The Effects of Intraguild Predation on Phytoplankton Assemblage Composition and Diversity: A Mesocosm Experiment
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Sampling and Processing
2.3. High-Throughput Sequencing
2.4. Statistics Analyses
3. Results
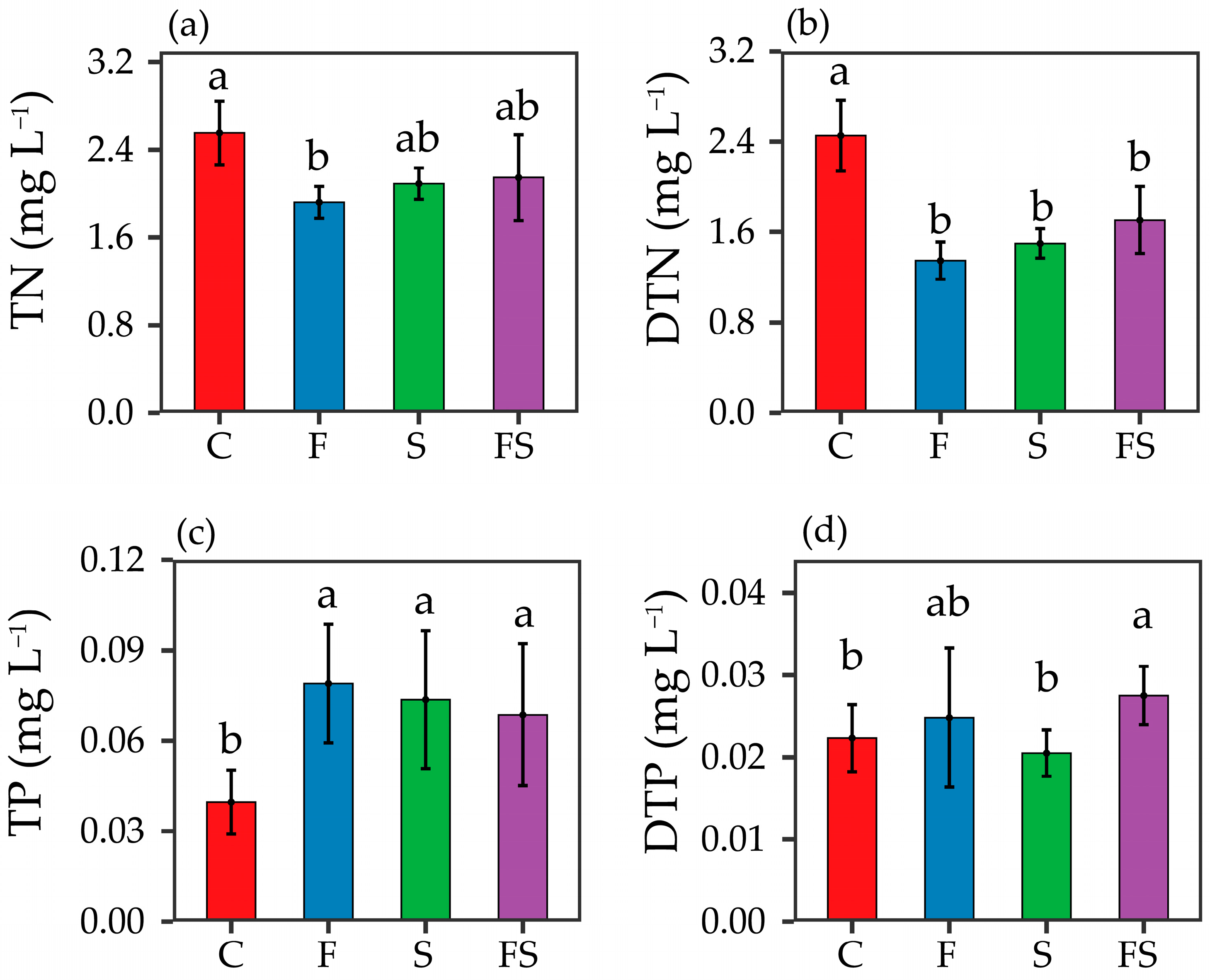
3.1. Environmental Factors
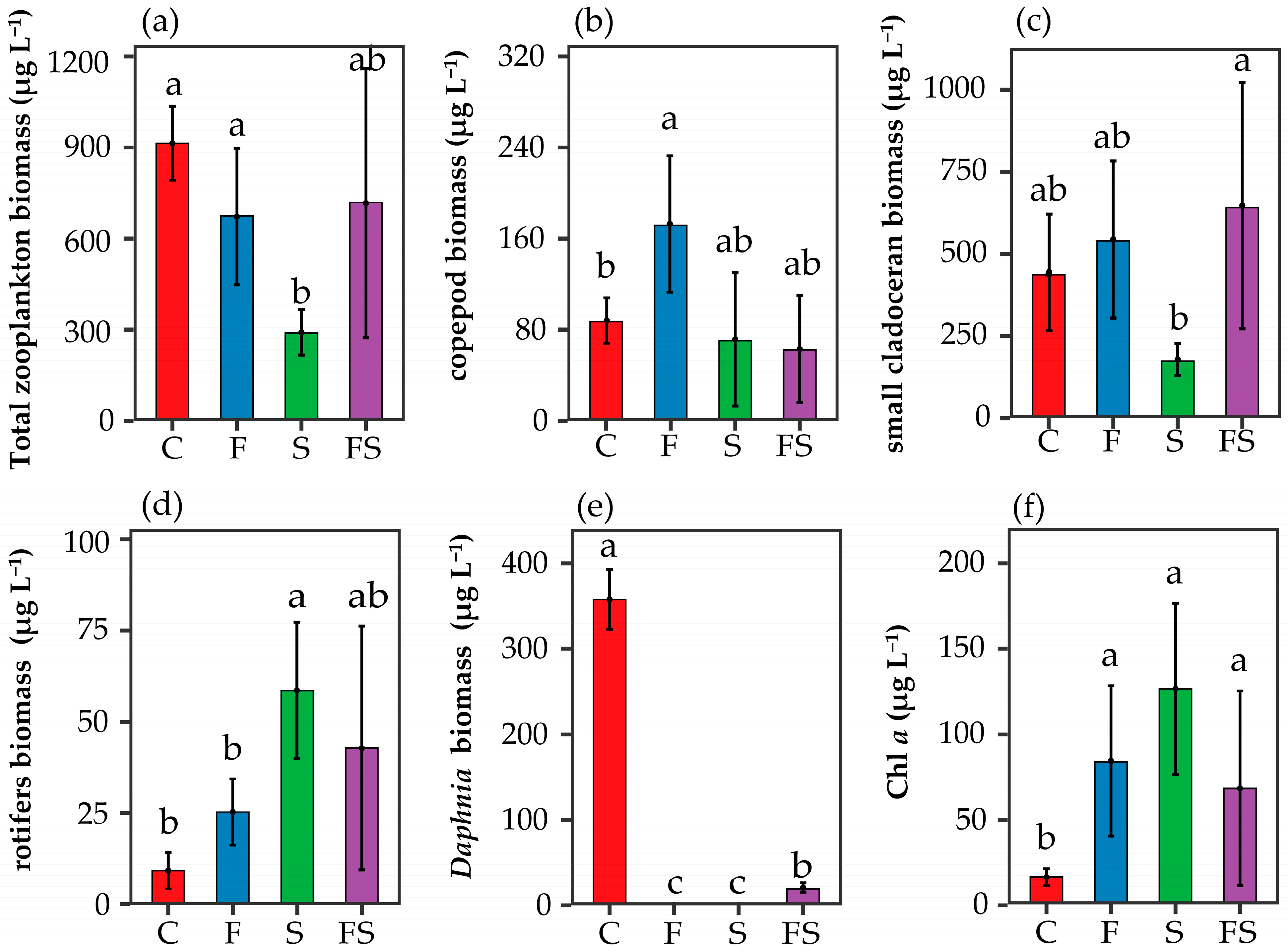
3.2. Trophic Cascade
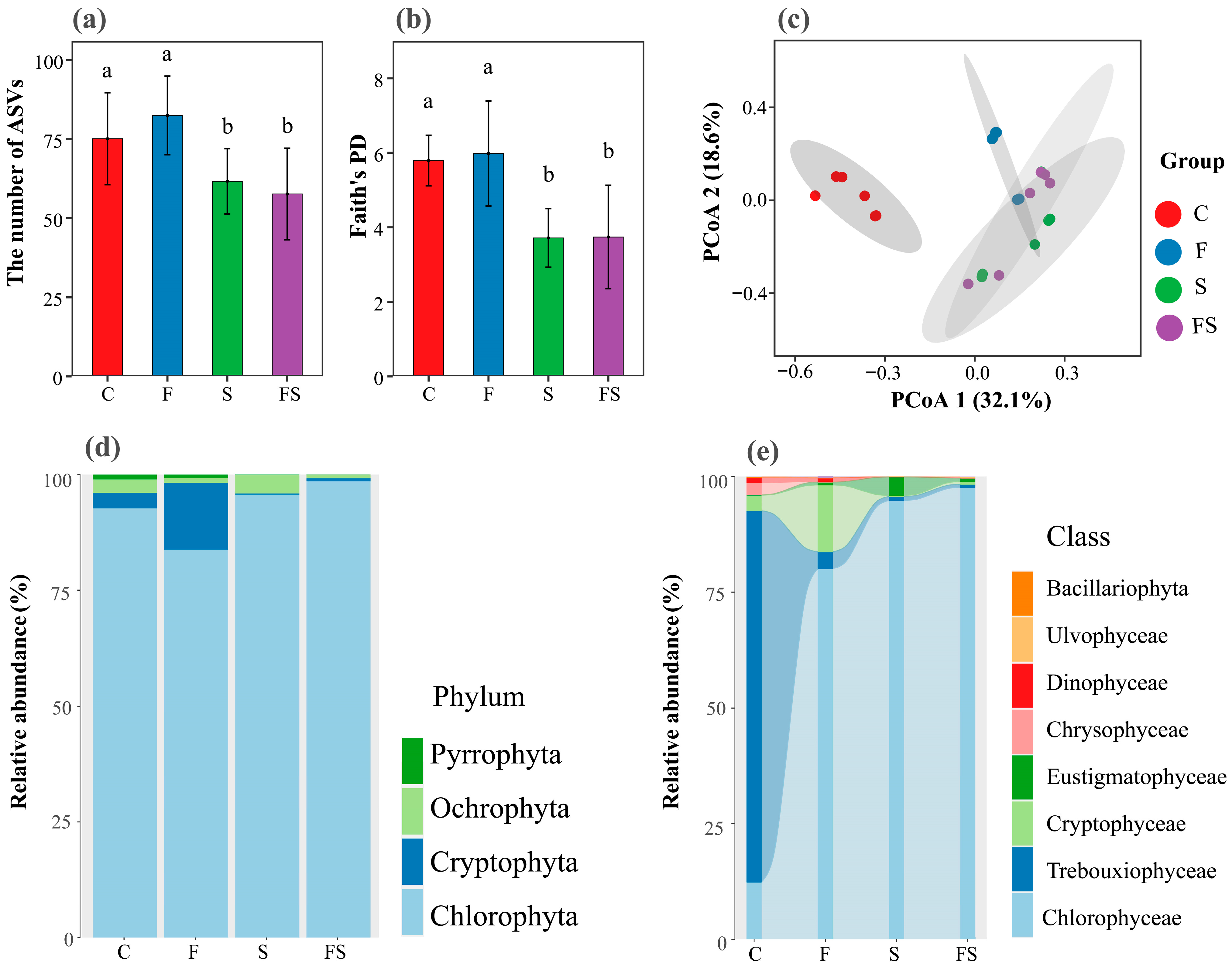
3.3. Diversity and Assemblage Composition of Phytoplankton
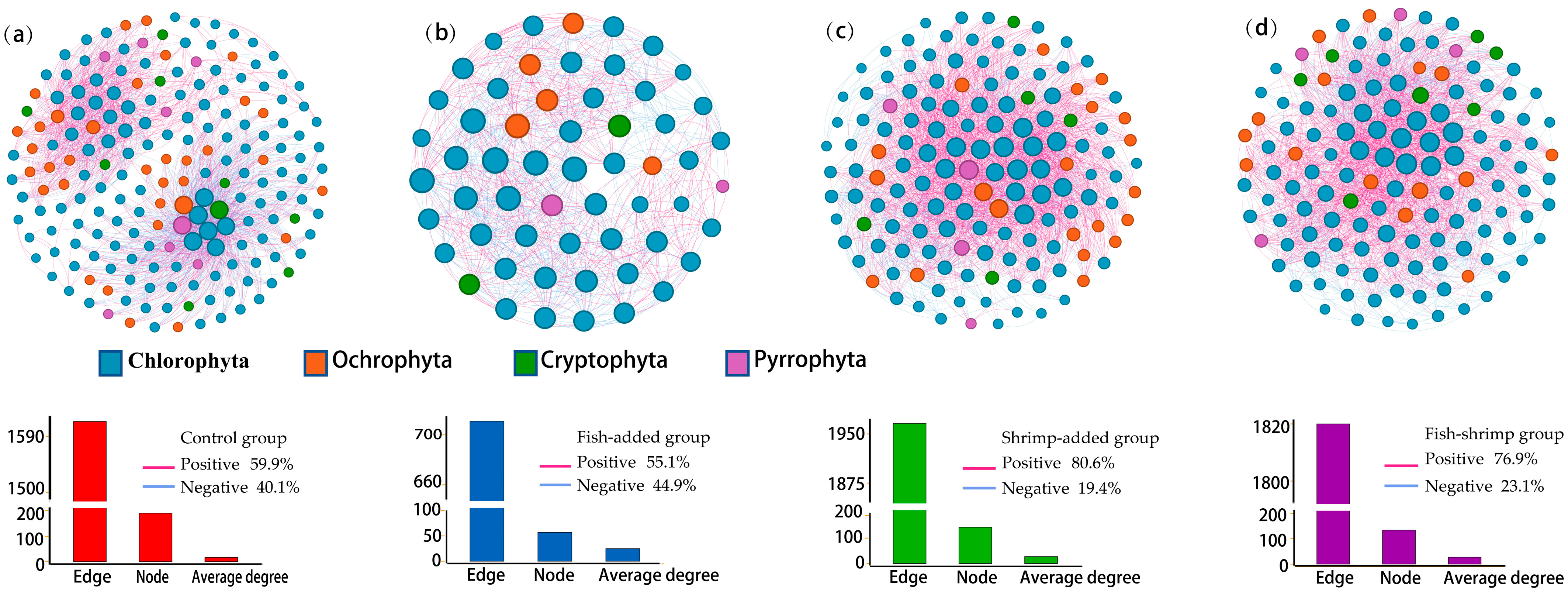
3.4. Interactions among Phytoplankton
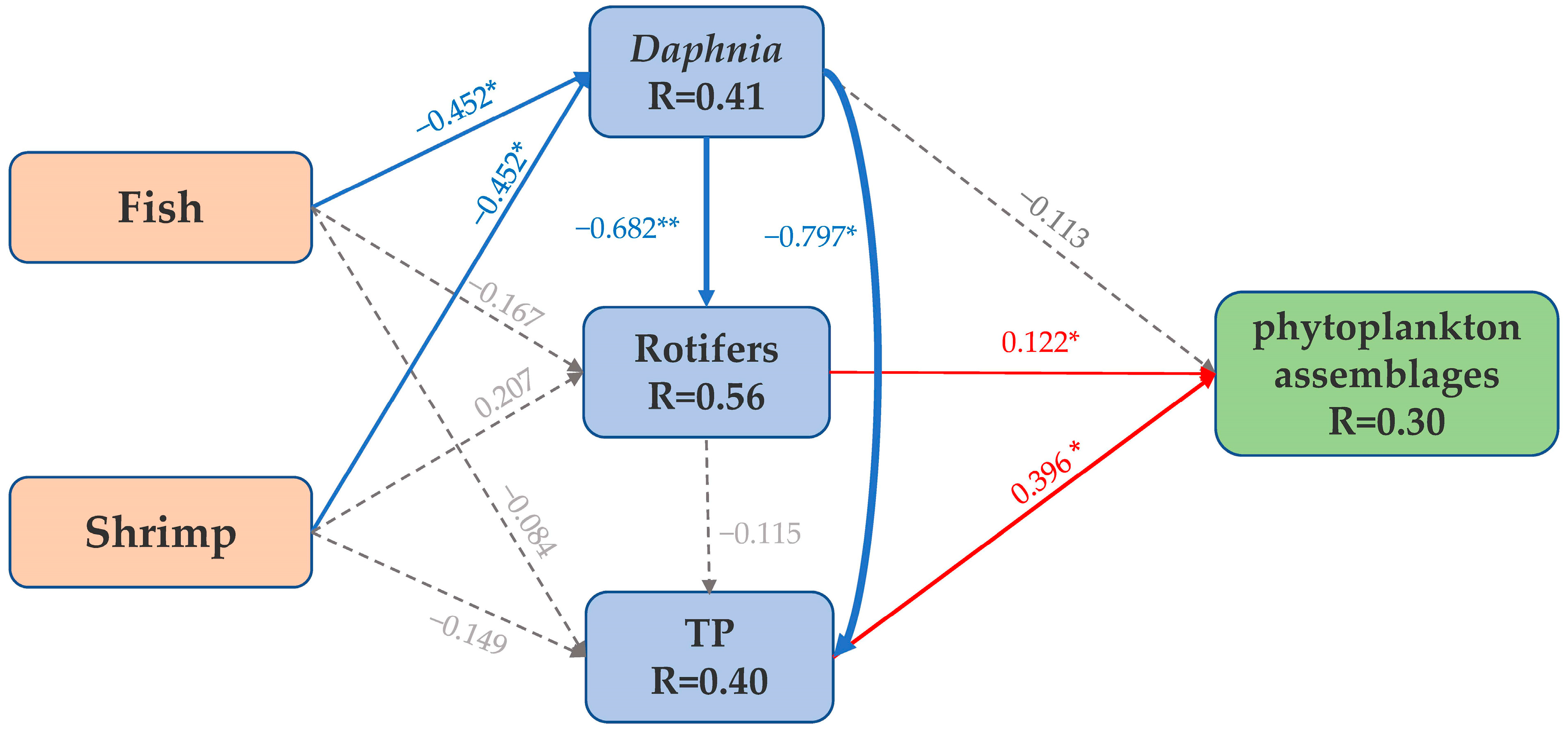
3.5. Environmental Factors Affecting Phytoplankton Diversity and Assemblages Composition
4. Discussion
4.1. Cascade Impact on Nutrient Cascades
4.2. Intraguild Predation Impact on Nutrient Cascades
4.3. The Presence of Shrimp Shapes Phytoplankton Diversity and Assemblage Composition through Top-Down and Bottom-Up Effects
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Makler-Pick, V.; Hipsey, M.R.; Zohary, T.; Carmel, Y.; Gal, G. Intraguild Predation Dynamics in a Lake Ecosystem Based on a Coupled Hydrodynamic-Ecological Model: The Example of Lake Kinneret (Israel). Biology 2017, 6, 22. [Google Scholar] [CrossRef]
- Jeppesen, E.; Søndergaard, M.; Lauridsen, T.L.; Davidson, T.A.; Liu, Z.; Mazzeo, N.; Trochine, C.; Özkan, K.; Jensen, H.S.; Trolle, D.; et al. Chapter 6—Biomanipulation as a Restoration Tool to Combat Eutrophication: Recent Advances and Future Challenges. In Advances in Ecological Research; Woodward, G., Jacob, U., O’Gorman, E.J., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 47, pp. 411–488. [Google Scholar]
- Lukić, D.; Ptacnik, R.; Vad, C.F.; Pόda, C.; Horváth, Z. Environmental constraint of intraguild predation: Inorganic turbidity modulates omnivory in fairy shrimps. Freshw. Biol. 2020, 65, 226–239. [Google Scholar] [CrossRef]
- He, H.; Ning, X.; Chen, K.; Li, Q.; Li, K.; Liu, Z.; Jeppesen, E. Intraguild predation dampens trophic cascades in shallow aquatic mesocosms in the subtropics: Implications for lake restoration by biomanipulation. Freshw. Biol. 2021, 66, 1571–1580. [Google Scholar] [CrossRef]
- Carpenter, S.R.; Kitchell, J.F.; Cottingham, K.L.; Schindler, D.E.; Christense, D.L.; Post, D.M.; Voichick, N. Chlorophyll Variability, Nutrient Input, and Grazing: Evidence from Whole- Lake Experiments. Ecology 1996, 77, 725–735. [Google Scholar] [CrossRef]
- Lemmens, P.; Declerck, S.A.J.; Tuytens, K.; Vanderstukken, M.; De Meester, L. Bottom-Up Effects on Biomass Versus Top-Down Effects on Identity: A Multiple-Lake Fish Community Manipulation Experiment. Ecosystems 2018, 21, 166–177. [Google Scholar] [CrossRef]
- Jeppesen, E.; Jensen, J.P.; Søndergaard, M.; Fenger-Grøn, M.; Bramm, M.E.; Sandby, K.; Møller, P.H.; Rasmussen, H.U. Impact of fish predation on cladoceran body weight distribution and zooplankton grazing in lakes during winter. Freshw. Biol. 2004, 49, 432–447. [Google Scholar] [CrossRef]
- Compte, J.; Brucet, S.; Gascón, S.; Boix, D.; Sala, J.; López-Flores, R.; Quintana, X.D. Impact of different developmental stages of Daphnia magna (Straus) on the plankton community under different trophic conditions. Hydrobiologia 2009, 635, 45–56. [Google Scholar] [CrossRef]
- Berta, C.; Gyulai, I.; Szabó, J.L.; Simon, E.; Nagy, A.S.; Somlyai, I.; Grigorszky, I. Cladocerans as indicators in the importance of passive nature conservation. Biologia 2018, 73, 875–884. [Google Scholar] [CrossRef]
- Tumurtogoo, U.; Figler, A.; Korponai, J.; Sajtos, Z.; Grigorszky, I.; Berta, C.; Gyulai, I. Density and Diversity Differences of Contemporary and Subfossil Cladocera Assemblages: A Case Study in an Oxbow Lake. Water 2022, 14, 2149. [Google Scholar] [CrossRef]
- Vanni, M.J.; Layne, C.D. Nutrient recycling and herbivory as mechanisms in the “top–down” effect of fish on algae in lakes. Ecology 1997, 78, 21–40. [Google Scholar]
- He, H.; Qian, T.; Shen, R.; Yu, J.; Li, K.; Liu, Z.; Jeppesen, E. Piscivore stocking significantly suppresses small fish but does not facilitate a clear-water state in subtropical shallow mesocosms: A biomanipulation experiment. Sci. Total Environ. 2022, 842, 156967. [Google Scholar] [CrossRef]
- Berta, C.; Tóthmérész, B.; Wojewódka, M.; Augustyniuk, O.; Korponai, J.; Bertalan-Balázs, B.; Nagy, A.S.; Grigorszky, I.; Gyulai, I. Community Response of Cladocera to Trophic Stress by Biomanipulation in a Shallow Oxbow Lake. Water 2019, 11, 929. [Google Scholar] [CrossRef]
- Sterner, R.W.; Elser, J.J.; Hessen, D.O. Stoichiometric relationships among producers, consumers and nutrient cycling in pelagic ecosystems. Biogeochemistry 1992, 17, 49–67. [Google Scholar] [CrossRef]
- MacIsaac, H.J.; Gilbert, J.J. Competition between rotifers and cladocerans of different body sizes. Oecologia 1989, 81, 295–301. [Google Scholar] [CrossRef]
- Lin, Q.; Zeng, D.; Guo, T.; Peng, L. Filter-feeding fish (Hypophthalmichthys molitrix) mediated phosphorus recycling versus grazing pressure as drivers of the trophic cascade in large enclosures subsidized by allochthonous detritus. Water Res. 2021, 204, 117579. [Google Scholar] [CrossRef]
- Liu, Y.; Song, S.; Chen, T.; Li, C. The diversity and structure of marine protists in the coastal waters of China revealed by morphological observation and 454 pyrosequencing. Estuar. Coast. Shelf Sci. 2017, 189, 143–155. [Google Scholar] [CrossRef]
- Bruno, J.F.; O’Connor, M.I. Cascading effects of predator diversity and omnivory in a marine food web. Ecol. Lett. 2005, 8, 1048–1056. [Google Scholar] [CrossRef]
- Hart, D.R. Intraguild Predation, Invertebrate Predators, and Trophic Cascades in Lake Food Webs. J. Theor. Biol. 2002, 218, 111–128. [Google Scholar] [CrossRef]
- Eland, L.E.; Davenport, R.; Mota, C.R. Evaluation of DNA extraction methods for freshwater eukaryotic microalgae. Water Res. 2012, 46, 5355–5364. [Google Scholar] [CrossRef]
- Tzafesta, E.; Saccomanno, B.; Zangaro, F.; Vadrucci, M.R.; Specchia, V.; Pinna, M. DNA Barcode Gap Analysis for Multiple Marker Genes for Phytoplankton Species Biodiversity in Mediterranean Aquatic Ecosystems. Biology 2022, 11, 1277. [Google Scholar] [CrossRef]
- De Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahé, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I.; et al. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Ji, Q.; Li, G.; Yin, K.; Gong, J. Diversity and distribution of bacterioplankton in the coastal upwelling waters off Hainan Island, China. Acta Oceanol. Sin. 2022, 41, 76–85. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, L.; Tang, Y.; Li, A.; Liu, C.; Xie, C.; Xiao, L.; Lu, S. Phytoplankton community and HAB species in the South China Sea detected by morphological and metabarcoding approaches. Harmful Algae 2022, 118, 102297. [Google Scholar] [CrossRef]
- Guijun, Y.; Boqiang, Q.; Xiangming, T.; Zhijun, G.; Chunni, Z.; Hua, Z.; Xiaodong, W. Contrasting zooplankton communities of two bays of the large, shallow, eutrophic Lake Taihu, China: Their relationship to environmental factors. J. Great Lakes Res. 2012, 38, 299–308. [Google Scholar] [CrossRef]
- Yu, N.; Lu, Q.P.; Zhou, G. Studies on the growth characteristics and food habits of Pseudobagrus fulvidraco (Richardson). J. Aquac. 1996, 3, 19–20. [Google Scholar]
- Chen, S.B.; Liao, C.S.; Zhao, X.J.; Ye, S.W.; Li, Z.J.; Zhang, T.L.; Liu, J.S. The growth and the reproductive biology of Exopalaemon modestus (heller, 1862) (crustacea, decapoda, palaemonidae) in the three gorges reservoir of china. Acta Hydrobiol. Sin. 2015, 39, 989–996. [Google Scholar]
- Paerl, H.W.; Xu, H.; McCarthy, M.J.; Zhu, G.; Qin, B.; Li, Y.; Gardner, W.S. Controlling harmful cyanobacterial blooms in a hyper-eutrophic lake (Lake Taihu, China): The need for a dual nutrient (N & P) management strategy. Water Res. 2011, 45, 1973–1983. [Google Scholar]
- Jin, X.C.; Tu, Q.Y. The Standard Methods for Observation and Analysis in Lake Eutrophication, 2nd ed.; Environmental Science Press: Beijing, China, 1990. [Google Scholar]
- Dumont, H.J.; Van de Velde, I.; Dumont, S. The dry weight estimate of biomass in a selection of Cladocera, Copepoda and Rotifera from the plankton, periphyton and benthos of continental waters. Oecologia 1975, 19, 75–97. [Google Scholar] [CrossRef]
- Li, H.; Xing, P.; Chen, M.; Bian, Y.; Wu, Q.L. Short-term bacterial community composition dynamics in response to accumulation and breakdown of Microcystis blooms. Water Res. 2011, 45, 1702–1710. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar]
- Díez, B.; Pedrós-Alió, C.; Marsh, T.L.; Massana, R. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 2001, 67, 2942–2951. [Google Scholar] [CrossRef]
- Li, H.; Zeng, J.; Ren, L.; Wang, J.; Xing, P.; Wu, Q.L. Contrasting patterns of diversity of abundant and rare bacterioplankton in freshwater lakes along an elevation gradient. Limnol. Oceanogr. 2017, 62, 1570–1585. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Owen, N.R.; Gumbs, R.; Gray, C.L.; Faith, D.P. Global conservation of phylogenetic diversity captures more than just functional diversity. Nat. Commun. 2019, 10, 859. [Google Scholar] [CrossRef] [PubMed]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Wang, J.; Pan, F.; Soininen, J.; Heino, J.; Shen, J. Nutrient enrichment modifies temperature-biodiversity relationships in large-scale field experiments. Nat. Commun. 2016, 7, 13960. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, X.; Wang, Y.; Fu, Y.; Yan, H.; Qian, S.; Cheng, L. Drivers of phenology shifts and their effect on productivity in northern grassland of China during 1984–2017—Evidence from long-term observational data. Int. J. Biometeorol. 2021, 65, 527–539. [Google Scholar] [CrossRef]
- Liu, L.; Chen, H.; Liu, M.; Yang, J.R.; Xiao, P.; Wilkinson, D.M.; Yang, J. Response of the eukaryotic plankton community to the cyanobacterial biomass cycle over 6 years in two subtropical reservoirs. ISME J. 2019, 13, 2196–2208. [Google Scholar] [CrossRef] [PubMed]
- Ang, Z.Q.; Sun, X.J.; Cao, X.Y.; Zhou, Q.; Guan, B.H.; Zeng, J. Diversity and network structure of epiphytic bacterial communities on different submerged macrophytes. J. Lake Sci. 2022, 34, 1234–1249. [Google Scholar]
- Carpenter, S.R.; Cole, J.J.; Hodgson, J.R.; Kitchell, J.F.; Pace, M.L.; Bade, D.; Cottingham, K.L.; Essington, T.E.; Houser, J.N.; Schindler, D.E. Trophic cascades, nutrients, and lake productivity: Whole-lake experiments. Ecol. Monogr. 2001, 71, 163–186. [Google Scholar] [CrossRef]
- Skov, C.; Perrow, M.R.; Berg, S.; Skovgaard, H. Changes in the fish community and water quality during seven years of stocking piscivorous fish in a shallow lake. Freshw. Biol. 2002, 47, 2388–2400. [Google Scholar] [CrossRef]
- Olin, M.; Rask, M.; Ruuhijärvi, J.; Keskitalo, J.; Horppila, J.; Tallberg, P.; Taponen, T.; Lehtovaara, A.; Sammalkorpi, I. Effects of Biomanipulation on Fish and Plankton Communities in Ten Eutrophic Lakes of Southern Finland. Hydrobiologia 2006, 553, 67–88. [Google Scholar] [CrossRef]
- Shi, W.G.; Yan, X.M.; Bing, X.W. Biology and feeding habit of Palaemon modestus (Heller) in Taihu Lake. J. Ecol. Monogr. 1995, 7, 69–76. [Google Scholar]
- Yin, C.; He, W.; Guo, L.; Gong, L.; Yang, Y.; Yang, J.; Ni, L.; Chen, Y.; Jeppesen, E. Can top-down effects of planktivorous fish removal be used to mitigate cyanobacterial blooms in large subtropical highland lakes? Water Res. 2022, 218, 118483. [Google Scholar] [CrossRef] [PubMed]
- Ripple, W.J.; Estes, J.A.; Schmitz, O.J.; Constant, V.; Kaylor, M.J.; Lenz, A.; Motley, J.L.; Self, K.E.; Taylor, D.S.; Wolf, C. What is a Trophic Cascade? Trends Ecol. Evol. 2016, 31, 842–849. [Google Scholar] [CrossRef]
- Min, C.; Johansson, L.S.; Søndergaard, M.; Lauridsen, T.L.; Chen, F.; Sh, T.; Jeppesen, E. Copepods as environmental indicator in lakes: Special focus on changes in the proportion of calanoids along nutrient and pH gradients. Aquat. Ecol. 2021, 55, 1241–1252. [Google Scholar] [CrossRef]
- Søndergaard, M.; Jeppesen, E.; Berg, S. Pike (Esox lucius L.) stocking as a biomanipulation tool 2. Effects on lower trophic levels in Lake Lyng, Denmark. Hydrobiologia 1997, 342, 319–325. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, P.; Tao, M.; Guo, L.; Chen, J.; Li, L.; Xuezhen, Z.; Zhang, L. The impact of fish predation and cyanobacteria on zooplankton size structure in 96 subtropical lakes. PLoS ONE 2013, 8, e76378. [Google Scholar] [CrossRef]
- Daewel, U.; Hjøllo, S.S.; Huret, M.; Ji, R.; Maar, M.; Niiranen, S.; Travers-Trolet, M.; Peck, M.A.; van de Wolfshaar, K.E. Predation control of zooplankton dynamics: A review of observations and models. ICES J. Mar. Sci. 2014, 71, 254–271. [Google Scholar] [CrossRef]
- Li, Y.; Wang, R.; Su, H.; Wang, J.; Xie, P.; Chen, F. Eutrophication and predation mediate zooplankton diversity and network structure. Limnol. Oceanogr. 2022, 67, S133–S145. [Google Scholar] [CrossRef]
- Lampert, W.; Fleckner, W.; Rai, H.; Taylor, B.E. Phytoplankton control by grazing zooplankton: A study on the spring clear-water phase1. Limnol. Oceanogr. 1986, 31, 478–490. [Google Scholar] [CrossRef]
- Sommer, U.; Adrian, R.; De Senerpont Domis, L.; Elser, J.J.; Gaedke, U.; Ibelings, B.; Jeppesen, E.; Lürling, M.; Molinero, J.C.; Mooij, W.M.; et al. Beyond the Plankton Ecology Group (PEG) Model: Mechanisms Driving Plankton Succession. Annu. Rev. Ecol. Evol. Syst. 2012, 43, 429–448. [Google Scholar] [CrossRef]
- Feniova, I.; Dawidowicz, P.; Ejsmont-Karabin, J.; Gladyshev, M.; Kalinowska, K.; Karpowicz, M.; Kostrzewska-Szlakowska, I.; Majsak, N.; Petrosyan, V.; Razlutskij, V.; et al. Effects of zebra mussels on cladoceran communities under eutrophic conditions. Hydrobiologia 2018, 822, 37–54. [Google Scholar] [CrossRef]
- Bowes, M.J.; Gozzard, E.; Johnson, A.C.; Scarlett, P.M.; Roberts, C.; Read, D.S.; Armstrong, L.K.; Harman, S.A.; Wickham, H.D. Spatial and temporal changes in chlorophyll-a concentrations in the River Thames basin, UK: Are phosphorus concentrations beginning to limit phytoplankton biomass? Sci. Total Environ. 2012, 426, 45–55. [Google Scholar] [CrossRef]
- Elser, J.J.; Urabe, J. The stoichiometry of consumer-driven nutrient recycling: Theory, observations, and consequences. Ecology 1999, 80, 735–751. [Google Scholar] [CrossRef]
- Snyder, M.N.; Small, G.E.; Pringle, C.M. Diet-switching by omnivorous freshwater shrimp diminishes differences in nutrient recycling rates and body stoichiometry across a food quality gradient. Freshw. Biol. 2015, 60, 526–536. [Google Scholar] [CrossRef]
- Sommer, U.; Sommer, F. Cladocerans versus copepods: The cause of contrasting top-down controls on freshwater and marine phytoplankton. Oecologia 2006, 147, 183–194. [Google Scholar] [CrossRef]
- Havens, K.E. Zooplankton structure and potential food web interactions in the plankton of a subtropical chain-of-lakes. ScientificWorldJournal 2002, 2, 926–942. [Google Scholar] [CrossRef] [PubMed]
- Jagadeesan, L.; Jyothibabu, R.; Arunpandi, N.; Parthasarathi, S. Copepod grazing and their impact on phytoplankton standing stock and production in a tropical coastal water during the different seasons. Environ. Monit. Assess. 2017, 189, 105. [Google Scholar] [CrossRef] [PubMed]
- Sommer, U.; Aberle, N.; Lengfellner, K.; Lewandowska, A. The Baltic Sea spring phytoplankton bloom in a changing climate: An experimental approach. Mar. Biol. 2012, 159, 2479–2490. [Google Scholar] [CrossRef]
- Ning, X.Y.; Zhang, L.; Chen, K.Q.; Han, C.A.; Li, Q.S.; Li, K.Y.; He, H.; Zhao, B.Y. Effects of Exopalaemon modestus on nutrient levels and plankton communities in spring and Summer subtropical lakes and reservoirs: A microcosm experiment. J. Lake Sci. 2022, 34, 582–589. [Google Scholar]
- Wu, X.; Griffin, J.N.; Sun, S. Cascading effects of predator-detritivore interactions depend on environmental context in a Tibetan alpine meadow. J. Anim. Ecol. 2014, 83, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Q.; Peng, Z.; Hu, Z.; Xue, J.; Wang, W. Rotifer community structure and assessment of water quality in Yangcheng Lake. Chin. J. Oceanol. Limnol. 2012, 30, 47–58. [Google Scholar] [CrossRef]
- Olsen, Y.; Vaddtein, O.; Andersen, T.; Jensen, A. Competition between Staurastrum luetkemuellerii (chlorophyceae) and Microcystis aeruginosa (cyanophyceae) under varying modes of phosphate supply1. J. Phycol. 1989, 25, 499–508. [Google Scholar] [CrossRef]
- Agasild, H.; Zingel, P.; Tõnno, I.; Haberman, J.; Nõges, T. Contribution of different zooplankton groups in grazing on phytoplankton in shallow eutrophic Lake Võrtsjärv (Estonia). Hydrobiologia 2007, 584, 167–177. [Google Scholar] [CrossRef]
- Gong, D.; Guo, Z.; Wei, W.; Bi, J.; Wang, Z.; Ji, X. Phytoplankton Community Structure and Its Relationship with Environmental Factors in Nanhai Lake. Diversity 2022, 14, 927. [Google Scholar] [CrossRef]
- Elser, J.J.; Andersen, T.; Baron, J.S.; Bergström, A.-K.; Jansson, M.; Kyle, M.; Nydick, K.R.; Steger, L.; Hessen, D.O. Shifts in Lake N:P Stoichiometry and Nutrient Limitation Driven by Atmospheric Nitrogen Deposition. Science 2009, 326, 835–837. [Google Scholar] [CrossRef]
- Hernandez, D.J.; David, A.S.; Menges, E.S.; Searcy, C.A.; Afkhami, M.E. Environmental stress destabilizes microbial networks. ISME J. 2021, 15, 1722–1734. [Google Scholar] [CrossRef] [PubMed]

| Response Variable (Unit) | Variable | Estimates | SE | z-Value | p |
|---|---|---|---|---|---|
| TN (mg L−1) | I | 2.56 | 0.15 | 16.6 | <0.001 |
| F | −0.63 | 0.22 | −2.91 | 0.009 | |
| S | −0.46 | 0.22 | −2.13 | 0.046 | |
| F × S | 0.69 | 0.31 | 2.24 | 0.037 | |
| TP (mg L−1) | I | 0.04 | 0.01 | 4.89 | <0.001 |
| F | 0.04 | 0.01 | 3.43 | 0.003 | |
| S | 0.03 | 0.01 | 2.96 | 0.008 | |
| F × S | −0.04 | 0.02 | −2.73 | 0.013 | |
| Chl-a (μg L−1) | I | 0.83 | 0.15 | 5.65 | <0.001 |
| F | 0.93 | 0.21 | 4.46 | <0.001 | |
| S | 0.87 | 0.21 | 4.2 | <0.001 | |
| F × S | −1 | 0.29 | −3.41 | 0.003 | |
| Zooplankton biomass (μg L−1) | I | 2.81 | 0.08 | 35.85 | <0.001 |
| S | −0.37 | 0.11 | −3.3 | 0.004 | |
| F × S | 0.45 | 0.16 | 2.89 | 0.009 | |
| Copepod biomass (μg L−1) | I | 1.69 | 0.24 | 7.05 | <0.001 |
| Daphnia biomass (μg L−1) | I | 2.55 | 0.17 | 15.13 | <0.001 |
| F | −2.55 | 0.24 | −10.7 | <0.001 | |
| S | −2.55 | 0.24 | −10.7 | <0.001 | |
| F × S | 3.08 | 0.34 | 9.14 | <0.001 | |
| Small cladoceran biomass (μg L−1) | I | 2.26 | 0.13 | 18.02 | <0.001 |
| S | −0.55 | 0.18 | −3.12 | 0.005 | |
| F × S | 0.74 | 0.25 | 2.94 | 0.008 |
| ADONIS | ANOSIM | |||
|---|---|---|---|---|
| R | p | R | p | |
| C vs. F | 0.203 | 0.004 | 0.596 | 0.001 |
| C vs. S | 0.217 | 0.007 | 0.796 | 0.004 |
| C vs. SF | 0.216 | 0.003 | 0.743 | 0.001 |
| F vs. S | 0.175 | 0.001 | 0.496 | 0.005 |
| F vs. SF | 0.165 | 0.003 | 0.359 | 0.004 |
| S vs. SF | 0.093 | 0.397 | 0.002 | 0.417 |
| Treatment | Observation Network | Random Network | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Node | Edge | Positive Correlation | Modularity | Average Clustering Coefficient | Average Path Length | Network Diameter | Average Degree | Modularity (SD) | |
| C | 191 | 1612 | 59.9% | 0.485 | 0.423 | 2.573 | 8 | 16.88 | 0.152 (0.008) |
| F | 57 | 711 | 55.1% | 0.18 | 0.227 | 1.926 | 3 | 24.95 | 0.111 (0.007) |
| S | 143 | 1966 | 80.6% | 0.182 | 0.214 | 1.95 | 4 | 27.5 | 0.110 (0.005) |
| SF | 134 | 1820 | 76.9% | 0.18 | 0.227 | 1.926 | 4 | 27.16 | 0.111 (0.007) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Da, J.; Xi, Y.; Cheng, Y.; He, H.; Liu, Y.; Li, H.; Wu, Q.L. The Effects of Intraguild Predation on Phytoplankton Assemblage Composition and Diversity: A Mesocosm Experiment. Biology 2023, 12, 578. https://doi.org/10.3390/biology12040578
Da J, Xi Y, Cheng Y, He H, Liu Y, Li H, Wu QL. The Effects of Intraguild Predation on Phytoplankton Assemblage Composition and Diversity: A Mesocosm Experiment. Biology. 2023; 12(4):578. https://doi.org/10.3390/biology12040578
Chicago/Turabian StyleDa, Jun, Yilong Xi, Yunshan Cheng, Hu He, Yanru Liu, Huabing Li, and Qinglong L. Wu. 2023. "The Effects of Intraguild Predation on Phytoplankton Assemblage Composition and Diversity: A Mesocosm Experiment" Biology 12, no. 4: 578. https://doi.org/10.3390/biology12040578
APA StyleDa, J., Xi, Y., Cheng, Y., He, H., Liu, Y., Li, H., & Wu, Q. L. (2023). The Effects of Intraguild Predation on Phytoplankton Assemblage Composition and Diversity: A Mesocosm Experiment. Biology, 12(4), 578. https://doi.org/10.3390/biology12040578






