Date Palm Waste Compost Application Increases Soil Microbial Community Diversity in a Cropping Barley (Hordeum vulgare L.) Field
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site and Design
2.2. Soil Sampling
2.3. DNA Extraction, Amplicon Library Preparation and Sequencing
2.4. Quantitative Real-Time PCR (qPCR) Analysis
2.5. Data Analysis and Statistics
3. Results
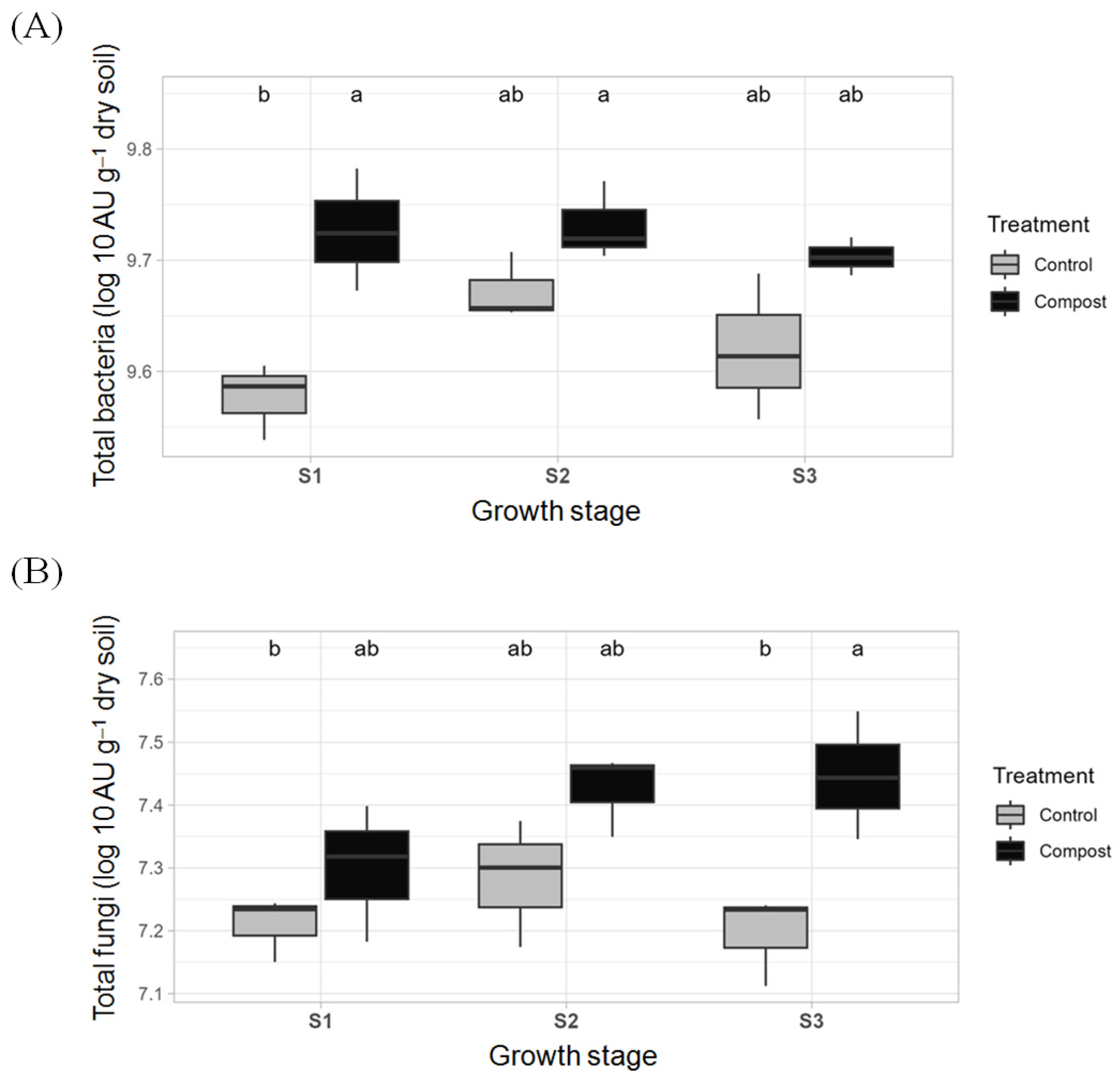
3.1. Changes in the Soil’s Bacteria and Fungi Abundances
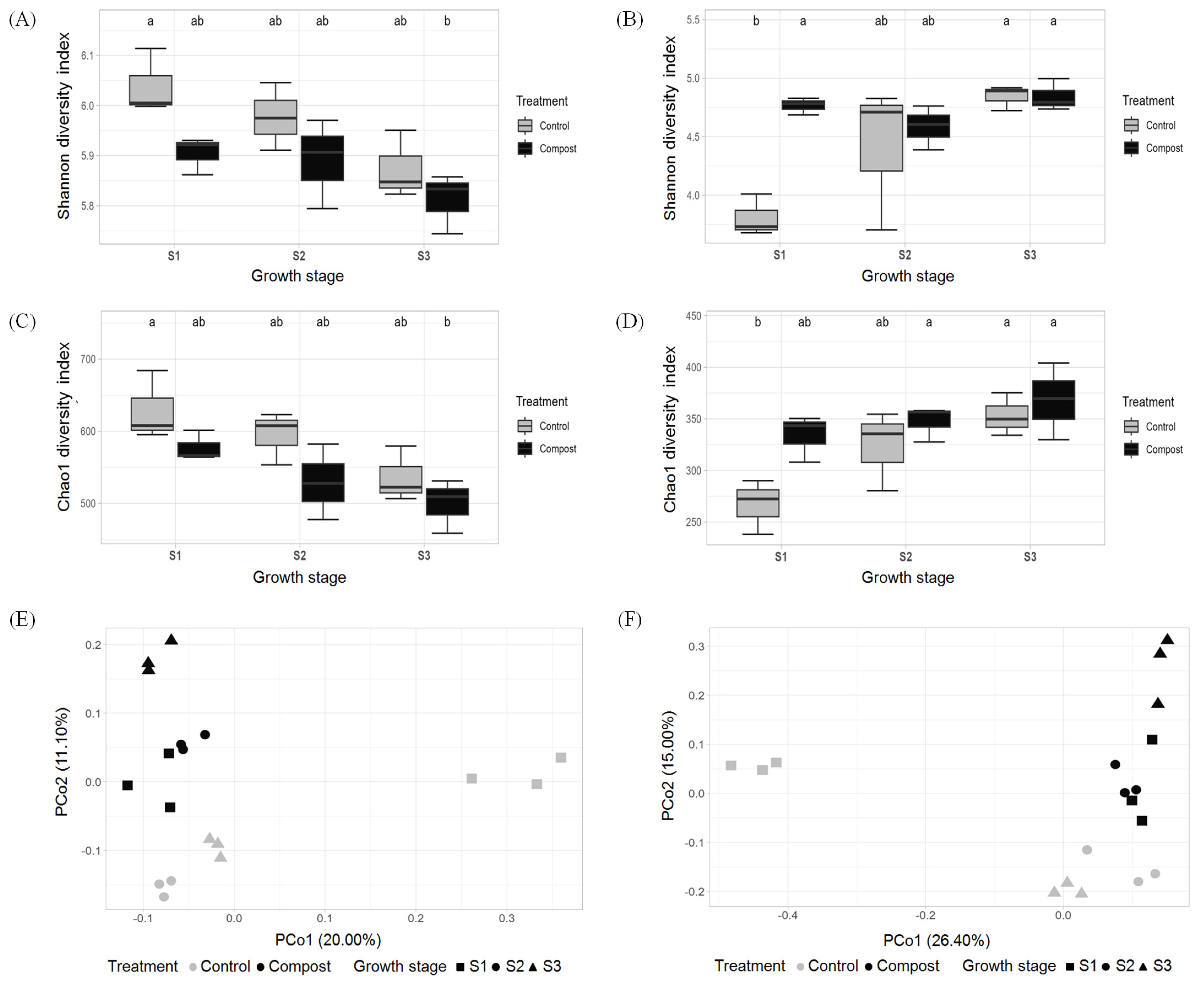
3.2. Effect of Compost Application on Bacterial and Fungal Diversity in Soil
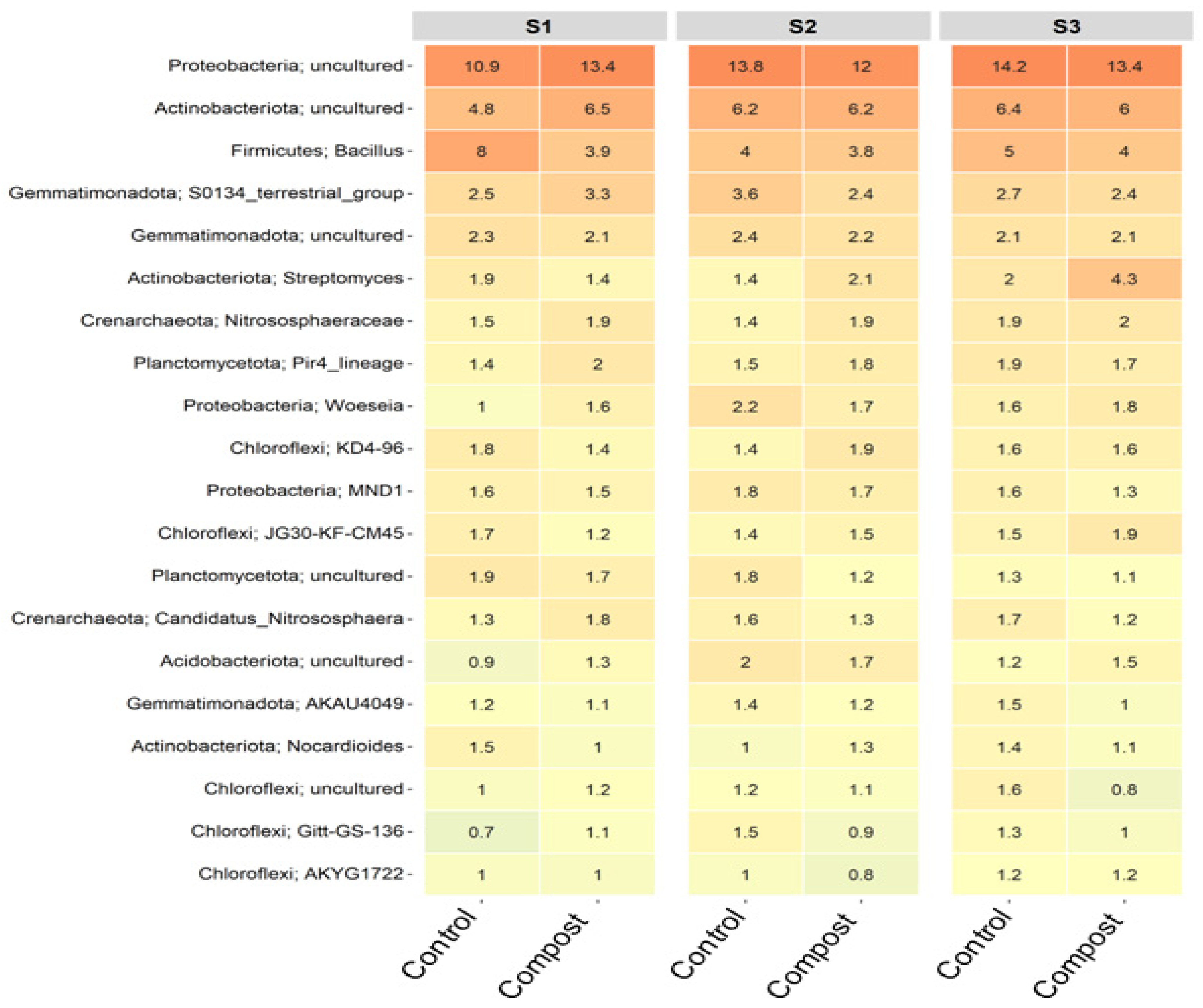
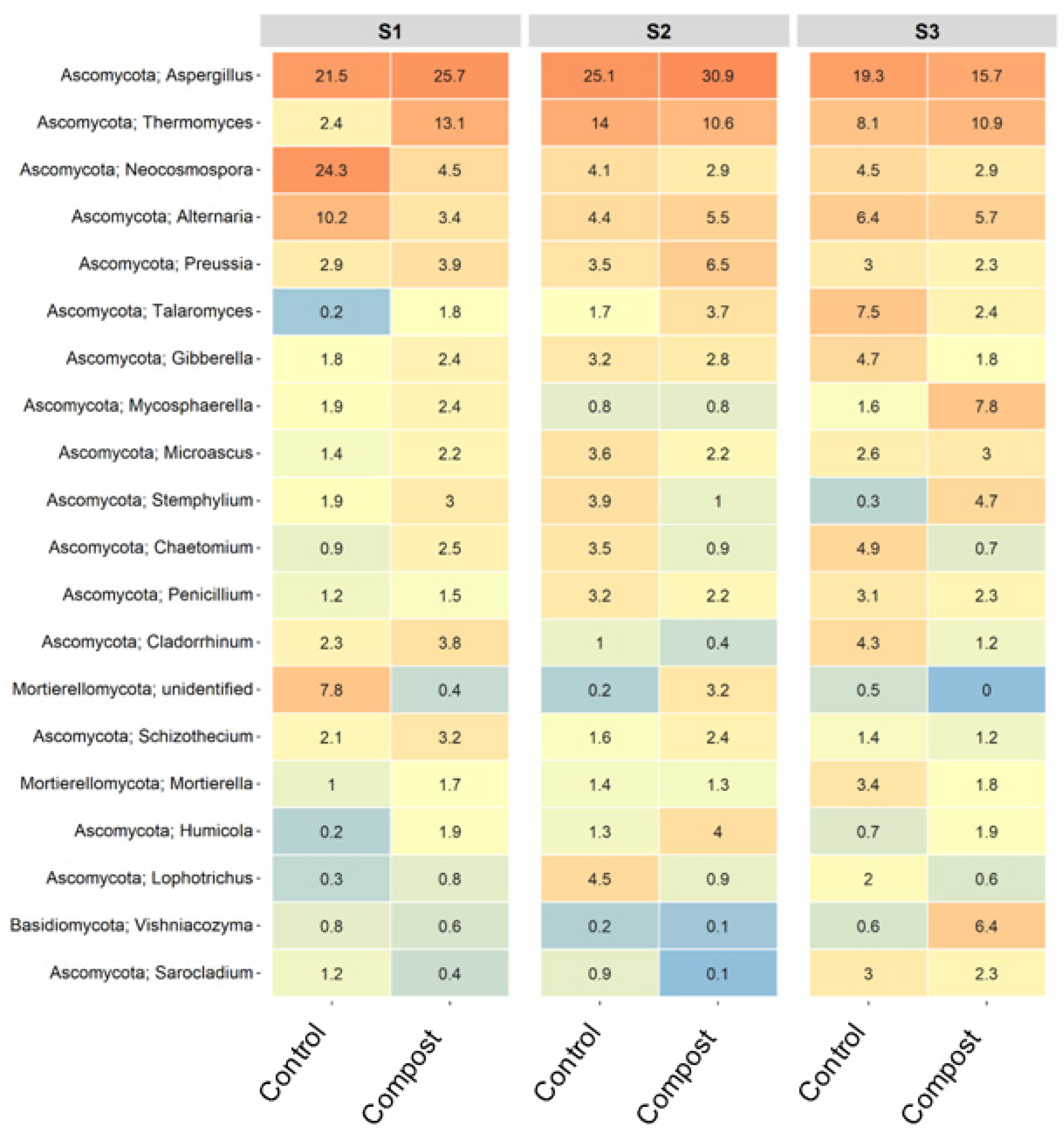
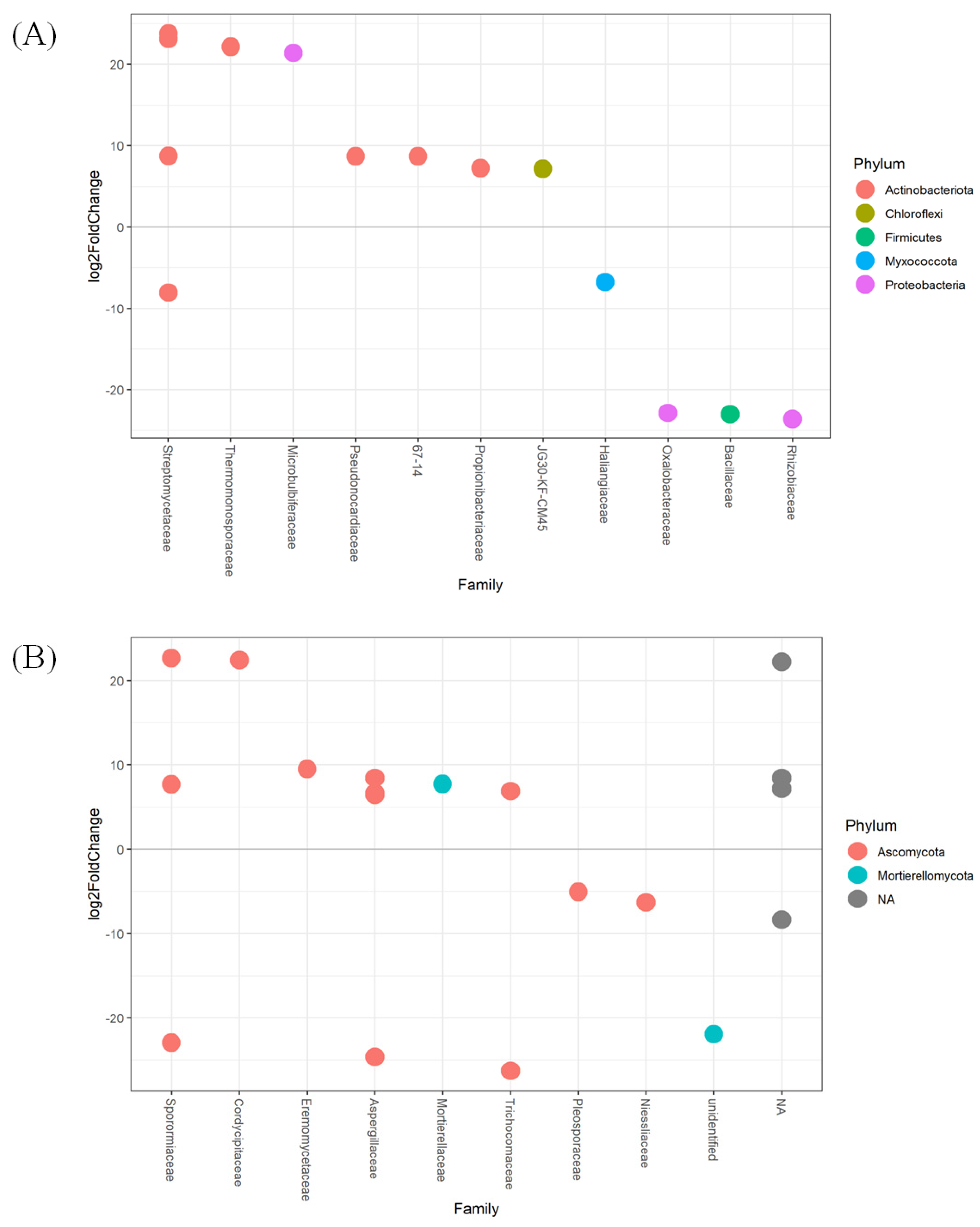
3.3. Taxonomic Changes to Bacterial and Fungal Populations in Soil
3.4. Major ASVs within Barley Soil Microbial Communities
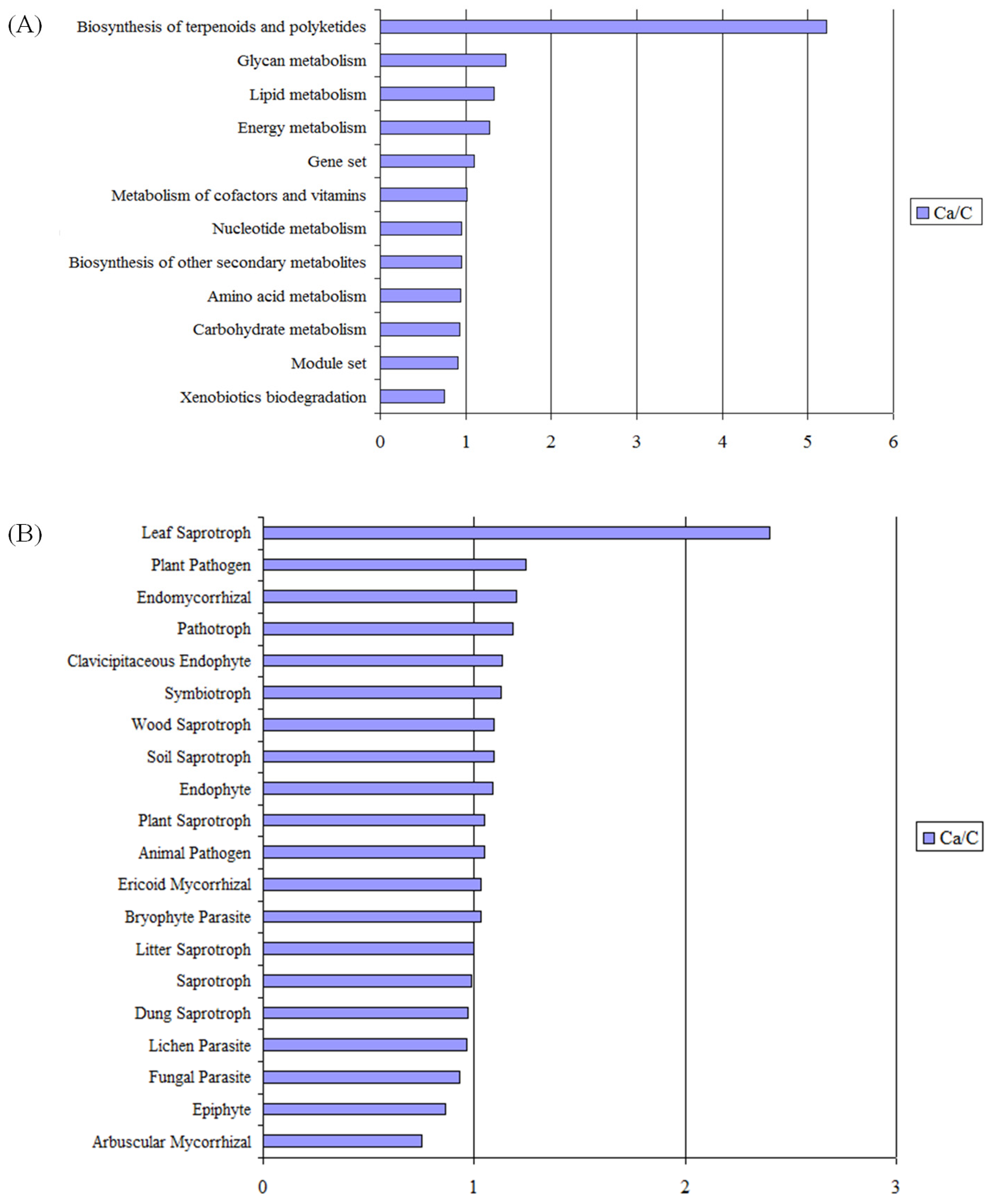
3.5. Metabolic Functional Features of the Microbial Community Present in Barley Rhizosphere
4. Discussion
4.1. Application of Compost Affects Soil Microbial Community Abundance and Diversity
4.2. Application of Compost Affects Bacterial and Fungal Community Composition
4.3. Effect of Date Palm Waste Compost Application on the Overall Function of Soil Microbial Community
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Asaye, Z.; Kim, D.G.; Yimer, F.; Prost, K.; Obsa, O.; Tadesse, M.; Gebrehiwot, M.; Brüggemann, N. Effects of combined application of compost and mineral fertilizer on soil carbon and nutrient content, yield, and agronomic nitrogen use efficiency in maize-potato cropping systems in southern Ethiopia. Land 2022, 11, 784. [Google Scholar] [CrossRef]
- Ho, T.T.K.; Tra, V.T.; Le, T.H.; Nguyen, N.K.Q.; Tran, G.S.; Nguyen, P.T.; Vo, T.D.H.; Thai, V.N.; Bui, X.T. Compost to improve sustainable soil cultivation and crop productivity. Case Stud. Chem. Environ. Eng. 2022, 6, 100211. [Google Scholar] [CrossRef]
- Heisey, S.; Ryals, R.; Maaz, T.M.; Nguyen, N.H. A single application of compost can leave lasting impacts on soil microbial community structure and alter cross-domain interaction networks. Front. Soil Sci. 2022, 2, 749212. [Google Scholar] [CrossRef]
- Ghouili, E.; Hidri, Y.; Cheikh M’Hamed, H.; Somenahally, A.; Xue, Q.; El Akram Znaïdi, I.; Jebara, M.; Nefissi Ouertani, R.; Muhovski, Y.; Riahi, J.; et al. Date palm waste compost promotes plant growth and nutrient transporter genes expression in barley (Hordeum vulgare L.). S. Afr. J. Bot. 2022, 149, 247–257. [Google Scholar] [CrossRef]
- Adomako, M.O.; Roiloa, S.; Yu, F.H. Potential roles of soil microorganisms in regulating the effect of soil nutrient heterogeneity on plant performance. Microorganisms 2022, 10, 2399. [Google Scholar] [CrossRef]
- Lòpez-Fernàndez, S.; Compant, S.; Vrhovsek, U.; Bianchedi, P.L.; Sessitsch, A.; Pertot, I.; Campisano, A. Grapevine colonization by endophytic bacteria shifts secondary metabolism and suggests activation of defense pathways. Plant Soil 2016, 405, 155–175. [Google Scholar] [CrossRef]
- Lau, J.A.; Lennon, J.T. Evolutionary ecology of plant–microbe interactions: Soil microbial structure alters selection on plant traits. New Phytol. 2011, 192, 215–224. [Google Scholar] [CrossRef]
- Chaney, L.; Baucom, R.S. The soil microbial community alters patterns of selection on flowering time and fitness-related traits in Ipomoea purpurea. Am. J. Bot. 2020, 107, 186–194. [Google Scholar] [CrossRef]
- Sharaf, H.; Thompson, A.A.; Williams, M.A.; Peck, G.M. Compost applications increase bacterial community diversity in the apple rhizosphere. Soil Sci. Soc. Am. J. 2021, 85, 1105–1121. [Google Scholar] [CrossRef]
- Zhen, Z.; Liu, H.; Wang, N.; Guo, L.; Meng, J.; Ding, N.; Wu, G.; Jiang, G. Effects of manure compost application on soil microbial community diversity and soil microenvironments in a temperate cropland in china. PLoS ONE 2014, 9, e108555. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, S.; Li, N.; Chen, H.; Jia, H.; Song, X.; Liu, G.; Ni, C.; Wang, Z.; Shao, H.; et al. Metagenomic insights into effects of wheat straw compost fertiliser application on microbial community composition and function in tobacco rhizosphere soil. Sci. Rep. 2019, 9, 6168. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shi, Y.; Kong, L.; Tong, L.; Cao, H.; Zhou, H.; Lv, Y. Long-term application of bio-compost increased soil microbial community diversity and altered its composition and network. Microorganisms 2022, 10, 462. [Google Scholar] [CrossRef] [PubMed]
- Benabderrahim, M.A.; Elfalleh, W.; Belayadi, H.; Haddad, M. Effect of date palm waste compost on forage alfalfa growth, yield, seed yield and minerals uptake. Int. J. Recycl. Org. Waste Agricult. 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Brahim, N.; Ibrahim, H.; Mlih, R.; Bouajila, A.; Karbout, N.; Bol, R. Soil OC and N stocks in the saline soil of Tunisian gataaya oasis eight years after application of manure and compost. Land 2022, 11, 442. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Mills, D.A. Improved selection of internal transcribed spacerspecific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 2013, 79, 2519–2526. [Google Scholar] [CrossRef]
- Jeanne, T.; D’Astous-Pagé, J.; Hogue, R. Spatial, temporal and technical variability in the diversity of prokaryotes and fungi in agricultural soils. Front. Soil Sci. 2022, 2, 945888. [Google Scholar] [CrossRef]
- Overbeek, W.; Jeanne, T.; Hogue, R.; Smith, D.L. Effects of microbial consortia, applied as fertilizer coating, on soil and rhizosphere microbial communities and potato yield. Front. Agron. 2021, 3, 714700. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jackson, B.; Jackson, R.B. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 2005, 71, 4117–4120. [Google Scholar] [CrossRef]
- Emerson, J.B.; Keady, P.B.; Brewer, T.E.; Clements, N.; Morgan, E.E.; Awerbuch, J.; Miller, S.L.; Fierer, N. Impacts of flood damage on airborne bacteria and fungi in homes after the 2013 Colorado front range flood. Environ. Sci. Technol. 2015, 49, 2675–2684. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Chao, A.; Shen, T.J. Nonparametric estimation of shannon’s index of diversity when there are unseen species in sample. Environ. Ecol. Stat. 2003, 10, 429–443. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell. System. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.S.; Kirkegaard, R.H.; Karst, S.M.; Albertsen, M. Ampvis2: An R package to analyse and visualise 16S rRNA amplicon data. BioRxiv 2018, 299537. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; HUTTENHOWER, C.; Langille, M.G. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic. Acids Res. 2022, 51, D587–D592. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Khmelevtsova, L.E.; Sazykin, I.S.; Azhogina, T.N.; Sazykina, M.A. Influence of agricultural practices on bacterial community of cultivated soils. Agriculture 2022, 12, 371. [Google Scholar] [CrossRef]
- Su, J.Y.; Liu, C.H.; Tampus, K.; Lin, Y.C.; Huang, C.H. Organic amendment types influence soil properties, the soil bacterial microbiome, and tomato growth. Agronomy 2022, 12, 1236. [Google Scholar] [CrossRef]
- Bhattacharyya, S.S.; Ros, G.H.; Furtak, K.; Iqbal, H.M.M.; Parra-Saldívar, R. Soil carbon sequestration–An interplay between soil microbial community and soil organic matter dynamics. Sci. Total Environ. 2022, 815, 152928. [Google Scholar] [CrossRef]
- Wu, J.; Shi, Z.; Zhu, J.; Cao, A.; Fang, W.; Yan, D.; Wang, Q.; Li, Y. Taxonomic response of bacterial and fungal populations to biofertilizers applied to soil or substrate in greenhouse-grown cucumber. Sci. Rep. 2022, 12, 18522. [Google Scholar] [CrossRef]
- Gao, W.; Gao, K.; Guo, Z.; Liu, Y.; Jiang, L.; Liu, C.; Liu, X.; Wang, G. Different responses of soil bacterial and fungal communities to 3 years of biochar amendment in an alkaline soybean soil. Front. Microbiol. 2021, 12, 630418. [Google Scholar] [CrossRef]
- Su, L.; Bai, T.; Qin, X.; Yu, H.; Wu, G.; Zhao, Q.; Tan, L. Organic manure induced soil food web of microbes and nematodes drive soil organic matter under jackfruit planting. Appl. Soil Ecol. 2021, 166, 103994. [Google Scholar] [CrossRef]
- Azeem, M.; Hale, L.; Montgomery, J.; Crowley, D.; McGiffen, M.E., Jr. Biochar and compost effects on soil microbial communities and nitrogen induced respiration in turfgrass soils. PLoS ONE 2020, 15, e0242209. [Google Scholar] [CrossRef]
- Tao, R.; Hu, B.; Chu, G. Impacts of organic fertilization with a drip irrigation system on bacterial and fungal communities in cotton field. Agric. Syst. 2020, 182, 102820. [Google Scholar] [CrossRef]
- Luo, P.; Han, X.; Wang, Y.; Han, M.; Shi, H.; Liu, N.; Bai, H. Influence of long-term fertilization on soil microbial biomass, dehydrogenase activity, and bacterial and fungal community structure in a brown soil of Northeast China. Ann. Microbiol. 2015, 65, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Ruan, Y.; Chao, X.; Zhang, J.; Li, R.; Shen, Q. Rhizosphere microbial community manipulated by 2 years of consecutive biofertilizer application associated with banana Fusarium wilt disease suppression. Biol. Fertil. Soils 2015, 51, 553–562. [Google Scholar] [CrossRef]
- Luo, S.; Wang, S.; Tian, L.; Li, S.; Li, X.; Shen, Y.; Tian, C. Long-term biochar application influences soil microbial community and its potential roles in semiarid farmland. Appl. Soil Ecol. 2017, 117, 10–15. [Google Scholar] [CrossRef]
- Van Elsas, J.D.; Garbeva, P.; Salles, J. Effects of agronomical measures on the microbial diversity of soils as related to the suppression of soil-borne plant pathogens. Biodegradation 2002, 13, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Wang, Y.; Jin, X.; Zhao, Y.; Yan, H.; Zhang, H.; Zhou, X.; Lu, G.; Deng, Y. Application of organic fertilizer changes the rhizosphere microbial communities of a gramineous grass on Qinghai–Tibet plateau. Microorganisms 2022, 10, 1148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.G.; Zhou, Z.B.; Xiao, T. Trichoderma biofertilizer facilitating Leymus chinensis production in different growth stages is strongly linked to dynamically altered soil microbiomes. Agric. Ecosyst. Environ. 2020, 324, 107706. [Google Scholar] [CrossRef]
- Fontaine, S.; Mariotti, A.; Abbadie, L. The priming effect of organic matter: A question of microbial competition? Soil Biol. Biochem. 2003, 35, 837–843. [Google Scholar] [CrossRef]
- Pan, H.; Chen, M.M.; Feng, H.J.; Wei, M.; Song, F.P.; Lou, Y.H.; Cui, X.M.; Wang, H.; Zhuge, Y.P. Organic and inorganic fertilizers respectively drive bacterial and fungal community compositions in a fluvo-aquic soil in northern China. Soil Till. Res. 2020, 198, 104540. [Google Scholar] [CrossRef]
- Yin, C.; Hulbert, S.H.; Schroeder, K.L.; Mavrodi, O.; Mavrodi, D.; Dhingra, A.; Schillinger, W.F.; Paulitz, T.C. Role of bacterial communities in the natural suppression of rhizoctonia solani bare patch disease of wheat (Triticum aestivum L.). Appl. Environ. Microbiol. 2013, 79, 7428–7438. [Google Scholar] [CrossRef]
- Gupta, A.; Dutta, A.; Sarkar, J.; Panigrahi, M.K.; Sar, P. Low-abundance members of the firmicutes facilitate bioremediation of soil impacted by highly acidic mine drainage from the malanjkhand copper project, India. Front. Microbiol. 2018, 9, 2882. [Google Scholar] [CrossRef] [PubMed]
- Pang, G.; Cai, F.; Li, R.X.; Zhao, Z.; Li, R.; Gu, X.L.; Shen, Q.R.; Chen, W. Trichoderma-enriched organic fertilizer can mitigate microbiome degeneration of monocropped soil to maintain better plant growth. Plant Soil. 2017, 416, 181–192. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Y.; Mao, W.; Wang, C.; Yin, S. Functional potential differences between Firmicutes and Proteobacteria in response to manure amendment in a reclaimed soil. Can. J. Microbiol. 2020, 66, 689–697. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, Q.; Han, X. Soil bacterial communities respond to mowing and nutrient addition in a steppe ecosystem. PLoS ONE 2013, 8, e84210. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Lu, L.; Peng, C.; Li, H.; Zhang, J.; Li, R.; Zhou, C. High-yield grass Pennisetum sinese Roxb plantation and organic manure alter bacterial and fungal communities structure in an ecological agriculture farm. AMB Expr. 2020, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, A.; Soares, T.; Rossetto, R.; Van Veen, J.A.; Tsai, S.M.; Kuramae, E.E. Verrucomicrobial community structure and abundance as indicators for changes in chemical factors linked to soil fertility. Antonie Van Leeuwenhoek 2015, 108, 741–752. [Google Scholar] [CrossRef]
- Kielak, A.M.; Barreto, C.; Kowalchuk, G.A.; Van Veen, J.A.; Kuramae, E.E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef]
- Bogino, P.C.; Oliva, M.D.L.M.; Sorroche, F.G.; Giordano, W. The role of bacterial biofilms and surface components in plant bacterial associations. Int. J. Mol. Sci. 2013, 14, 15838–15859. [Google Scholar] [CrossRef]
- Kielak, A.M.; Cipriano, M.A.; Kuramae, E.E. Acidobacteria strains from subdivision 1 act as plant growth-promoting bacteria. Arch. Microbiol. 2016, 198, 987–993. [Google Scholar] [CrossRef]
- Alami, M.M.; Xue, J.; Ma, Y.; Zhu, D.; Abbas, A.; Gong, Z.; Wang, X. Structure, function, diversity, and composition of fungal communities in rhizospheric soil of Coptis chinensis Franch under a successive cropping system. Plants 2020, 9, 244. [Google Scholar] [CrossRef]
- Steindorff, A.S.; Serra, L.A.; Formighieri, E.F.; de Faria, F.P.; Poças-Fonseca, M.J.; de Almeida, J.R.M. Insights into the lignocellulose-degrading enzyme system of Humicola grisea var. thermoidea based on genome and transcriptome analysis. Microbiol. Spectr. 2021, 9, e01088-21. [Google Scholar] [CrossRef]
- Kumar, R.; Kundu, A.; Dutta, A.; Saha, S.; Das, A.; Bhowmik, A. Chemo-profiling of bioactive metabolites from Chaetomium globosum for biocontrol of Sclerotinia rot and plant growth promotion. Fungal Biol. 2021, 125, 167–176. [Google Scholar] [CrossRef]
- De Corato, U.; Patruno, L.; Avella, N.; Lacolla, G.; Cucci, G. Composts from green sources show an increased suppressiveness to soilborne plant pathogenic fungi: Relationships between physicochemical properties, disease suppression, and the microbiome. Crop Prot. 2019, 124, 104870. [Google Scholar] [CrossRef]
- Nikolic, M.; Savic, I.; Nikolic, A.; Stankovic, G.; Stankovic, S. Maize resistance to ear rot caused by aspergillus parasiticus. Genetika 2019, 51, 357–363. [Google Scholar] [CrossRef]
- Riaz, M.; Akhtar, N.; Msimbira, L.; Antar, M.; Ashraf, S.; Khan, S.N.; Smith, D.L. Neocosmospora rubicola, a stem rot disease in potato: Characterization, distribution and management. Front. Microbiol. 2022, 13, 953097. [Google Scholar] [CrossRef]
- Naraghi, L.; Heydari, A.; Rezaee, S.; Razavi, M. Biocontrol agent Talaromyces flavus stimulates the growth of cotton and potato. J. Plant Growth Regul. 2012, 31, 471–477. [Google Scholar] [CrossRef]
- Yamagiwa, Y.; Inagaki, Y.; Ichinose, Y.; Toyoda, K.; Hyakumachi, M.; Shiraishi, T. Talaromyces wortmannii FS2 emits β-caryphyllene, which promotes plant growth and induces resistance. J. Gen. Plant Pathol. 2011, 77, 336–341. [Google Scholar] [CrossRef]
- Ozimek, E.; Hanaka, A. Mortierella species as the plant growth-promoting fungi present in the agricultural soils. Agriculture 2021, 11, 7. [Google Scholar] [CrossRef]
- Sun, R.B.; Dsouza, M.; Gilbert, J.A.; Guo, X.S.; Wang, D.Z.; Guo, Z.B.; Ni, Y.Y.; Chu, H.Y. Fungal community composition in soils subjected to long-term chemical fertilization is most influenced by the type of organic matter. Environ. Microbiol. 2016, 18, 5137–5150. [Google Scholar] [CrossRef]
- Jones, R.T.; Robeson, M.S.; Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 2009, 3, 442–453. [Google Scholar] [CrossRef]
- Qiao, H.; Sun, X.R.; Wu, X.Q.; Li, G.E.; Wang, Z.; Li, D.W. The phosphate-solubilizing ability of Penicillium guanacastense and its effects on the growth of Pinus massoniana in phosphate-limiting conditions. Biol. Open. 2019, 8, bio046797. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, R.; Kang, S.M.; Baek, I.Y.; Lee, I.J. Characterization of plant growth-promoting traits of Penicillium species against the effects of high soil salinity and root disease. J. Plant Interact. 2014, 9, 754–762. [Google Scholar] [CrossRef]
- Mącik, M.; Gryta, A.; Sas-Paszt, L.; Frąc, M. The status of soil microbiome as affected by the application of phosphorus biofertilizer: Fertilizer enriched with beneficial bacterial strains. Int. J. Mol. Sci. 2020, 21, 8003. [Google Scholar] [CrossRef] [PubMed]
- Oluseyi Osunmakinde, C.; Selvarajan, R.; Mamba, B.B.; Msagati, T. Profiling bacterial diversity and potential pathogens in wastewater treatment plants using high-throughput sequencing analysis. Microorganisms 2019, 7, 506. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ji, L.; Wan, Q.; Li, H.; Li, R.; Yang, Y. Short-term effects of bio-organic fertilizer on soil fertility and bacterial community composition in tea plantation soils. Agronomy 2022, 12, 2168. [Google Scholar] [CrossRef]
- Song, D.; Huo, T.; Zhang, Z.; Cheng, L.; Wang, L.; Ming, K.; Liu, H.; Li, M.; Du, X. Metagenomic analysis reveals the response of microbial communities and their functions in lake sediment to environmental factors. Int. J. Environ. Res. Public Health 2022, 19, 16870. [Google Scholar] [CrossRef]
- Nie, S.A.; Lei, X.M.; Zhao, L.X.; Brookes, P.C.; Wang, F.; Chen, C.R.; Yang, W.H.; Xing, S.H. Fungal communities and functions response to long-term fertilization in paddy soils. Appl. Soil Ecol. 2018, 130, 251–258. [Google Scholar] [CrossRef]

| Parameters | Value |
|---|---|
| Clay (%) | 5.50 |
| Silt (%) | 8.30 |
| Sand (%) | 84.40 |
| Soil texture | Sandy |
| pH | 7.50 |
| EC (dS m−1) | 4.02 |
| Organic matter (%) | 0.91 |
| Total organic carbon (%) | 0.53 |
| Total N (mg kg−1 soil) | 280 |
| Available P (mg kg−1 soil) | 4.92 |
| Exchange K (mg kg−1 soil) | 292 |
| Total coliforms (MPN g DW−1 soil) | 24 × 102 |
| Fecal coliforms (MPN g DW−1 soil) | <0.3 |
| Escherichia coli (MPN g DW−1 soil) | <0.3 |
| Fecal Streptococci (MPN g DW−1 soil) | 39 × 102 |
| Parameters | Value |
|---|---|
| Total organic carbon (%) | 18.58 |
| Total N (%) | 1.21 |
| C/N | 15.36 |
| P (%) | 0.54 |
| K (%) | 0.95 |
| Ca (%) | 8.18 |
| Mg (%) | 1.05 |
| Na (%) | 0.42 |
| Alkalinity (% CaCO3) | 11.50 |
| Zn (mg kg−1 DW compost) | 70.10 |
| Fe (g kg−1 DW compost) | 70 |
| Mn (mg kg−1 DW compost) | 130 |
| Cu (mg kg−1 DW compost) | 11.60 |
| Cd (mg kg−1 DW compost) | 0.20 |
| Pb (mg kg−1 DW compost) | 4.15 |
| Cr (mg kg−1 DW compost) | 11.50 |
| Ni (mg kg−1 DW compost) | 5.88 |
| Total coliforms (MPN g−1 DW compost) | 143.33 ± 5.77 |
| Fecal coliforms (MPN g−1 DW compost) | 120 ± 17.32 |
| Escherichia coli (MPN g−1 DW compost) | 114 ± 23.79 |
| Fecal Streptococci (MPN g−1 DW compost) | 114.33 ± 23.80 |
| Salmonella spp. (MPN g−1 DW compost) | <0.3 |
| Shigella spp. (MPN g−1 DW compost) | <0.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghouili, E.; Abid, G.; Hogue, R.; Jeanne, T.; D’Astous-Pagé, J.; Sassi, K.; Hidri, Y.; M’Hamed, H.C.; Somenahally, A.; Xue, Q.; et al. Date Palm Waste Compost Application Increases Soil Microbial Community Diversity in a Cropping Barley (Hordeum vulgare L.) Field. Biology 2023, 12, 546. https://doi.org/10.3390/biology12040546
Ghouili E, Abid G, Hogue R, Jeanne T, D’Astous-Pagé J, Sassi K, Hidri Y, M’Hamed HC, Somenahally A, Xue Q, et al. Date Palm Waste Compost Application Increases Soil Microbial Community Diversity in a Cropping Barley (Hordeum vulgare L.) Field. Biology. 2023; 12(4):546. https://doi.org/10.3390/biology12040546
Chicago/Turabian StyleGhouili, Emna, Ghassen Abid, Richard Hogue, Thomas Jeanne, Joël D’Astous-Pagé, Khaled Sassi, Yassine Hidri, Hatem Cheikh M’Hamed, Anil Somenahally, Qingwu Xue, and et al. 2023. "Date Palm Waste Compost Application Increases Soil Microbial Community Diversity in a Cropping Barley (Hordeum vulgare L.) Field" Biology 12, no. 4: 546. https://doi.org/10.3390/biology12040546
APA StyleGhouili, E., Abid, G., Hogue, R., Jeanne, T., D’Astous-Pagé, J., Sassi, K., Hidri, Y., M’Hamed, H. C., Somenahally, A., Xue, Q., Jebara, M., Nefissi Ouertani, R., Riahi, J., de Oliveira, A. C., & Muhovski, Y. (2023). Date Palm Waste Compost Application Increases Soil Microbial Community Diversity in a Cropping Barley (Hordeum vulgare L.) Field. Biology, 12(4), 546. https://doi.org/10.3390/biology12040546





