Individual Proportion Loss of Functional Connectivity Strength: A Novel Individual Functional Connectivity Biomarker for Subjective Cognitive Decline Populations
Abstract
Simple Summary
Abstract
1. Introduction
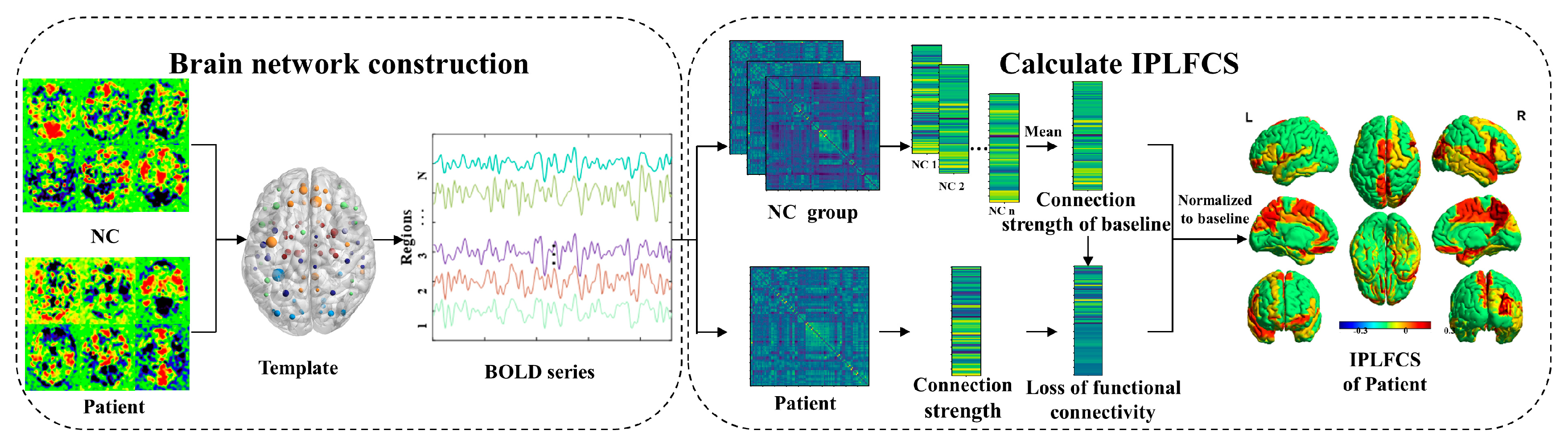
2. IPLFCS Analytical Framework
2.1. Brain Network Construction
2.2. Calculation of IPLFCS
3. Materials and Methods
3.1. Subjects
3.2. Inclusion Criterion
3.3. Standard Protocol Approvals, Registrations, and Patient Consents
3.4. Image Acquisition Protocol
3.5. Image Pre-Processing
3.6. Statistical Analysis
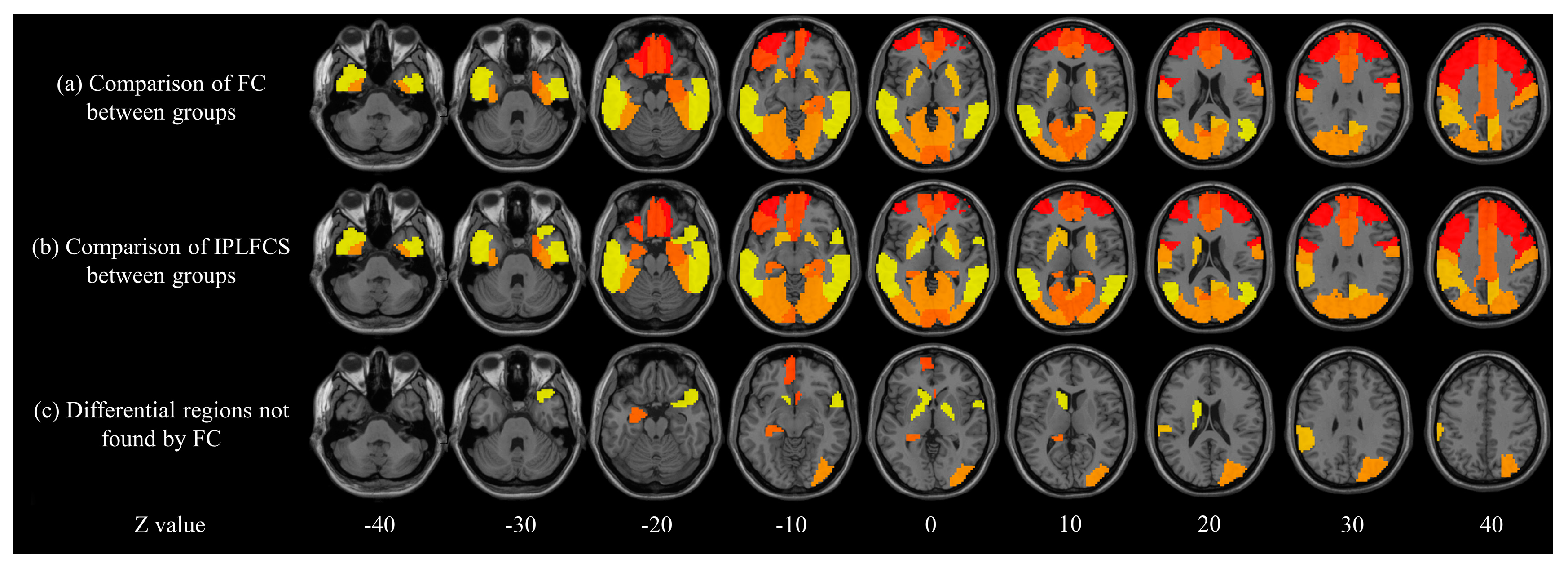
3.6.1. Comparison between FC and IPLFCS
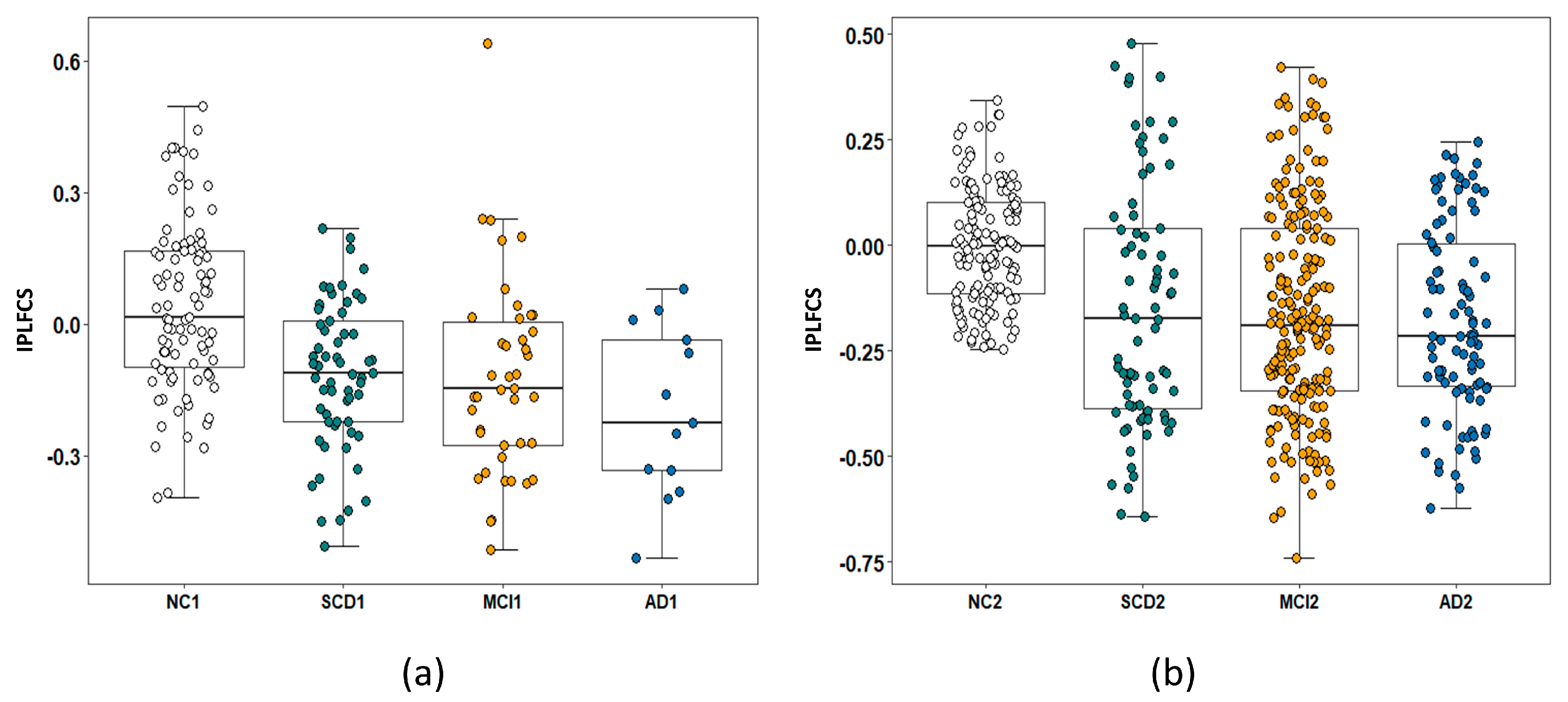
3.6.2. Identification of IPLFCS Biomarkers for SCD
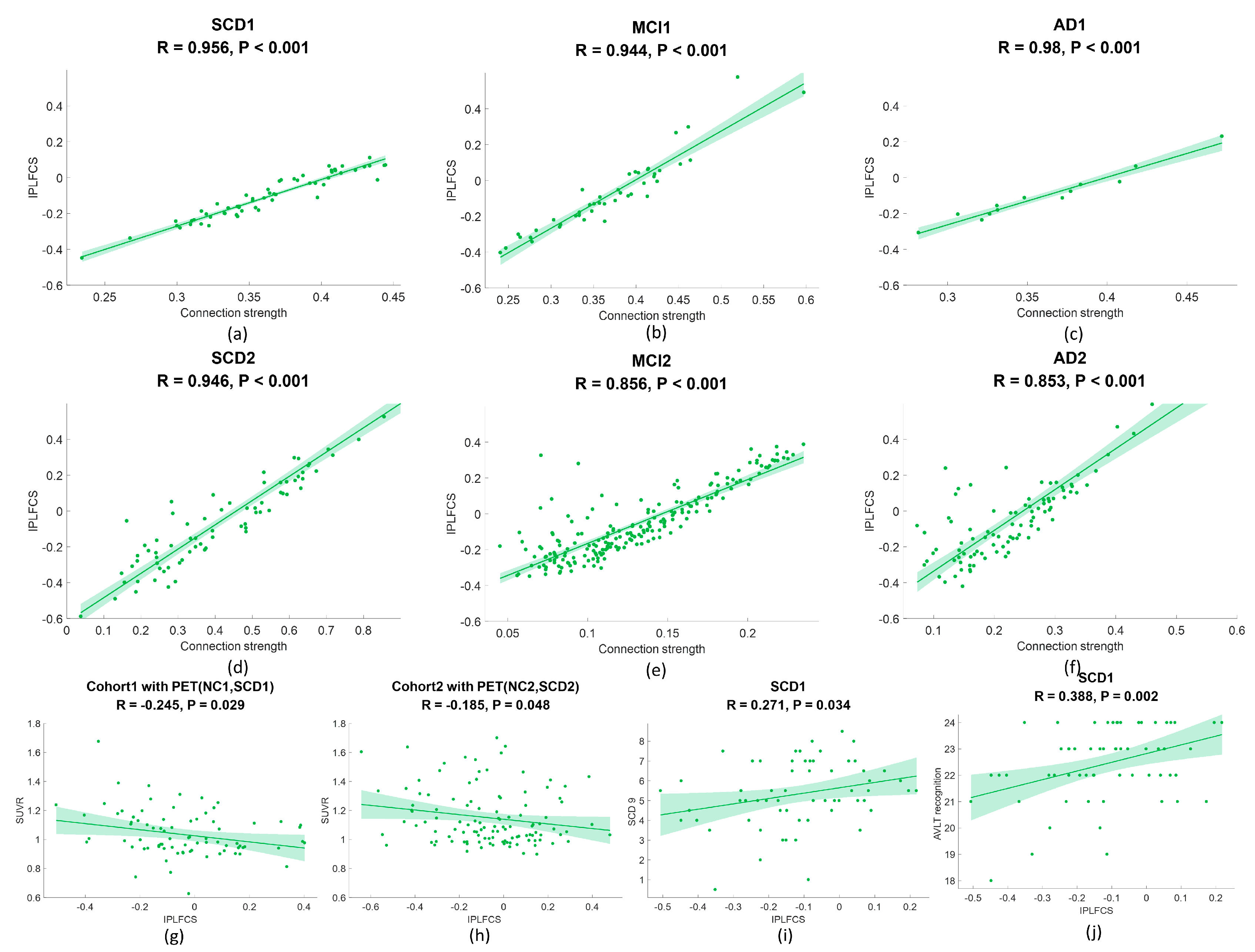
3.6.3. Correlation between IPLFCS and FC Strength
3.6.4. Correlation Analysis of Amyloid SUVR and IPLFCS
3.6.5. Correlation Analysis of Clinical Scale and IPLFCS
3.6.6. Receiver Operating Characteristic Curve
4. Results
4.1. Demographic Findings
4.2. Comparison between FC and IPLFCS
4.3. IPLFCS Biomarkers for SCD
4.4. Correlation between IPLFCS and FC
4.5. Correlation Analysis of Amyloid SUVR and IPLFCS
4.6. Correlation Analysis of Clinical Scale and IPLFCS
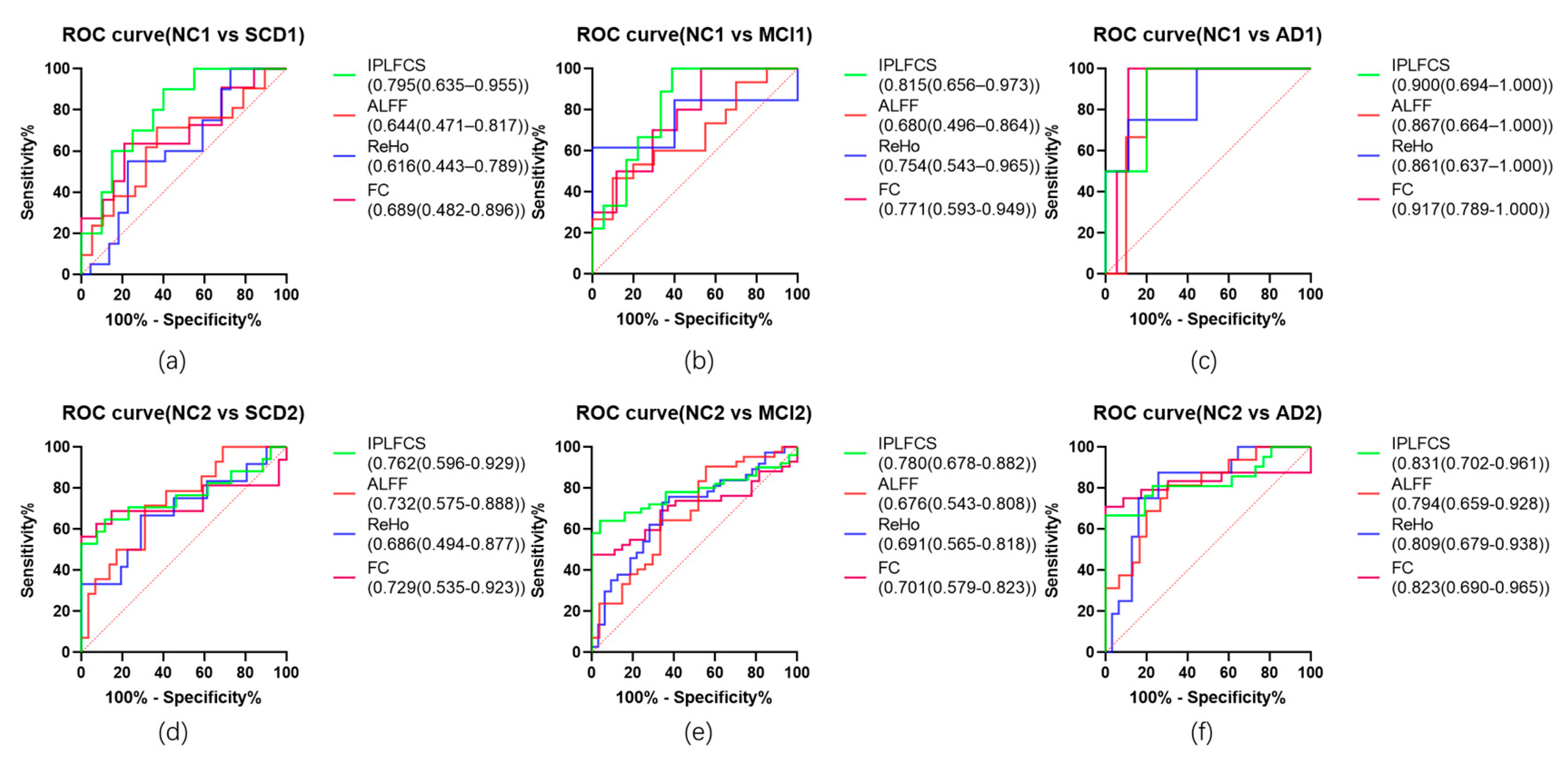
4.7. Receiver Operating Characteristic Curve
5. Discussion
6. Limitations
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, G.; Yang, K.; Du, W.; Hu, X.; Han, Y. Clinical Characteristics in Subjective Cognitive Decline with and without Worry: Baseline Investigation of the SILCODE Study. J. Alzheimers. Dis. 2019, 72, 443–454. [Google Scholar] [CrossRef]
- Palmqvist, S.; Tideman, P.; Cullen, N.; Zetterberg, H.; Blennow, K.; Dage, J.L.; Stomrud, E.; Janelidze, S.; Mattsson-Carlgren, N.; Hansson, O. Prediction of Future Alzheimer’s Disease Dementia Using Plasma Phospho-Tau Combined with Other Accessible Measures. Nat. Med. 2021, 27, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- Jessen, F.; Amariglio, R.E.; Van Boxtel, M.; Breteler, M.; Ceccaldi, M.; Chételat, G.; Dubois, B.; Dufouil, C.; Ellis, K.A.; Van Der Flier, W.M.; et al. A Conceptual Framework for Research on Subjective Cognitive Decline in Preclinical Alzheimer’s Disease. Alzheimer’s Dement. 2014, 10, 844–852. [Google Scholar] [CrossRef] [PubMed]
- Bessi, V.; Mazzeo, S.; Padiglioni, S.; Piccini, C.; Nacmias, B.; Sorbi, S.; Bracco, L. From Subjective Cognitive Decline to Alzheimer’s Disease: The Predictive Role of Neuropsychological Assessment, Personality Traits, and Cognitive Reserve. A 7-Year Follow-Up Study. J. Alzheimer’s Dis. 2018, 63, 1523–1535. [Google Scholar] [CrossRef] [PubMed]
- Ribaldi, F.; Palomo, R.; Altomare, D.; Garibotto, V.; Chicherio, C.; Frisoni, G.B. The Taxonomy of Subjective Cognitive Decline, Preliminary Evidence from the Geneva Memory Clinic Cohort. AD/PD 2023 Alzheimer’s Park. Dis. Conf. 2023, 1–15. [Google Scholar] [CrossRef]
- Bessi, V.; Mazzeo, S.; Bagnoli, S.; Padiglioni, S.; Carraro, M.; Piaceri, I.; Bracco, L.; Sorbi, S.; Nacmias, B. The Implication of BDNF Val66Met Polymorphism in Progression from Subjective Cognitive Decline to Mild Cognitive Impairment and Alzheimer’s Disease: A 9-Year Follow-up Study. Eur. Arch. Psychiatry Clin. Neurosci. 2020, 270, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Ávila-Villanueva, M.; Marcos Dolado, A.; Gómez-Ramírez, J.; Fernández-Blázquez, M. Brain Structural and Functional Changes in Cognitive Impairment Due to Alzheimer’s Disease. Front. Psychol. 2022, 13, 2959. [Google Scholar] [CrossRef] [PubMed]
- Rabin, L.A.; Smart, C.M.; Amariglio, R.E. Subjective Cognitive Decline in Preclinical Alzheimer’s Disease. Annu. Rev. Clin. Psychol. 2017, 13, 369–396. [Google Scholar] [CrossRef]
- Yang, L.; Yan, Y.; Wang, Y.; Hu, X.; Lu, J.; Chan, P.; Yan, T.; Han, Y. Gradual Disturbances of the Amplitude of Low-Frequency Fluctuations (ALFF) and Fractional ALFF in Alzheimer Spectrum. Front. Neurosci. 2018, 12, 975. [Google Scholar] [CrossRef]
- Yan, T.; Wang, Y.; Weng, Z.; Du, W.; Liu, T.; Chen, D.; Li, X.; Wu, J.; Han, Y. Early-Stage Identification and Pathological Development of Alzheimer’s Disease Using Multimodal MRI. J. Alzheimer’s Dis. 2019, 68, 1013–1027. [Google Scholar] [CrossRef]
- Dong, Q.Y.; Li, T.R.; Jiang, X.Y.; Wang, X.N.; Han, Y.; Jiang, J.H. Glucose Metabolism in the Right Middle Temporal Gyrus Could Be a Potential Biomarker for Subjective Cognitive Decline: A Study of a Han Population. Alzheimer’s Res. Ther. 2021, 13, 1–12. [Google Scholar] [CrossRef]
- Li, S.; Daamen, M.; Scheef, L.; Gaertner, F.C.; Buchert, R.; Buchmann, M.; Buerger, K.; Catak, C.; Dobisch, L.; Drzezga, A.; et al. Abnormal Regional and Global Connectivity Measures in Subjective Cognitive Decline Depending on Cerebral Amyloid Status. J. Alzheimers. Dis. 2021, 79, 493–509. [Google Scholar] [CrossRef]
- Cai, C.; Huang, C.; Yang, C.; Lu, H.; Hong, X.; Ren, F.; Hong, D.; Ng, E. Altered Patterns of Functional Connectivity and Causal Connectivity in Salience Subnetwork of Subjective Cognitive Decline and Amnestic Mild Cognitive Impairment. Front. Neurosci. 2020, 14, 288. [Google Scholar] [CrossRef]
- López-Sanz, D.; Bruña, R.; Garcés, P.; Martín-Buro, M.C.; Walter, S.; Delgado, M.L.; Montenegro, M.; Higes, R.L.; Marcos, A.; Maestú, F. Functional Connectivity Disruption in Subjective Cognitive Decline and Mild Cognitive Impairment: A Common Pattern of Alterations. Front. Aging Neurosci. 2017, 9, 109. [Google Scholar] [CrossRef]
- Chen, B.; Wang, Q.; Zhong, X.; Mai, N.; Zhang, M.; Zhou, H.; Haehner, A.; Chen, X.; Wu, Z.; Auber, L.A.; et al. Structural and Functional Abnormalities of Olfactory-Related Regions in Subjective Cognitive Decline, Mild Cognitive Impairment, and Alzheimer’s Disease. Int. J. Neuropsychopharmacol. 2022, 25, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Risacher, S.L.; West, J.D.; McDonald, B.C.; Magee, T.R.; Farlow, M.R.; Gao, S.; O’Neill, D.P.; Saykin, A.J. Altered Default Mode Network Connectivity in Older Adults with Cognitive Complaints and Amnestic Mild Cognitive Impairment. J. Alzheimer’s Dis. 2013, 35, 751–760. [Google Scholar] [CrossRef]
- Chiesa, P.A.; Cavedo, E.; Grothe, M.J.; Houot, M.; Teipel, S.J.; Potier, M.C.; Habert, M.O.; Lista, S.; Dubois, B.; Hampel, H. Relationship between Basal Forebrain Resting-State Functional Connectivity and Brain Amyloid-b Deposition in Cognitively Intact Older Adults with Subjective Memory Complaints. Radiology 2019, 290, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zang, F.; Wang, Q.; Zhang, Q.; Tan, C.; Zhang, S.; Hu, T.; Qi, L.; Xu, S.; Ren, Q.; et al. Connectome-Based Model Predicts Episodic Memory Performance in Individuals with Subjective Cognitive Decline and Amnestic Mild Cognitive Impairment. Behav. Brain Res. 2021, 411, 113387. [Google Scholar] [CrossRef]
- Xue, C.; Yuan, B.; Yue, Y.; Xu, J.; Wang, S.; Wu, M.; Ji, N.; Zhou, X.; Zhao, Y.; Rao, J.; et al. Distinct Disruptive Patterns of Default Mode Subnetwork Connectivity Across the Spectrum of Preclinical Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 307. [Google Scholar] [CrossRef]
- Damoiseaux, J.S.; Prater, K.E.; Miller, B.L.; Greicius, M.D. Functional Connectivity Tracks Clinical Deterioration in Alzheimer’s Disease. Neurobiol. Aging 2012, 33, 828.e19–828.e30. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Xie, C.; Shu, H.; Liao, W.; Wang, Z.; Liu, D.; Zhang, Z. Differential Effects of APOE Genotypes on the Anterior and Posterior Subnetworks of Default Mode Network in Amnestic Mild Cognitive Impairment. J. Alzheimer’s Dis. 2016, 54, 1409–1423. [Google Scholar] [CrossRef]
- Niu, H.; Zhu, Z.; Wang, M.; Li, X.; Yuan, Z.; Sun, Y.; Han, Y. Abnormal Dynamic Functional Connectivity and Brain States in Alzheimer’s Diseases: Functional near-Infrared Spectroscopy Study. Neurophotonics 2019, 6, 025010. [Google Scholar] [CrossRef]
- Xue, J.; Guo, H.; Gao, Y.; Wang, X.; Cui, H.; Chen, Z.; Wang, B.; Xiang, J. Altered Directed Functional Connectivity of the Hippocampus in Mild Cognitive Impairment and Alzheimer’s Disease: A Resting-State FMRI Study. Front. Aging Neurosci. 2019, 11, 326. [Google Scholar] [CrossRef] [PubMed]
- Hojjati, S.H.; Ebrahimzadeh, A.; Khazaee, A.; Babajani-Feremi, A. Predicting Conversion from MCI to AD by Integrating Rs-FMRI and Structural MRI. Comput. Biol. Med. 2018, 102, 30–39. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, K.R.A.; Sabuncu, M.R.; Buckner, R.L. The Influence of Head Motion on Intrinsic Functional Connectivity MRI. Neuroimage 2012, 59, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Diao, Y.; Yin, T.; Gruetter, R.; Jelescu, I.O. PIRACY: An Optimized Pipeline for Functional Connectivity Analysis in the Rat Brain. Front. Neurosci. 2021, 15, 602170. [Google Scholar] [CrossRef]
- Birn, R.M. The Role of Physiological Noise in Resting-State Functional Connectivity. Neuroimage 2012, 62, 864–870. [Google Scholar] [CrossRef]
- Teeuw, J.; Hulshoff Pol, H.E.; Boomsma, D.I.; Brouwer, R.M. Reliability Modelling of Resting-State Functional Connectivity. Neuroimage 2021, 231, 117842. [Google Scholar] [CrossRef]
- Power, J.D.; Mitra, A.; Laumann, T.O.; Snyder, A.Z.; Schlaggar, B.L.; Petersen, S.E. Methods to Detect, Characterize, and Remove Motion Artifact in Resting State FMRI. Neuroimage 2014, 84, 320–341. [Google Scholar] [CrossRef]
- Murphy, K.; Birn, R.M.; Bandettini, P.A. Resting-State FMRI Confounds and Cleanup. Neuroimage 2013, 80, 349–359. [Google Scholar] [CrossRef]
- Rittman, T.; Rubinov, M.; Vértes, P.E.; Patel, A.X.; Ginestet, C.E.; Ghosh, B.C.P.; Barker, R.A.; Spillantini, M.G.; Bullmore, E.T.; Rowe, J.B. Regional Expression of the MAPT Gene Is Associated with Loss of Hubs in Brain Networks and Cognitive Impairment in Parkinson Disease and Progressive Supranuclear Palsy. Neurobiol. Aging 2016, 48, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Achard, S.; Delon-Martin, C.; Vértes, P.E.; Renard, F.; Schenck, M.; Schneider, F.; Heinrich, C.; Kremer, S.; Bullmore, E.T. Hubs of Brain Functional Networks Are Radically Reorganized in Comatose Patients. Proc. Natl. Acad. Sci. USA 2012, 109, 20608–20613. [Google Scholar] [CrossRef]
- Cope, T.E.; Rittman, T.; Borchert, R.J.; Jones, P.S.; Vatansever, D.; Allinson, K.; Passamonti, L.; Vazquez Rodriguez, P.; Bevan-Jones, W.R.; O’Brien, J.T.; et al. Tau Burden and the Functional Connectome in Alzheimer’s Disease and Progressive Supranuclear Palsy. Brain 2018, 141, 550–567. [Google Scholar] [CrossRef] [PubMed]
- Tzourio-Mazoyer, N.; Landeau, B.; Papathanassiou, D.; Crivello, F.; Etard, O.; Delcroix, N.; Mazoyer, B.; Joliot, M. Automated Anatomical Labeling of Activations in SPM Using a Macroscopic Anatomical Parcellation of the MNI MRI Single-Subject Brain. Neuroimage 2002, 15, 273–289. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Su, L.; Hu, X.; Han, Y. Sino Longitudinal Study on Cognitive Decline (SILCODE): Protocol for a Chinese Longitudinal Observational Study to Develop Risk Prediction Models of Conversion to Mild Cognitive Impairment in Individuals with Subjective Cognitive Decline. BMJ Open 2019, 9, e028188. [Google Scholar] [CrossRef]
- Li, T.R.; Wu, Y.; Jiang, J.J.; Lin, H.; Han, C.L.; Jiang, J.H.; Han, Y. Radiomics Analysis of Magnetic Resonance Imaging Facilitates the Identification of Preclinical Alzheimer’s Disease: An Exploratory Study. Front. Cell Dev. Biol. 2020, 8, 605734. [Google Scholar] [CrossRef]
- Pet, F.; Fakhry-darian, D.; Patel, N.H.; Khan, S.; Barwick, T.; Svensson, W.; Win, Z. Optimisation and Usefulness of Quantitative Analysis Of 18F-florbetapir PET. Br. J. Radiol. 2019, 92, 1101. [Google Scholar] [CrossRef]
- Bondi, M.W.; Edmonds, E.C.; Jak, A.J.; Clark, L.R.; Delano-Wood, L.; McDonald, C.R.; Nation, D.A.; Libon, D.J.; Au, R.; Galasko, D.; et al. Neuropsychological Criteria for Mild Cognitive Impairment Improves Diagnostic Precision, Biomarker Associations, and Progression Rates. J. Alzheimer’s Dis. 2014, 42, 275–289. [Google Scholar] [CrossRef] [PubMed]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The Diagnosis of Dementia Due to Alzheimer’s Disease: Recommendations from the National Institute on Aging-Alzheimer’s Association Workgroups on Diagnostic Guidelines for Alzheimer’s Disease. Alzheimer’s Dement. 2011, 7, 263–269. [Google Scholar] [CrossRef]
- Ashton, N.J.; Leuzy, A.; Karikari, T.K.; Mattsson-Carlgren, N.; Dodich, A.; Boccardi, M.; Corre, J.; Drzezga, A.; Nordberg, A.; Ossenkoppele, R.; et al. The Validation Status of Blood Biomarkers of Amyloid and Phospho-Tau Assessed with the 5-Phase Development Framework for AD Biomarkers. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 2140–2156. [Google Scholar] [CrossRef]
- Jack, C.R.; Hampel, H.J.; Universities, S.; Cu, M.; Petersen, R.C. A New Classification System for AD, Independent of Cognition A/T/N: An Unbiased Descriptive Classification Scheme for Alzheimer Disease Biomarkers. Neurology 2016, 87, 539–547. [Google Scholar] [CrossRef]
- Blennow, K.; Mattsson, N.; Schöll, M.; Hansson, O.; Zetterberg, H. Amyloid Biomarkers in Alzheimer’s Disease. Trends Pharmacol. Sci. 2015, 36, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Lane, C.A.; Hardy, J.; Schott, J.M. Alzheimer’s Disease. Eur. J. Neurol. 2018, 25, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.J.; Lee, J.-H. Subjective Cognitive Decline and Alzheimer’s Disease Spectrum Disorder. Dement. Neurocognitive Disord. 2017, 16, 40. [Google Scholar] [CrossRef]
- Hu, X.; Teunissen, C.E.; Spottke, A.; Heneka, M.T.; Düzel, E.; Peters, O.; Li, S.; Priller, J.; Buerger, K.; Teipel, S.; et al. Smaller Medial Temporal Lobe Volumes in Individuals with Subjective Cognitive Decline and Biomarker Evidence of Alzheimer’s Disease—Data from Three Memory Clinic Studies. Alzheimer’s Dement. 2019, 15, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Yushkevich, P.A.; Pluta, J.B.; Wang, H.; Xie, L.; Ding, S.L.; Gertje, E.C.; Mancuso, L.; Kliot, D.; Das, S.R.; Wolk, D.A. Automated Volumetry and Regional Thickness Analysis of Hippocampal Subfields and Medial Temporal Cortical Structures in Mild Cognitive Impairment. Hum. Brain Mapp. 2015, 36, 258–287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xi, J.; Xu, G.; Shu, H.; Wang, X.; Li, P. Cortical Dynamics of Acoustic and Phonological Processing in Speech Perception. PLoS ONE 2011, 6, e20963. [Google Scholar] [CrossRef]
- Onitsuka, T.; Shenton, M.E.; Salisbury, D.F.; Dickey, C.C.; Kasai, K.; Toner, S.K.; Frumin, M.; Kikinis, R.; Jolesz, F.A.; McCarley, R.W. Middle and Inferior Temporal Gyrus Gray Matter Volume Abnormalities in Chronic Schizophrenia: An MRI Study. Am. J. Psychiatry 2004, 161, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Kann, M.; Hedrich, K.; Vieregge, P.; Jacobs, H.; Müller, B.; Kock, N.; Schwinger, E.; Klein, C.; Marder, K.; Harris, J.; et al. The Parkin Gene Is Not Involved in Late-Onset Parkinson’s Disease. Neurology 2002, 58, 835. [Google Scholar] [CrossRef]
- Busatto, G.F.; Garrido, G.E.J.; Almeida, O.P.; Castro, C.C.; Camargo, C.H.P.; Cid, C.G.; Buchpiguel, C.A.; Furuie, S.; Bottino, C.M. A Voxel-Based Morphometry Study of Temporal Lobe Gray Matter Reductions in Alzheimer’s Disease. Neurobiol. Aging 2003, 24, 221–231. [Google Scholar] [CrossRef]
- Ci, H.; van Graan, A.; Gonzálvez, G.; Thompson, P.; Hill, A.; Duncan, J.S. Mandarin Functional MRI Language Paradigms. Brain Behav. 2016, 6, e00525. [Google Scholar] [CrossRef] [PubMed]
- Park, D.C.; Reuter-Lorenz, P. The Adaptive Brain: Aging and Neurocognitive Scaffolding. Annu. Rev. Psychol. 2009, 60, 173–196. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Maraganore, D.M.; Ioannidis, J.P.A.; Riess, O.; Aasly, J.O.; Annesi, G.; Abahuni, N.; Bentivoglio, A.R.; Brice, A.; Van Broeckhoven, C.; et al. Role of Sepiapterin Reductase Gene at the PARK3 Locus in Parkinson’s Disease. Neurobiol. Aging 2011, 32, 2108.e1–2108.e5. [Google Scholar] [CrossRef] [PubMed]
- Hammerschmidt, W.; Kagan, I.; Kulke, L.; Schacht, A. Implicit Reward Associations Impact Face Processing: Time-Resolved Evidence from Event-Related Brain Potentials and Pupil Dilations. Neuroimage 2018, 179, 557–569. [Google Scholar] [CrossRef] [PubMed]
| Cohort 1 | Cohort 2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NC (91) | SCD (61) | MCI (44) | AD (13) | p Value | NC (140) | SCD (75) | MCI (205) | AD (94) | p Value | |
| Age (years) | 65.5 ± 5.7 | 66.7 ± 5.9 | 66.9 ± 8.4 | 71.4 ± 8.9 | 0.204 b | 72.0 ± 5.9 | 71.6 ± 5.5 | 71.3 ± 7.2 | 74.0 ± 7.8 | 0.560 b |
| Education (years) | 12.5 ± 3.0 | 12.8 ± 2.7 | 10.6 ± 3.9 | 11.5 ± 4.2 | 0.488 b | 16.2 ± 2.2 | 16.9 ± 2.5 | 16.0 ± 2.7 | 14.9 ± 2.5 | 0.035 b,* |
| Gender (male/female) | 38/53 | 12/49 | 19/25 | 3/10 | 0.004 a,* | 59/81 | 32/43 | 100/105 | 45/49 | 0.941 a |
| Aβ(+/−) | 24/20 | 22/14 | - | - | 0.555 a | 45/19 | 31/19 | - | - | 0.350 a |
| HAMD | 2.8 ± 3.8 | 5.3 ± 1.7 | 6.4 ± 6.8 | 4.7 ± 5.7 | 0.044 b,* | - | - | - | - | - |
| HAMA | 3.3 ± 2.2 | 5.3 ± 4.5 | 5.0 ± 4.9 | 4.5 ± 4.4 | 0.002 b,* | - | - | - | - | - |
| SCD-9 | 3.5 ± 2.2 | 5.3 ± 1.7 | 2.0 ± 1.8 | 2.8 ± 0.8 | <0.001 b,* | - | - | - | - | - |
| MMSE | 28.9 ± 1.4 | 28.9 ± 1.3 | 26.0 ± 2.7 | 17.0 ± 5.3 | 0.861 b | 28.7 ± 4.6 | 29.0 ± 0.9 | 27.3 ± 2.5 | 21.1 ± 3.5 | 0.119 b |
| AVLT-N5 | 7.5 ± 1 | 7.1 ± 1.9 | 3.8 ± 2.4 | 0.9 ± 0.4 | 0.248 b | - | - | - | - | - |
| AVLT- Recognition | 22.4 ± 1.5 | 22.4 ± 1.4 | 19.7 ± 2.6 | 15.7 ± 2.6 | 0.881 b | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Lin, H.; Zhang, Q.; Shi, R.; Xu, H.; Yang, F.; Jiang, X.; Wang, L.; Han, Y.; Jiang, J. Individual Proportion Loss of Functional Connectivity Strength: A Novel Individual Functional Connectivity Biomarker for Subjective Cognitive Decline Populations. Biology 2023, 12, 564. https://doi.org/10.3390/biology12040564
Li Z, Lin H, Zhang Q, Shi R, Xu H, Yang F, Jiang X, Wang L, Han Y, Jiang J. Individual Proportion Loss of Functional Connectivity Strength: A Novel Individual Functional Connectivity Biomarker for Subjective Cognitive Decline Populations. Biology. 2023; 12(4):564. https://doi.org/10.3390/biology12040564
Chicago/Turabian StyleLi, Zhuoyuan, Hua Lin, Qi Zhang, Rong Shi, Huanyu Xu, Fan Yang, Xueyan Jiang, Luyao Wang, Ying Han, and Jiehui Jiang. 2023. "Individual Proportion Loss of Functional Connectivity Strength: A Novel Individual Functional Connectivity Biomarker for Subjective Cognitive Decline Populations" Biology 12, no. 4: 564. https://doi.org/10.3390/biology12040564
APA StyleLi, Z., Lin, H., Zhang, Q., Shi, R., Xu, H., Yang, F., Jiang, X., Wang, L., Han, Y., & Jiang, J. (2023). Individual Proportion Loss of Functional Connectivity Strength: A Novel Individual Functional Connectivity Biomarker for Subjective Cognitive Decline Populations. Biology, 12(4), 564. https://doi.org/10.3390/biology12040564







