CrossRef]
- Neto, A.M.P.; de Souza, R.A.S.; Leon-Nino, A.D.; da Costa, J.D.a.A.; Tiburcio, R.S.; Nunes, T.A.; de Mello, T.C.S.; Kanemoto, F.T.; Saldanha-Corrêa, F.M.P.; Gianesella, S.M.F. Improvement in microalgae lipid extraction using a sonication-assisted method. Renew. Energy 2013, 55, 525–531. [Google Scholar] [CrossRef]
- Bundhoo, Z.M. Microwave-assisted conversion of biomass and waste materials to biofuels. Renew. Sustain. Energy Rev. 2018, 82, 1149–1177. [Google Scholar] [CrossRef]
- Silve, A.; Papachristou, I.; Wüstner, R.; Sträßner, R.; Schirmer, M.; Leber, K.; Guo, B.; Interrante, L.; Posten, C.; Frey, W. Extraction of lipids from wet microalga Auxenochlorella protothecoides using pulsed electric field treatment and ethanol-hexane blends. Algal Res. 2018, 29, 212–222. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, Y.; Chen, Y.; Kong, X.; Hua, Y. Effects of pH on protein components of extracted oil bodies from diverse plant seeds and endogenous protease-induced oleosin hydrolysis. Food Chem. 2016, 200, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ono, T. Simple extraction method of non-allergenic intact soybean oil bodies that are thermally stable in an aqueous medium. J. Agric. Food Chem. 2010, 58, 7402–7407. [Google Scholar] [CrossRef]
- Kapchie, V.N.; Towa, L.T.; Hauck, C.; Murphy, P.A. Evaluation of enzyme efficiency for soy oleosome isolation and ultrastructural aspects. Food Res. Int. 2010, 43, 241–247. [Google Scholar] [CrossRef]
- Maffei, G.; Bracciale, M.P.; Broggi, A.; Zuorro, A.; Santarelli, M.L.; Lavecchia, R. Effect of an enzymatic treatment with cellulase and mannanase on the structural properties of Nannochloropsis microalgae. Bioresour. Technol. 2018, 249, 592–598. [Google Scholar] [CrossRef]
- Taher, H.; Al-Zuhair, S.; Al-Marzouqi, A.H.; Haik, Y.; Farid, M. Effective extraction of microalgae lipids from wet biomass for biodiesel production. Biomass Bioenergy 2014, 66, 159–167. [Google Scholar] [CrossRef]
- Chen, B.; McClements, D.J.; Gray, D.A.; Decker, E.A. Physical and oxidative stability of pre-emulsified oil bodies extracted from soybeans. Food Chem. 2012, 132, 1514–1520. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, Y.; Yan, Z.; Kong, X.; Hua, Y. Physicochemical and rheological properties and oxidative stability of oil bodies recovered from soybean aqueous extract at different pHs. Food Hydrocoll. 2016, 61, 685–694. [Google Scholar] [CrossRef]
- Hou, J.; Feng, X.; Jiang, M.; Wang, Q.; Cui, C.; Sun, C.; Hussain, M.A.; Jiang, L.; Jiang, Z.; Li, A. Effect of NaCl on oxidative stability and protein properties of oil bodies from different oil crops. LWT 2019, 113, 108263. [Google Scholar] [CrossRef]
- Mantzouridou, F.T.; Naziri, E.; Kyriakidou, A.; Paraskevopoulou, A.; Tsimidou, M.Z.; Kiosseoglou, V. Oil bodies from dry maize germ as an effective replacer of cow milk fat globules in yogurt-like product formulation. LWT 2019, 105, 48–56. [Google Scholar] [CrossRef]
- Nikiforidis, C.V.; Biliaderis, C.G.; Kiosseoglou, V. Rheological characteristics and physicochemical stability of dressing-type emulsions made of oil bodies–egg yolk blends. Food Chem. 2012, 134, 64–73. [Google Scholar] [CrossRef]
- Yi, S.; Yang, J.; Huang, J.; Guan, L.; Du, L.; Guo, Y.; Zhai, F.; Wang, Y.; Lu, Z.; Wang, L. Expression of bioactive recombinant human fibroblast growth factor 9 in oil bodies of Arabidopsis thaliana. Protein Expr. Purif. 2015, 116, 127–132. [Google Scholar] [CrossRef]
- Van Rooijen, G.J.; Motoney, M.M. Plant seed oil-bodies as carriers for foreign proteins. Bio/Technol 1995, 13, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Wen, R.; Li, W.; Wang, X.; Tian, H.; Yi, S.; Zhang, L.; Li, X.; Jiang, C.; Li, H. Oil body bound oleosin-rhFGF9 fusion protein expressed in safflower (Carthamus tinctorius L.) stimulates hair growth and wound healing in mice. BMC Biotechnol. 2018, 18, 1–11. [Google Scholar] [CrossRef]
- Schryvers, A.; Hutchins, W.; Moloney, M.; Alcantra, J. Use of Plant Oil-Bodies in Vaccine Delivery Systems. U.S. Patent 10/297,585, 29 April 2004. [Google Scholar]
- Nykiforuk, C.L.; Boothe, J.G.; Murray, E.W.; Keon, R.G.; Goren, H.J.; Markley, N.A.; Moloney, M.M. Transgenic expression and recovery of biologically active recombinant human insulin from Arabidopsis thaliana seeds. Plant Biotechnol. J. 2006, 4, 77–85. [Google Scholar] [CrossRef]
- Zubair, M.; Ahmad, J. Role of growth factors and cytokines in diabetic foot ulcer healing: A detailed review. Rev. Endocr. Metab. Disord. 2019, 20, 207–217. [Google Scholar] [CrossRef]
- Shi, G.-J.; Shi, G.-R.; Zhou, J.-y.; Zhang, W.-j.; Gao, C.-y.; Jiang, Y.-p.; Zi, Z.-G.; Zhao, H.-h.; Yang, Y.; Yu, J.-Q. Involvement of growth factors in diabetes mellitus and its complications: A general review. Biomed. Pharmacother. 2018, 101, 510–527. [Google Scholar] [CrossRef]
- Zafar, M.I.; Mills, K.; Ye, X.; Blakely, B.; Min, J.; Kong, W.; Zhang, N.; Gou, L.; Regmi, A.; Hu, S.Q. Association between the expression of vascular endothelial growth factors and metabolic syndrome or its components: A systematic review and meta-analysis. Diabetol. Metab. Syndr. 2018, 10, 1–17. [Google Scholar] [CrossRef]
- Li, W.; Li, L.; Li, K.; Lin, J.; Sun, X.; Tang, K. Expression of biologically active human insulin-like growth factor 1 in Arabidopsis thaliana seeds via oleosin fusion technology. Biotechnol. Appl. Biochem. 2011, 58, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yang, J.; Guan, L.; Yi, S.; Du, L.; Tian, H.; Guo, Y.; Zhai, F.; Lu, Z.; Li, H. Expression of bioactive recombinant human fibroblast growth factor 10 in Carthamus tinctorius L. seeds. Protein Expr. Purif. 2017, 138, 7–12. [Google Scholar] [CrossRef]
- Qiang, W.; Feng, X.; Li, Y.; Lan, X.; Ji, K.; Sun, X.; Chen, X.; Li, H.; Du, L.; Yang, J. Expression of a functional recombinant vascular endothelial growth factor 165 (VEGF165) in Arabidopsis thaliana. Turk. J. Biochem. 2019, 44, 254–260. [Google Scholar] [CrossRef]
- Guo, Y.; Li, Y.; Wu, Q.; Lan, X.; Chu, G.; Qiang, W.; Noman, M.; Gao, T.; Guo, J.; Han, L. Optimization of the extraction conditions and dermal toxicity of oil body fused with acidic fibroblast growth factor (OLAF). Cutan. Ocul. Toxicol. 2021, 40, 221–231. [Google Scholar] [CrossRef]
- Lundquist, P.K.; Shivaiah, K.-K.; Espinoza-Corral, R. Lipid droplets throughout the evolutionary tree. Prog. Lipid Res. 2020, 78, 101029. [Google Scholar] [CrossRef]
- Allen, D.; Ruan, C.-H.; King, B.; Ruan, K.-H. Recent advances and near future of insulin production and therapy. Future Med. Chem. 2019, 11, 1513–1517. [Google Scholar] [CrossRef] [PubMed]
- Schillberg, S.; Raven, N.; Spiegel, H.; Rasche, S.; Buntru, M. Critical analysis of the commercial potential of plants for the production of recombinant proteins. Front. Plant Sci. 2019, 10, 720. [Google Scholar] [CrossRef]
- Abookazemi, K.; Jalali, J.M.; Mohebodini, M.; Vaseghi, A. Transfer of human proinsulin gene into Cucumber (Cucumis sativus L.) via agrobacterium method. Genetika 2017, 49, 717–728. [Google Scholar] [CrossRef]
- Kashani, K.; Javaran, M.J.; Mohebodini, M.; Moieni, A.; Deh Abadi, M.S. Regeneration and ‘Agrobacterium’-mediated transformation of three potato cultivars (‘Solanum tuberosum’ cv. Desiree, agria and marfona) by human proinsulin gene. Aust. J. Crop Sci. 2012, 6, 1212–1220. [Google Scholar]
- Shanmugaraj, B.; Bulaon, C.J.I.; Phoolcharoen, W. Plant molecular farming: A viable platform for recombinant biopharmaceutical production. Plants 2020, 9, 842. [Google Scholar] [CrossRef]
- McNulty, M.J.; Silberstein, D.Z.; Kuhn, B.T.; Padgett, H.S.; Nandi, S.; McDonald, K.A.; Cross, C.E. Alpha-1 antitrypsin deficiency and recombinant protein sources with focus on plant sources: Updates, challenges and perspectives. Free Radic. Biol. Med. 2021, 163, 10–30. [Google Scholar] [CrossRef] [PubMed]
- Kjeldsen, T.; Balschmidt, P.; Diers, I.; Hach, M.; Kaarsholm, N.C.; Ludvigsen, S. Expression of insulin in yeast: The importance of molecular adaptation for secretion and conversion. Biotechnol. Genet. Eng. Rev. 2001, 18, 89–121. [Google Scholar] [CrossRef]
- Tenchov, R.; Bird, R.; Curtze, A.E.; Zhou, Q. Lipid Nanoparticles—From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15, 16982–17015. [Google Scholar] [CrossRef]
- Maherani, B.; Arab-Tehrany, E.; Mozafari, M.R.; Gaiani, C.; Linder, M. Liposomes: A review of manufacturing techniques and targeting strategies. Curr. Nanosci. 2011, 7, 436–452. [Google Scholar] [CrossRef]
- Machado, A.R.; de Assis, L.M.; Machado, M.I.R.; de Souza-Soares, L.A. Importance of lecithin for encapsulation processes. Afr. J. Food Sci. 2014, 8, 176–183. [Google Scholar]
- Pattni, B.S.; Chupin, V.V.; Torchilin, V.P. New developments in liposomal drug delivery. Chem. Rev. 2015, 115, 10938–10966. [Google Scholar] [CrossRef]
- Deshpande, P.P.; Biswas, S.; Torchilin, V.P. Current trends in the use of liposomes for tumor targeting. Nanomedicine 2013, 8, 1509–1528. [Google Scholar] [CrossRef]
- Weissmann, G.; Bloomgarden, D.; Kaplan, R.; Cohen, C.; Hoffstein, S.; Collins, T.; Gotlieb, A.; Nagle, D. A general method for the introduction of enzymes, by means of immunoglobulin-coated liposomes, into lysosomes of deficient cells. Proc. Natl. Acad. Sci. USA 1975, 72, 88–92. [Google Scholar] [CrossRef]
- Li, Y.; Tenchov, R.; Smoot, J.; Liu, C.; Watkins, S.; Zhou, Q. A comprehensive review of the global efforts on COVID-19 vaccine development. ACS Cent. Sci. 2021, 7, 512–533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, W.; Nguyen, G.N.; Zhang, C.; Zeng, C.; Yan, J.; Du, S.; Hou, X.; Li, W.; Jiang, J. Functionalized lipid-like nanoparticles for in vivo mRNA delivery and base editing. Sci. Adv. 2020, 6, eabc2315. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.E.; Gargano, J.W.; Marin, M.; Wallace, M.; Curran, K.G.; Chamberland, M.; McClung, N.; Campos-Outcalt, D.; Morgan, R.L.; Mbaeyi, S. The advisory committee on immunization practices’ interim recommendation for use of Pfizer-BioNTech COVID-19 vaccine—United States, December 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 1922. [Google Scholar] [CrossRef]
- Deckers, H.M.; Van Rooijen, G.; Boothe, J.; Goll, J.; Moloney, M.M.; Schryvers, A.B.; Alcantara, J.; Hutchins, W.A. Immunogenic Formulations Comprising Oil Bodies. U.S. Patent No 6,761,914, 13 July 2004. [Google Scholar]
- Higuera-Ciapara, I.; Felix-Valenzuela, L.; Goycoolea, F. Astaxanthin: A review of its chemistry and applications. Crit. Rev. Food Sci. Nutr. 2006, 46, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Ambati, R.R.; Phang, S.-M.; Ravi, S.; Aswathanarayana, R.G. Astaxanthin: Sources, extraction, stability, biological activities and its commercial applications—A review. Mar. Drugs 2014, 12, 128–152. [Google Scholar] [CrossRef]
- Rodriguez-Ruiz, V.; Salatti-Dorado, J.Á.; Barzegari, A.; Nicolas-Boluda, A.; Houaoui, A.; Caballo, C.; Caballero-Casero, N.; Sicilia, D.; Bastias Venegas, J.; Pauthe, E. Astaxanthin-loaded nanostructured lipid carriers for preservation of antioxidant activity. Molecules 2018, 23, 2601. [Google Scholar] [CrossRef]
- Park, J.S.; Chyun, J.H.; Kim, Y.K.; Line, L.L.; Chew, B.P. Astaxanthin decreased oxidative stress and inflammation and enhanced immune response in humans. Nutr. Metab. 2010, 7, 1–10. [Google Scholar] [CrossRef]
- Sangsuriyawong, A.; Limpawattana, M.; Siriwan, D.; Klaypradit, W. Properties and bioavailability assessment of shrimp astaxanthin loaded liposomes. Food Sci. Biotechnol. 2019, 28, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Wang, H.; Gu, K. Nanoliposomes as vehicles for astaxanthin: Characterization, in vitro release evaluation and structure. Molecules 2018, 23, 2822. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Zhang, S.; Gu, K.; Zhang, N. Preparation of astaxanthin-loaded liposomes: Characterization, storage stability and antioxidant activity. Cyta-J. Food 2018, 16, 607–618. [Google Scholar] [CrossRef]
- Hama, S.; Uenishi, S.; Yamada, A.; Ohgita, T.; Tsuchiya, H.; Yamashita, E.; Kogure, K. Scavenging of hydroxyl radicals in aqueous solution by astaxanthin encapsulated in liposomes. Biol. Pharm. Bull. 2012, 35, 2238–2242. [Google Scholar] [CrossRef]
- Acevedo, F.; Rubilar, M.; Jofré, I.; Villarroel, M.; Navarrete, P.; Esparza, M.; Romero, F.; Vilches, E.A.; Acevedo, V.; Shene, C. Oil bodies as a potential microencapsulation carrier for astaxanthin stabilisation and safe delivery. J. Microencapsul. 2014, 31, 488–500. [Google Scholar] [CrossRef]
- Demirbas, A. Biofuels sources, biofuel policy, biofuel economy and global biofuel projections. Energy Convers. Manag. 2008, 49, 2106–2116. [Google Scholar] [CrossRef]
- Sati, H.; Mitra, M.; Mishra, S.; Baredar, P. Microalgal lipid extraction strategies for biodiesel production: A review. Algal Res. 2019, 38, 101413. [Google Scholar] [CrossRef]
- Mueller, S.P.; Krause, D.M.; Mueller, M.J.; Fekete, A. Accumulation of extra-chloroplastic triacylglycerols in Arabidopsis seedlings during heat acclimation. J. Exp. Bot. 2015, 66, 4517–4526. [Google Scholar] [CrossRef]
- Xu, K.; Zou, X.; Wen, H.; Xue, Y.; Qu, Y.; Li, Y. Effects of multi-temperature regimes on cultivation of microalgae in municipal wastewater to simultaneously remove nutrients and produce biomass. Appl. Microbiol. Biotechnol. 2019, 103, 8255–8265. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Zou, X.; Wen, H.; Xue, Y.; Zhao, S.; Li, Y. Buoy-bead flotation harvesting of the microalgae Chlorella vulgaris using surface-layered polymeric microspheres: A novel approach. Bioresour. Technol. 2018, 267, 341–346. [Google Scholar] [CrossRef]
- Lee, C.S.; Oh, H.-S.; Oh, H.-M.; Kim, H.-S.; Ahn, C.-Y. Two-phase photoperiodic cultivation of algal–bacterial consortia for high biomass production and efficient nutrient removal from municipal wastewater. Bioresour. Technol. 2016, 200, 867–875. [Google Scholar] [CrossRef] [PubMed]
- Taleb, A.; Pruvost, J.; Legrand, J.; Marec, H.; Le-Gouic, B.; Mirabella, B.; Legeret, B.; Bouvet, S.; Peltier, G.; Li-Beisson, Y. Development and validation of a screening procedure of microalgae for biodiesel production: Application to the genus of marine microalgae Nannochloropsis. Bioresour. Technol. 2015, 177, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.S.; Miranda, A.L.; de Morais, M.G.; Costa, J.A.V.; Druzian, J.I. Microalgae as source of polyhydroxyalkanoates (PHAs)—A review. Int. J. Biol. Macromol. 2019, 131, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Devadas, V.V.; Khoo, K.S.; Chia, W.Y.; Chew, K.W.; Munawaroh, H.S.H.; Lam, M.-K.; Lim, J.-W.; Ho, Y.-C.; Lee, K.T.; Show, P.L. Algae biopolymer towards sustainable circular economy. Bioresour. Technol. 2021, 325, 124702. [Google Scholar] [CrossRef]
- Chong, J.W.R.; Yew, G.Y.; Khoo, K.S.; Ho, S.-H.; Show, P.L. Recent advances on food waste pretreatment technology via microalgae for source of polyhydroxyalkanoates. J. Environ. Manag. 2021, 293, 112782. [Google Scholar] [CrossRef]
- Samui, A.B.; Kanai, T. Polyhydroxyalkanoates based copolymers. Int. J. Biol. Macromol. 2019, 140, 522–537. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Chen, J.-C.; Wu, Q.; Chen, G.-Q. Polyhydroxyalkanoates as a source of chemicals, polymers, and biofuels. Curr. Opin. Biotechnol. 2011, 22, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Vea, E.B.; Fabbri, S.; Spierling, S.; Owsianiak, M. Inclusion of multiple climate tipping as a new impact category in life cycle assessment of polyhydroxyalkanoate (PHA)-based plastics. Sci. Total Environ. 2021, 788, 147544. [Google Scholar] [CrossRef] [PubMed]
- Mehrpouya, M.; Vahabi, H.; Barletta, M.; Laheurte, P.; Langlois, V. Additive manufacturing of polyhydroxyalkanoates (PHAs) biopolymers: Materials, printing techniques, and applications. Mater. Sci. Eng. C 2021, 127, 112216. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.M.; Pratt, S.; Halley, P.; Richardson, D.; Werker, A.; Laycock, B.; Vandi, L.-J. Mechanical and physical stability of polyhydroxyalkanoate (PHA)-based wood plastic composites (WPCs) under natural weathering. Polym. Test. 2019, 73, 214–221. [Google Scholar] [CrossRef]
- Karan, H.; Funk, C.; Grabert, M.; Oey, M.; Hankamer, B. Green bioplastics as part of a circular bioeconomy. Trends Plant Sci. 2019, 24, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Beckstrom, B.D.; Wilson, M.H.; Crocker, M.; Quinn, J.C. Bioplastic feedstock production from microalgae with fuel co-products: A techno-economic and life cycle impact assessment. Algal Res. 2020, 46, 101769. [Google Scholar] [CrossRef]
- Anjum, A.; Zuber, M.; Zia, K.M.; Noreen, A.; Anjum, M.N.; Tabasum, S. Microbial production of polyhydroxyalkanoates (PHAs) and its copolymers: A review of recent advancements. Int. J. Biol. Macromol. 2016, 89, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Suriyamongkol, P.; Weselake, R.; Narine, S.; Moloney, M.; Shah, S. Biotechnological approaches for the production of polyhydroxyalkanoates in microorganisms and plants—A review. Biotechnol. Adv. 2007, 25, 148–175. [Google Scholar] [CrossRef]
- Norhafini, H.; Huong, K.-H.; Amirul, A. High PHA density fed-batch cultivation strategies for 4HB-rich P (3HB-co-4HB) copolymer production by transformant Cupriavidus malaysiensis USMAA1020. Int. J. Biol. Macromol. 2019, 125, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- García, G.; Sosa-Hernández, J.E.; Rodas-Zuluaga, L.I.; Castillo-Zacarías, C.; Iqbal, H.; Parra-Saldívar, R. Accumulation of PHA in the microalgae Scenedesmus sp. under nutrient-deficient conditions. Polymers 2021, 13, 131. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.S.; Miranda, A.L.; Andrade, B.B.; de Jesus Assis, D.; Souza, C.O.; de Morais, M.G.; Costa, J.A.V.; Druzian, J.I. Influence of nitrogen on growth, biomass composition, production, and properties of polyhydroxyalkanoates (PHAs) by microalgae. Int. J. Biol. Macromol. 2018, 116, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Chew, K.W.; Chia, S.R.; Show, P.L.; Ling, T.C.; Arya, S.S.; Chang, J.-S. Food waste compost as an organic nutrient source for the cultivation of Chlorella vulgaris. Bioresour. Technol. 2018, 267, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Zeller, M.A.; Hunt, R.; Jones, A.; Sharma, S. Bioplastics and their thermoplastic blends from Spirulina and Chlorella microalgae. J. Appl. Polym. Sci. 2013, 130, 3263–3275. [Google Scholar] [CrossRef]
- Singh, A.K.; Sharma, L.; Mallick, N.; Mala, J. Progress and challenges in producing polyhydroxyalkanoate biopolymers from cyanobacteria. J. Appl. Phycol. 2017, 29, 1213–1232. [Google Scholar] [CrossRef]

| Organisms | LD Size (μm) | Zeta Potential (mV) | Isoelectric Point | References | |
|---|---|---|---|---|---|
| Plants | Soybean | 0.2–0.5 | −20 to +12 | Around pH 4 | [28,54] |
| Peanuts | 0.6–5.4 | −18 to −8 | Around pH 5 | [55] | |
| Sunflower seed | 0.3–13 | −22 to −9 | pH 5–6 | [30,55] | |
| Maize | 0.95–2.55 | −16.4 to 23.3 | pH 4.6–4.8 | [56] | |
| Paeonia ostia seed | 0.4–1.2 | −50 to −35 | Na* | [57] | |
| Rapeseed | 0.2–6 | −65 to +55 | pH 5–7 | [58] | |
| Coconut | 1–20 | −33.8 to −13.0 | <pH 6.1 | [31] | |
| Microalgae | Eremosphaera viridis | 1–2.5 | Na* | Na* | [59] |
| Chlorella sp. | 0.1–5.0 | Na* | Na* | [60] | |
| Chlamydomonas reinhardtii | 1.7–2.5 | Na* | Na* | [61] | |
| Dunaliella salina | 0.5–0.8 | Na* | Na* | [33] | |
| Phaeodactylum tricornutum | 1–2.5 | Na* | Na* | [62] | |
| Thraustochytrid | 0.1–1 | −57 to +4 | Around pH 3 | [14] |
| Physical Methods | Advantages | Limitations |
|---|---|---|
| Grinding | Easy to operate | Low rupture efficiency; time-consuming |
| Pressing | Easy to set up and operate; applicable on a large scale | Low rupture efficiency |
| High-pressure homogenization | Low operational cost; possibility of upscaling; low risk of thermal degradation | High energy consumption |
| Ultrasound | Reduced extraction time | High energy consumption; lead to denaturing of the intracellular components; scaling up is not suitable |
| Microwave | Reduced extraction time; high efficiency with superior quality; possibility of upscaling | High energy costs of scaling up |
| Bead beating | Easy to operate; simplicity of the equipment | Low rupture efficiency; time-consuming; not suitable for scaling up is not suitable |
| Pulsed electric field | Low energy consumption; fast and efficient Possibility of up-scaling | Sensitive to the conductivity of the medium |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, K.; Zou, W.; Peng, B.; Guo, C.; Zou, X. Lipid Droplets from Plants and Microalgae: Characteristics, Extractions, and Applications. Biology 2023, 12, 594. https://doi.org/10.3390/biology12040594
Xu K, Zou W, Peng B, Guo C, Zou X. Lipid Droplets from Plants and Microalgae: Characteristics, Extractions, and Applications. Biology. 2023; 12(4):594. https://doi.org/10.3390/biology12040594
Chicago/Turabian StyleXu, Kaiwei, Wen Zou, Biao Peng, Chao Guo, and Xiaotong Zou. 2023. "Lipid Droplets from Plants and Microalgae: Characteristics, Extractions, and Applications" Biology 12, no. 4: 594. https://doi.org/10.3390/biology12040594
APA StyleXu, K., Zou, W., Peng, B., Guo, C., & Zou, X. (2023). Lipid Droplets from Plants and Microalgae: Characteristics, Extractions, and Applications. Biology, 12(4), 594. https://doi.org/10.3390/biology12040594





