Chlorophyll a Synthesis in Rhodobacter sphaeroides by Chlorophyll Synthase of Nicotiana tabacum
Simple Summary
Abstract
1. Introduction

2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Construction of Plasmids
2.3. Construction of R. sphaeroides Mutants
2.4. Isolation and Spectral Analysis of the Membrane Fraction from R. sphaeroides
2.5. Pigment Extraction from R. sphaeroides
2.6. Preparation of BChlide a, Chlide a, BChl aGG, and Chl aGG
2.7. HPLC Determination of BChl aGG, Chl aGG, BChl aP, and Chl aP
2.8. Heterologous Expression and Quantification of BchG and ChlG in E. coli
2.9. Kinetic Analysis of BchG and ChlG Activities
2.10. Determination of Intracellular ROS Levels
2.11. Statistical Analysis
3. Results
3.1. Angiosperm ChlGs Prenylated BChlide a
3.2. NtChlG Exhibited the Highest Catalytic Efficiency for BChlide a among the Angiosperm ChlGs Examined
3.3. Angiosperm ChlGs Showed Resistance to Inhibition by Bacteriochlorin
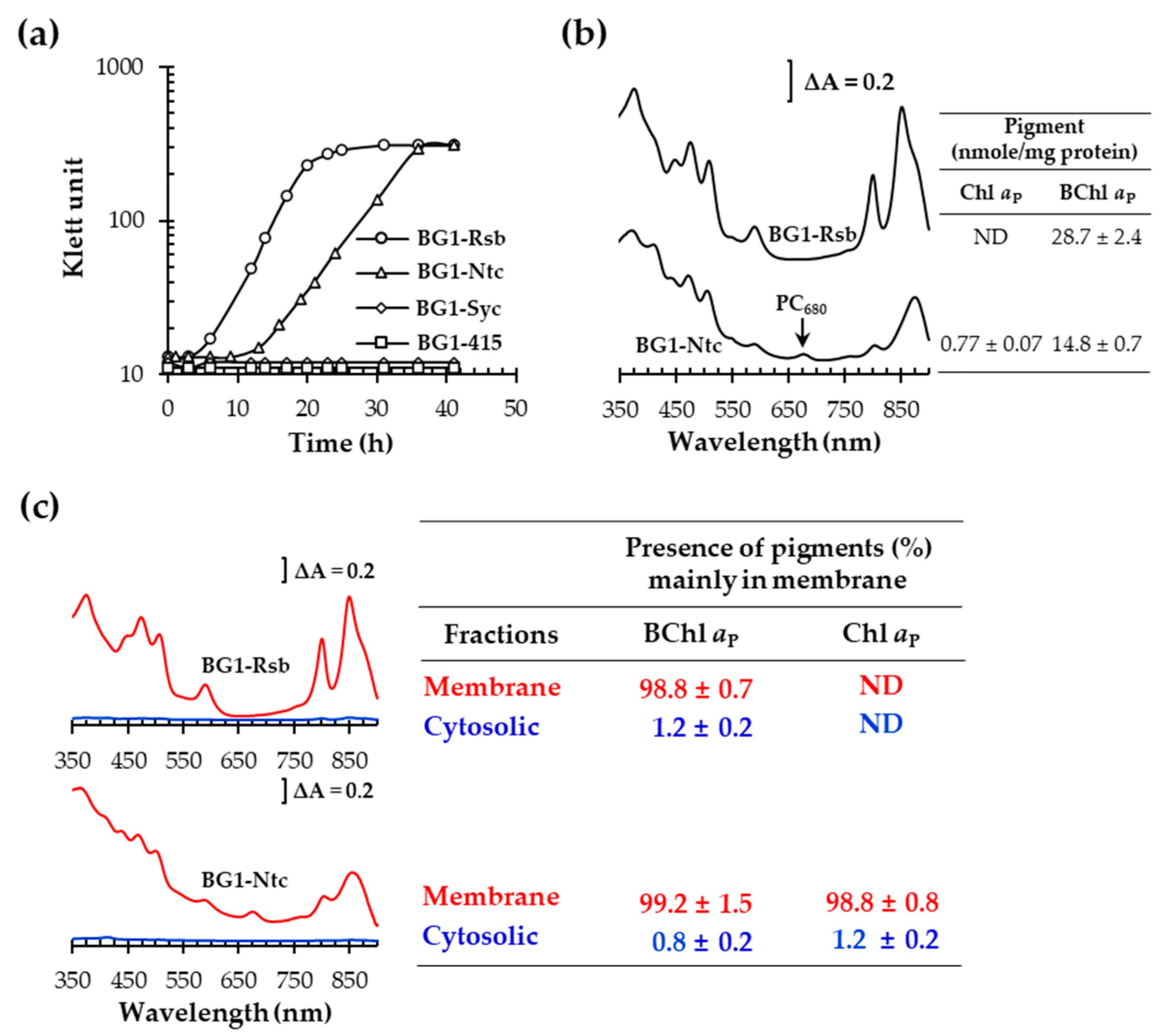
3.4. Heterologous Expression of NtchlG Resulted in the Formation of Free Chl aP in R. sphaeroides while Generating ROS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schmid, H.C.; Oster, U.; Kögel, J.; Lenz, S.; Rüdiger, W. Cloning and Characterisation of Chlorophyll Synthase from Avena sativa. Biol. Chem. 2001, 382, 903–911. [Google Scholar] [CrossRef]
- Kim, E.J.; Lee, J.K. Competitive inhibitions of the chlorophyll synthase of Synechocystis sp. strain PCC 6803 by bacteriochlorophyllide a and the bacteriochlorophyll synthase of Rhodobacter sphaeroides by chlorophyllide a. J. Bacteriol. 2010, 192, 198–207. [Google Scholar] [CrossRef]
- Oster, U.; Bauer, C.E.; Rüdiger, W. Characterization of Chlorophyll a and bacteriochlorophyll a Synthases by heterologous expression in Escherichia coli. J. Biol. Chem. 1997, 272, 9671–9676. [Google Scholar] [CrossRef]
- Proctor, M.S.; Sutherland, G.A.; Canniffe, D.P.; Hitchcock, A. The terminal enzymes of (bacterio)chlorophyll biosynthesis. R. Soc. Open Sci. 2022, 9, 211903. [Google Scholar] [CrossRef] [PubMed]
- Canniffe, D.P.; Hunter, C.N. Engineered biosynthesis of bacteriochlorophyll b in Rhodobacter sphaeroides. Biochim. Biophys. Acta Bioenerg. 2014, 1837, 1611–1616. [Google Scholar] [CrossRef] [PubMed]
- Tsukatani, Y.; Harada, J.; Nomata, J.; Yamamoto, H.; Fujita, Y.; Mizoguchi, T.; Tamiaki, H. Rhodobacter sphaeroides mutants overexpressing chlorophyllide a oxidoreductase of Blastochloris viridis elucidate functions of enzymes in late bacteriochlorophyll biosynthetic pathways. Sci. Rep. 2015, 5, 9741. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Ramos, M.; Canniffe, D.P.; Radle, M.I.; Hunter, C.N.; Bryant, D.A.; Golbeck, J.H. Engineered biosynthesis of bacteriochlorophyll gF in Rhodobacter sphaeroides. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 501–509. [Google Scholar] [CrossRef]
- Hitchcock, A.; Jackson, P.J.; Chidgey, J.W.; Dickman, M.J.; Hunter, C.N.; Canniffe, D.P. Biosynthesis of Chlorophyll a in a Purple Bacterial Phototroph and Assembly into a Plant Chlorophyll-Protein Complex. ACS Synth. Biol. 2016, 5, 948–954. [Google Scholar] [CrossRef]
- Benz, J.; Rüdiger, W. Chlorophyll Biosynthesis: Various Chlorophyllides as Exogenous Substrates for Chlorophyll Synthetase. Z. Nat. C 1981, 36, 51–57. [Google Scholar] [CrossRef]
- Cohen-Bazire, G.; Sistrom, W.R.; Stanier, R.Y. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell. Comp. Physiol. 1957, 49, 25–68. [Google Scholar] [CrossRef]
- Sistrom, W.R. A Requirement for Sodium in the Growth of Rhodopseudomonas spheroides. J. Gen. Microbiol. 1960, 22, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Donohue, T.J.; McEwan, A.G.; Kaplan, S. Cloning, DNA Sequence, and Expression of the Rhodobacter sphaeroides Cytochrome c2 Gene. J. Bacteriol. 1986, 168, 962–972. [Google Scholar] [CrossRef]
- Wasil, M.; Halliwell, B.; Grootveld, M.; Moorhouse, C.P.; Hutchison, D.C.S.; Baum, H. The specificity of thiourea, dimethylthiourea and dimethyl sulphoxide as scavengers of hydroxyl radicals Their protection of α1-antiproteinase against inactivation by hypochlorous acid. Biochem. J. 1987, 243, 867–870. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012. [Google Scholar]
- Kim, E.J.; Kim, J.S.; Kim, M.S.; Lee, J.K. Effect of changes in the level of light harvesting complexes of Rhodobacter sphaeroides on the photoheterotrophic production of hydrogen. Int. J. Hydrog. Energy 2006, 31, 531–538. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Kim, E.J.; Kim, J.S.; Lee, I.H.; Rhee, H.J.; Lee, J.K. Superoxide generation by chlorophyllide a reductase of Rhodobacter sphaeroides. J. Biol. Chem. 2008, 283, 3718–3730. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Kakuno, T.; Yamashita, J.; Horio, T. Purification and Properties of Chlorophyllase from Greened Rye Seedlings. J. Biochem. 1982, 92, 1763–1773. [Google Scholar] [CrossRef]
- Omata, T.; Murata, N. Preparation of Chlorophyll a, Chlorophyll b and Bacteriochlorophyll a by Column Chromatography with DEAE-Sepharose CL-6B and Sepharose CL-6B. Plant Cell Physiol. 1983, 24, 1093–1100. [Google Scholar] [CrossRef]
- Yan, X.; Fan, Y.; Wei, W.; Wang, P.; Liu, Q.; Wei, Y.; Zhang, L.; Zhao, G.; Yue, J.; Zhou, Z. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014, 24, 770–773. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.; Lee, J.K. Biochemical characterization of protoporphyrinogen dehydrogenase and protoporphyrin ferrochelatase of Vibrio vulnificus and the critical complex formation between these enzymes. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2674–2687. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Licht, M.K.; Nuss, A.M.; Volk, M.; Konzer, A.; Beckstette, M.; Berghoff, B.A.; Klug, G. Adaptation to Photooxidative Stress: Common and Special Strategies of the Alphaproteobacteria Rhodobacter sphaeroides and Rhodobacter capsulatus. Microorganisms 2020, 8, 283. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Kim, H.; Lee, J.K. The Photoheterotrophic Growth of Bacteriochlorophyll Synthase-Deficient Mutant of Rhodobacter sphaeroides Is Restored by I44F Mutant Chlorophyll Synthase of Synechocystis sp. PCC 6803. J. Microbiol. Biotechnol. 2016, 26, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Schmid, H.C.; Rassadina, V.; Oster, U.; Schoch, S.; Rüdiger, W. Pre-Loading of Chlorophyll Synthase with Tetraprenyl Diphosphate Is an Obligatory Step in Chlorophyll Biosynthesis. Biol. Chem. 2002, 383, 1769–1776. [Google Scholar] [CrossRef] [PubMed]
- Wittig, I.; Karas, M.; Schägger, H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteom. 2007, 6, 1215–1225. [Google Scholar] [CrossRef]
- Ipekoğlu, E.M.; Göçmen, K.; Öz, M.T.; Gürgan, M.; Yücel, M. Cloning and heterologous expression of chlorophyll a synthase in Rhodobacter sphaeroides. J. Basic Microbiol. 2017, 57, 238–244. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Rüdiger, W.; Böhm, S.; Helfrich, M.; Schulz, S.; Schoch, S. Enzymes of the Last Steps of Chlorophyll Biosynthesis: Modification of the Substrate Structure Helps To Understand the Topology of the Active Centers. Biochemistry 2005, 44, 10864–10872. [Google Scholar] [CrossRef]
- Shkuropatov, A.Y.; Shuvalov, V.A. Electron transfer in pheophytin a-modified reaction centers from Rhodobacter sphaeroides (R-26). FEBS Lett. 1993, 322, 168–172. [Google Scholar] [CrossRef]
- Shkuropatov, A.Y.; Proskuryakov, I.I.; Shkuropatova, V.A.; Zvereva, M.G.; Shuvalov, V.A. Formation of charge separated state P+QA- and triplet state 3P at low temperature in Rhodobacter sphaeroides (R-26) reaction centers in which bacteriopheophytin a is replaced by plant pheophytin a. FEBS Lett. 1994, 351, 249–252. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Franken, E.M.; Shkuropatov, A.Y.; Francke, C.; Neerken, S.; Gast, P.; Shuvalov, V.A.; Hoff, A.J.; Aartsma, T.J. Reaction centers of Rhodobacter sphaeroides R-26 with selective replacement of bacteriopheophytin by pheophytin a: I. Characterisation of steady-state absorbance and circular dichroism, and of the P+QA− state. Biochim. Biophys. Acta 1997, 1319, 242–250. [Google Scholar] [CrossRef]
- Franken, E.M.; Shkuropatov, A.Y.; Francke, C.; Neerken, S.; Gast, P.; Shuvalov, V.A.; Hoff, A.J.; Aartsma, T.J. Reaction centers of Rhodobacter sphaeroides R-26 with selective replacement of bacteriopheophytin a by pheophytin a: II. Temperature dependence of the quantum yield of P+QA− and 3P formation. Biochim. Biophys. Acta 1997, 1321, 1–9. [Google Scholar] [CrossRef]
- Kennis, J.T.M.; Shkuropatov, A.Y.; Stokkum, I.H.M.V.; Gast, P.; Hoff, A.J.; Shuvalov, V.A.; Aartsma, T.J. Formation of a Long-Lived P+BA− State in Plant Pheophytin-Exchanged Reaction Centers of Rhodobacter sphaeroides R26 at Low Temperature. Biochemistry 1997, 36, 16231–16238. [Google Scholar] [CrossRef] [PubMed]
- Swainsbury, D.J.K.; Faries, K.M.; Niedzwiedzki, D.M.; Martin, E.C.; Flinders, A.J.; Canniffe, D.P.; Shen, G.; Bryant, D.A.; Kirmaier, C.; Holten, D.; et al. Engineering of B800 bacteriochlorophyll binding site specificity in the Rhodobacter sphaeroides LH2 antenna. Biochim. Biophys. Acta Bioenerg. 2019, 1860, 209–223. [Google Scholar] [CrossRef]
- Niedzwiedzki, D.M.; Swainsbury, D.J.K.; Hunter, C.N. Carotenoid-to-(bacterio)chlorophyll energy transfer in LH2 antenna complexes from Rba. sphaeroides reconstituted with non-native (bacterio)chlorophylls. Photosynth. Res. 2020, 144, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Eraso, J.M.; Kaplan, S. prrA, a Putative Response Regulator Involved in Oxygen Regulation of Photosynthesis Gene Expression in Rhodobacter sphaeroides. J. Bacteriol. 1994, 176, 32–43. [Google Scholar] [CrossRef]
- Lenz, O.; Schwartz, E.; Dernedde, J.; Eitinger, M.; Friedrich, B. The Alcaligenes eutrophus H16 hoxX Gene Participates in Hydrogenase Regulation. J. Bacteriol. 1994, 176, 4385–4393. [Google Scholar] [CrossRef]
- Keen, N.T.; Tamaki, S.; Kobayashi, D.; Troilinger, D. Improved brood-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 1988, 70, 191–197. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Lee, J.K.; Kaplan, S. Transcriptional Regulation of puc Operon Expression in Rhodobacter sphaeroides. J. Biol. Chem. J. Biol. Chem. 1995, 270, 20453–20458. [Google Scholar] [CrossRef]


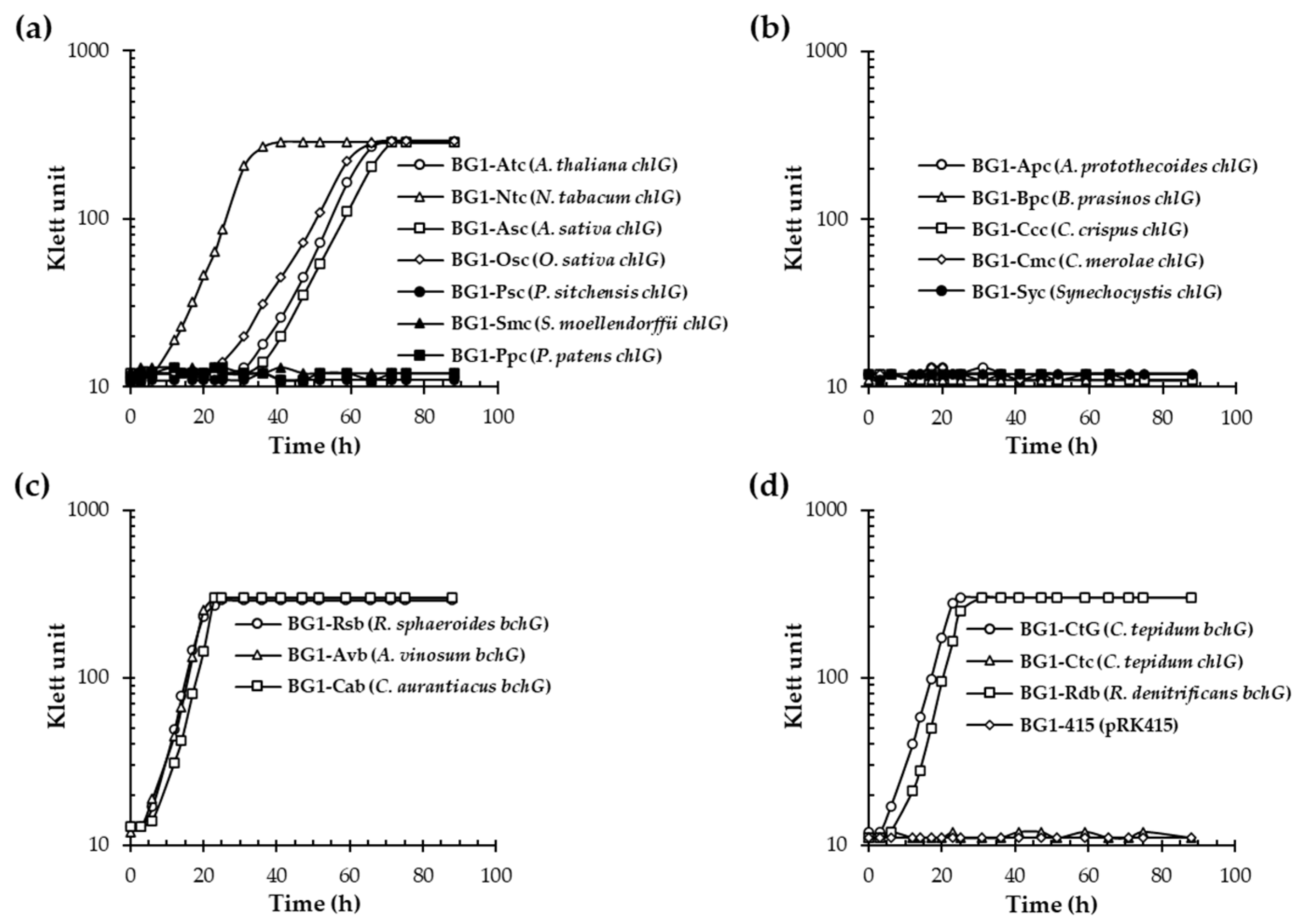

| Classification | Species | Enzyme | BchG Activity | ChlG Activity | ||
|---|---|---|---|---|---|---|
| Vascular plant | Angiosperm | Eudicot | Arabidopsis thaliana | ChlG | + | + |
| Nicotiana tabacum | ChlG | + | + | |||
| ChlG2 | + | + | ||||
| Monocot | Avena sativa | ChlG | + a | + | ||
| Oryza sativa | ChlG | + | + | |||
| Gymnosperm | Picea sitchensis | ChlG | − | + | ||
| Lycophyte | Selaginella moellendorffii | ChlG | − | + | ||
| Bryophyte | Moss | Physcomitrella patens | ChlG | − | + | |
| Algae | Chlorophyte (green algae) | Auxenochlorella protothecoides | ChlG | − | + | |
| Bathycoccus prasinos | ChlG | − | + | |||
| Rhodophyte (red algae) | Chondrus crispus | ChlG | − | + | ||
| Cyanidioschyzon merolae | ChlG | − | + | |||
| PS bacteria | Cyanobacteria | Synechocystis sp. PCC6803 | ChlG | − | + | |
| Purple nonsulfur bacteria | Rhodobacter sphaeroides | BchG | + | − | ||
| Purple sulfur bacteria | Allochromatium vinosum | BchG | + | − | ||
| Green nonsulfur bacteria | Chloroflexus aurantiacus | BchG | + | − | ||
| Green sulfur bacteria | Chlorobaculum tepidum | BchG | + | − | ||
| ChlG | − | + | ||||
| Aerobic anoxygenic PS bacteria | Roseobacter denitrificans | BchG | + | − | ||
| Enzyme a | First Substrate | Second Substrate | Km (μM) | kcat (min−1) | kcat/Km (mM−1 min−1) |
|---|---|---|---|---|---|
| BchG (R. sphaeroides) | GGPP | BChlide a | 23 ± 2 | 4.6 ± 0.2 | 201 ± 7 |
| PPP | 37 ± 2 | 3.4 ± 0.5 | 92 ± 11 | ||
| ChlG (Synechocystis) | GGPP | Chlide a | 14 ± 2 | 17.1 ± 0.9 | 1193 ± 59 |
| PPP | 47 ± 1 | 15.8 ± 0.3 | 337 ± 6 | ||
| ChlG (A. thaliana) | GGPP | BChlide a | 549 ± 34 | 2.2 ± 0.1 | 4 ± 1 |
| PPP | 611 ± 33 | 1.7 ± 0.3 | 3 ± 1 | ||
| GGPP | Chlide a | 10 ± 1 | 10.6 ± 0.3 | 1084 ± 32 | |
| PPP | 22 ± 1 | 9.0 ± 0.1 | 412 ± 4 | ||
| ChlG (N. tabacum) | GGPP | BChlide a | 180 ± 19 | 2.0 ± 0.1 | 11 ± 1 |
| PPP | 211 ± 30 | 1.6 ± 0.2 | 8 ± 1 | ||
| GGPP | Chlide a | 10 ± 2 | 9.3 ± 0.7 | 932 ± 68 | |
| PPP | 24 ± 1 | 7.8 ± 0.1 | 330 ± 2 | ||
| ChlG (A. sativa) | GGPP | BChlide a | 398 ± 17 | 2.2 ± 0.1 | 6 ± 1 |
| PPP | 483 ± 27 | 1.7 ± 0.3 | 4 ± 1 | ||
| GGPP | Chlide a | 12 ± 1 | 14.7 ± 0.5 | 1213 ± 39 | |
| PPP | 29 ± 3 | 11.6 ± 0.1 | 398 ± 4 | ||
| ChlG (O. sativa) | GGPP | BChlide a | 413 ± 17 | 2.4 ± 0.1 | 6 ± 1 |
| PPP | 621 ± 10 | 1.8 ± 0.3 | 3 ± 1 | ||
| GGPP | Chlide a | 9 ± 1 | 12.5 ± 0.3 | 1370 ± 34 | |
| PPP | 23 ± 1 | 10.5 ± 0.1 | 457 ± 5 |
| Enzyme a | First Substrate | Second Substrate | Ki (μM) b | |||||
|---|---|---|---|---|---|---|---|---|
| BChlide a | Chlide a | BChl aGG | Chl aGG | BChl aP | Chl aP | |||
| BchG (R. sphaeroides) | GGPP | BChlide a | NA c | 35 ± 3 d | 797 ± 96 | 75 ± 6 d | 673 ± 46 | 65 ± 4 d |
| PPP | NA | 73 ± 5 d | 623 ± 25 | 62 ± 5 d | 676 ± 17 | 100 ± 6 d | ||
| ChlG (Synechocystis) | GGPP | Chlide a | 30 ± 1 d | NA | 63 ± 4 d | 688 ± 20 | 40 ± 3 d | 583 ± 35 |
| PPP | 97 ± 4 d | NA | 43 ± 3 d | 596 ± 47 | 105 ± 4 d | 750 ± 67 | ||
| ChlG (A. thaliana) | GGPP | Chlide a | 412 ± 47 | NA | 318 ± 41 | 695 ± 85 | 248 ± 37 | 596 ± 41 |
| PPP | 587 ± 10 | NA | 201 ± 16 | 631 ± 66 | 265 ± 9 | 806 ± 20 | ||
| ChlG (N. tabacum) | GGPP | Chlide a | 502 ± 57 | NA | 395 ± 47 | 665 ± 59 | 360 ± 36 | 612 ± 28 |
| PPP | 681 ± 26 | NA | 348 ± 41 | 617 ± 35 | 484 ± 42 | 796 ± 17 | ||
| ChlG (A. sativa) | GGPP | Chlide a | 379 ± 37 | NA | 306 ± 31 | 808 ± 53 | 276 ± 24 | 785 ± 59 |
| PPP | 490 ± 36 | NA | 242 ± 19 | 661 ± 82 | 308 ± 21 | 723 ± 45 | ||
| ChlG (O. sativa) | GGPP | Chlide a | 375 ± 54 | NA | 319 ± 39 | 705 ± 22 | 265 ± 31 | 646 ± 51 |
| PPP | 561 ± 47 | NA | 271 ± 29 | 595 ± 47 | 341 ± 23 | 703 ± 20 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Lee, J.K.; Kim, E.-J. Chlorophyll a Synthesis in Rhodobacter sphaeroides by Chlorophyll Synthase of Nicotiana tabacum. Biology 2023, 12, 573. https://doi.org/10.3390/biology12040573
Kim J, Lee JK, Kim E-J. Chlorophyll a Synthesis in Rhodobacter sphaeroides by Chlorophyll Synthase of Nicotiana tabacum. Biology. 2023; 12(4):573. https://doi.org/10.3390/biology12040573
Chicago/Turabian StyleKim, June, Jeong K. Lee, and Eui-Jin Kim. 2023. "Chlorophyll a Synthesis in Rhodobacter sphaeroides by Chlorophyll Synthase of Nicotiana tabacum" Biology 12, no. 4: 573. https://doi.org/10.3390/biology12040573
APA StyleKim, J., Lee, J. K., & Kim, E.-J. (2023). Chlorophyll a Synthesis in Rhodobacter sphaeroides by Chlorophyll Synthase of Nicotiana tabacum. Biology, 12(4), 573. https://doi.org/10.3390/biology12040573






