Simple Summary
Molecular tools continue to reveal cryptic biodiversity within common and ecologically important species. The decorator worm Diopatra cuprea is an ecosystem engineer of intertidal beds of high-salinity estuaries of the Atlantic and Gulf of Mexico shorelines. Here, we sequenced mitochondrial cytochrome oxidase I (COI) in D. cuprea populations and discover evidence for several deep mitochondrial lineages, suggesting the presence of cryptic diversity.
Abstract
Marine annelid taxonomy is experiencing a period of rapid revision, with many previously “cosmopolitan” species being split into species with more limited geographic ranges. This is exemplified by the Diopatra genus, which has recently witnessed dozens of new species descriptions rooted in genetic analyses. In the northwestern Atlantic, the name D. cuprea (Bosc 1802) has been applied to populations from Cape Cod through the Gulf of Mexico, Central America, and Brazil. Here, we sequenced mitochondrial cytochrome oxidase I (COI) in D. cuprea populations from the Gulf of Mexico to Massachusetts. We find evidence for several deep mitochondrial lineages, suggesting that cryptic diversity is present in the D. cuprea complex from this coastline.
1. Introduction
Annelid taxonomy is experiencing a period of rapid revision at every level, from individual species to the entire phylum [1,2,3,4]. Historically, taxonomists of the 19th and early 20th centuries embraced the ‘cosmopolitan species concept’, believing that marine annelid faunas were dominated by a relatively small number of very widespread species. This view began to crumble in the face of increasingly detailed morphological work beginning in the 1970s, and has now fully collapsed under evidence from modern phylogenetic methods (reviewed by [3]). While we now understand that annelid diversity is vastly greater than was once thought, the work of fully understanding that diversity remains very much a work in progress.
The problem of refining “cosmopolitan” taxa is especially well-illustrated in the polychaete genus Diopatra. Historically, the two best-known species—D. neapolitana from western Europe and D. cuprea from eastern North America—were applied to far-flung onuphids from Africa to India, Southeast Asia, and Australia [5,6]. Systematists have been working at this problem since at least the 1980s [7,8], with over a dozen new Diopatra species described in the past decade alone, and more to come [9,10,11,12,13,14,15,16,17,18,19,20]. However, one of the ecologically best-known species, D. cuprea (Bosc 1802), has not yet been evaluated using modern phylogenetic methods.
D. cuprea occurs intertidally on the US east coast from Duxbury MA southward (the southern limit is not well established) [21,22]). D. cuprea provides one of the best-described examples of ecosystem engineering in estuarine sediments; the worm builds robust tubes that descend 1 m or more vertically into the sediment, with an above-sediment tube-cap emerging 2–5 cm above the sediment surface. This tube-cap is characteristically decorated with fragments of drift algae, shell, and other debris. The tube stabilizes sediment, alters water flow across the sediment surface, and physically excludes epibenthic predators. Together, these physical effects increase the diversity and abundance of nearby infauna, and of the epifauna living on the tube itself (reviewed by [22]). By attaching to algae, the worm creates an algal canopy in habitats where attached algae would otherwise be rare [23], and this behavior is facilitating the invasive red alga Gracilaria vermiculophylla throughout most of the US east coast [24,25,26,27]. D. cuprea’s emergent, decorated tube is typical of the genus, and our model for understanding the genus’ ecological role worldwide is largely informed by studies of D. cuprea [22].
The D. cuprea’s type locality is in South Carolina, but the name has been applied to Diopatra from Cape Cod through Brazil [10,20]. Diopatra from Brazil have recently been redescribed with four new species, none of which were D. cuprea [20], consistent with a non-cosmopolitan range for this species. The possible existence of cryptic Diopatra diversity in the northwestern Atlantic remains currently unexplored. D. cuprea is known to exhibit a curious latitudinal gradient in its tube-decorating behavior, with worms in Florida decorating far less than worms between Cape Cod and Georgia, even when offered the most commonly utilized algae in controlled conditions [21]. This biogeographic pattern in behavior raises the hypothesis that there may be hidden population-level genetic diversity in this region. Here, we sequenced a portion of the mitochondrial cytochrome oxidase I (COI) gene from D. cuprea populations from the Gulf of Mexico to Massachusetts in order to assess population genetic structure and any cryptic diversity.
2. Materials and Methods
Per site, 10–40 samples were collected by hand, placed into 95% ethanol and returned to the College of Charleston Marine Laboratory in Charleston SC in approximately 2009–2012 (Table 1). Samples were identified using morphological traits previously described [6,28], and extracted using a Nucleospin Tissue and Blood Kit (Maschery-Nagel), with the manufacturer’s recommended protocol. A portion of the COI gene was then PCR amplified using the protocol in [29]. These were cleaned with an EXO-SAP-IT protocol and sent for Sanger sequencing with these same primers at a private company.

Table 1.
Populations collected. We indicate the regions (see text), latitude and longitude, alternative IDs, collector (Sarah Berke or Tina Bell), sample size (n), COI haplotype clade frequencies, nucleotide diversity and proportion of unique haplotypes.
The sequences were trimmed and quality-checked with 4Peaks software v1.8 (nucleobytes.com) and saved as a fasta-formatted file. This yielded a dataset of 539 nucleotides across 153 individuals (GenBank Accession Numbers OQ700009—OQ700161). We used ape 5.0 [30] and a custom code in R v4.2.3 [31] to estimate the genetic distance between individuals, and assigned labels to all 15 unique haplotypes (Hap 1–15). To assess the placement of northwestern Atlantic D. cuprea into a wider phylogeographic context, we used MUSCLE v3.8.31 [32] with default parameters embedded in SeaView 5.0.5 [33] to align our dataset and sequences from published sources (Table 2). We ran PhyML 20120412 [34] as implemented in SeaView to estimate the phylogenetic relationships with default parameters (GTR + 4 rate classes as a model of evolution; 100 bootstrap replicates). We also generated a Bayesian analysis with MrBayes 3.2.7a [35] with 10,000 generations, a sampling frequency of 100, and a burn-in period of 250 generations. We used R::ape to visualize the Bayesian tree. We also used strataG 1.0 [36] to assess nucleotide diversity, pairwise Weir and Cockerham’s FST and the proportion of unique haplotypes within populations. Finally, we used linear regressions to assess the relationship between latitude and nucleotide diversity, and the latitude and proportion of unique haplotypes.

Table 2.
GenBank accession numbers of COI for Diopatra spp. and other polycheates.
3. Results
The within-population nucleotide diversity ranged from <0.001 to 0.101 and the proportion of unique haplotypes per populations ranged from 7 to 67% (Table 1). There was no linear relationship of nucleotide diversity with latitude (r2 = 0.317; F1,9 = 4.185; p = 0.071) nor proportion of unique haplotypes with latitude (r2 = 0.015; F1,9 = 0.1367; p = 0.720).
All the COI haplotypes sequenced from morphologically similar Diopatra cuprea from the east coast of the United States (Gulf of Mexico to Massachussetts) clustered into a single monophyletic clade, although the clade had minimal statistical support (i.e., ML bootstrap and Bayesian posterior probabilities were less than 90%). None of these samples clustered within Clades 1–3 from the work of Hektoen et al., 2022 [12]. Instead, samples were within a lineage labeled as Clade 4–5 of Hektoen et al., 2022 ([12]; Figure 1).
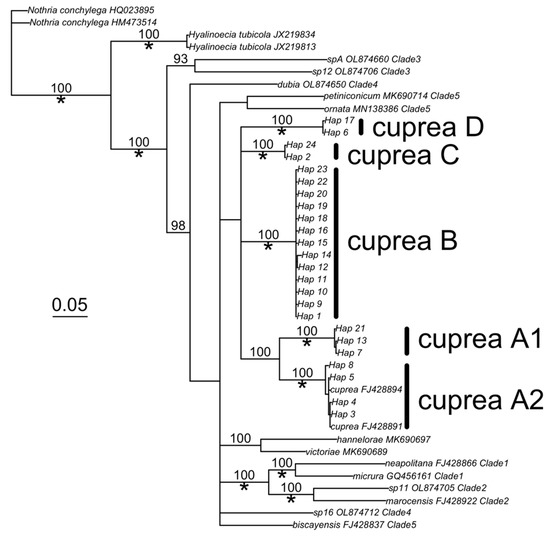
Figure 1.
Cryptic Diopatra cuprea genetic diversity. The Bayesian phylogeny was based on 550 bp of cytochrome c oxidase I. Numbers on edges indicate posterior probability, while asterisks indicate 90% or greater maximum likelihood bootstrap support (100 replicates). Clade designations (i.e., numbers after taxa) were defined using the work of [12]. An approximate 5% divergence between haplotypes is indicated by the segment.
Within the northwestern Atlantic D. cuprea samples, we detected five divergent lineages that differed between 14.6 and 20.5% (Table 3). We labeled these D. cuprea clades based on their relationship to each other and their geographic distribution (Figure 2; see clade frequencies in Table 1). A1 and A2 are a monophyletic group found mainly in Florida; A1 dominated the St. Thersa Beach population in the Gulf of Mexico (81% of haplotypes), while A2 dominated the southeastern coasts of Florida (100% of haplotypes from Fort Pierce and Chicken Island). Clade B dominates the mid-Atlantic states, from northeastern Florida (Amelia Island) to Virginia, and rarely occurred in other locations. Clade C was only found in North Carolina and Clade D dominated Massachusetts populations, and rarely occurred in mid-Atlantic states.

Table 3.
Mean percent difference between COI haplotype clades.
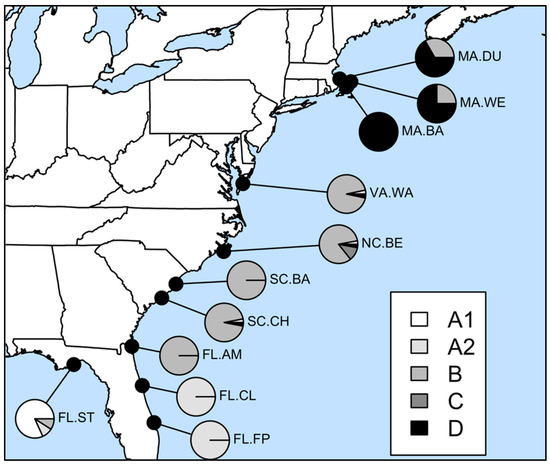
Figure 2.
Geographic distribution of COI clades (labeled A1-D; see Figure 1) of Diopatra cuprea.
The overall FST across all populations was 0.728 (p < 0.001) and the overall FST across the four regions (FL-Gulf; FL-Atlantic; mid-Atlantic; MA) was 0.826 (p < 0.001). Table 4 and Table 5 indicate the pairwise FST among the populations and regions, respectively.

Table 4.
Pairwise PhiST (below diagonal) and p-values (above diagonal) among populations.

Table 5.
Pairwise PhiST (below diagonal) and p-values (above diagonal) among regions.
4. Discussion
In morphological terms, all the individuals collected from the Gulf of Mexico through Massachusetts are consistent with the currently described Diopatra cuprea (Bosc 1802). While we found no statistical evidence for a monophyletic clade of D. cuprea in the northwestern Atlantic, all five COI clades were clustered away from the recently defined Clades 1, 2 and 3 in the work of Hektoen et al., 2022 [12]. This paper utilized two mitochondrial (COI and 16s rDNA) and one nuclear locus (28S rDNA) to assess phylogenetic relationships; it is likely that the application of these two other loci would allow greater resolution of the D. cuprea lineages and their exact placement within the wider phylogeny presented in the work of Hektoen et al., 2022 ([12]; see [11]). We also note that the type locality of D. cuprea was South Carolina, which is strongly dominated by Clade B [19].
The phylogeography of the widespread Diopatra cuprea mirrors that of several nearshore and estuarine species in North America. The monophyletic split between the A1 and A2 versus the others (B, C and D) likely reflects the Gulf of Mexico vs. eastern seaboard, which now have a secondary contact zone in eastern Florida near Cape Canaveral [40,41,42], and was driven by the separation of these water bodies during the Pliocene or Pleistocene. The split between Massachusetts Clade D from the mid-Atlantic clades (B and C) likely mirrors historical separation during the Pleistocene [43] and has a secondary contact zone near the New Jersey/New York border [44]. There are few species whose geographic range spans the Gulf of Mexico through New England; of these, one that displays similar phylogenetic breaks is the monocot Spartina alterniflora [45], which lives in close proximity to the high-salinity marshes that also constitute D. cuprea habitats.
The populations were not reciprocally monophyletic. Each population was dominated by one or two COI clades, but most populations harbored 1–2 other clades at low frequency. As examples, the Gulf of Mexico population has a low frequency of the mid-Atlantic B clade, while a North Carolina population has a low frequency of the Gulf of Mexico A1 clade and Massachusetts D clade. These may reflect low-frequency dispersal events between the regions, incomplete lineage sorting, or a mix of both.
Low-frequency dispersal events could occur naturally or via human-assisted transport. Unlike Diopatra in Europe, D. cuprea is not harvested for bait in the US (most likely because it is more difficult to obtain than D. neapolitana [22]). Transport as a bait worm is, therefore, unlikely. Diopatra’s larval period is quite short, making transport in ballast unlikely [46]. If human-assisted transport plays any role in these distributions, it is most likely via the transport of newly settled juveniles in mud associated with bivalve aquaculture. This is also the mechanism proposed for the D. biscayensis disjunct distribution in France [46]. Galaska et al. [46] found that individual D. biscayensis in the disjunct population were disproportionately found near the ropes used for mussel culture. None of our sampling sites were (to our knowledge) especially close to commercial aquaculture sites, but bivalve aquaculture certainly does occur throughout the eastern United States. We can tease apart the relative importance of lineage sorting and more recent dispersal by assessing the phylogeographic patterns in the nuclear genome (e.g., [38]). Moreover, future sampling efforts could also evaluate whether rare clades are associated with aquaculture sites.
Overall, our results provide preliminary evidence that cryptic diversity is indeed present in D. cuprea populations of the northwestern Atlantic. This is consistent with the explorations of other Diopatra populations in other parts of the world.
Author Contributions
Conceptualization, methodology, software and validation, E.E.S., T.B. and S.B.; formal analysis, E.E.S.; investigation and resources, T.B. and S.B.; data curation, writing—original draft preparation and writing—review and editing, visualization, supervision, project administration and funding acquisition, E.E.S. All authors have read and agreed to the published version of the manuscript.
Funding
This effort was funded by the National Science Foundation (DEB-0919064 and OCE-1924599).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Code and datasets can be found at https://github.com/esotka/DiopatraCOI.git. COI sequences were uploaded to GenBank.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Andrade, S.C.S.; Novo, M.; Kawauchi, G.Y.; Worsaae, K.; Pleijel, F.; Giribet, G.; Rouse, G.W. Articulating “Archiannelids”: Phylogenomics and Annelid Relationships, with Emphasis on Meiofaunal Taxa. Mol. Biol. Evol. 2015, 32, 2860–2875. [Google Scholar] [CrossRef] [PubMed]
- Carr, C.M.; Hardy, S.M.; Brown, T.M.; Macdonald, T.A.; Hebert, P.D.N. A tri-oceanic perspective: DNA barcoding reveals geographic structure and cryptic diversity in Canadian polychaetes. PLoS ONE 2011, 6, e22232. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, P.; Kupriyanova, E. Cosmopolitan polychaetes—Fact or fiction? Personal and historical perspectives. Invertebr. Syst. 2018, 32, 1–9. [Google Scholar] [CrossRef]
- Weigert, A.; Bleidorn, C. Current status of annelid phylogeny. Org. Divers. Evol. 2016, 16, 345–362. [Google Scholar] [CrossRef]
- Day, J.H. Polychaeta of Southern Africa Parts 1: Errantia and 2: Sedentaria; Trustees of the British Museum (Natural History): London, UK, 1967. [Google Scholar]
- Hartman, O. Polychaetous Annelids. Part V. Eunicea. In Allan Hancock Pacific Expeditions; The University of Southern California Press: Los Angeles, CA, USA, 1944; Volume 10, pp. 1–237. Available online: https://www.biodiversitylibrary.org/page/4679950 (accessed on 15 March 2023).
- Kirkegaard, J.B. The Polychaeta of West Africa Part II. Errant Species. 2. Nephtyidae to Dorvilleidae. Atlantide Rep. 1988, 14, 7–89. [Google Scholar]
- Paxton, H. Diopatra Audouin and Milne Edwards (Polychaeta: Onuphidae) from Australia, with a discussion of developmental patterns in the genus. Beagle Rec. North. Territ. Mus. Arts Sci. 1993, 10, 115–154. [Google Scholar] [CrossRef]
- Arias, A.; Paxton, H. The cryptogenic bait worm Diopatra biscayensis Fauchald et al., 2012 (Annelida: Onuphidae)—Revisiting its history, biology and ecology. Estuar. Coast. Shelf Sci. 2015, 163, 22–36. [Google Scholar] [CrossRef]
- Budaeva, N.; Fauchald, K. Diopatra tuberculantennata, a new species of Onuphidae (Polychaeta) from Belize with a key to onuphids from the Caribbean Sea. Zootaxa 2008, 1795, 29045. [Google Scholar] [CrossRef]
- Elgetany, A.H.; van Rensburg, H.; Hektoen, M.; Matthee, C.; Budaeva, N.; Simon, C.A.; Struck, T.H. Species delineation in the speciation grey zone—The case of Diopatra (Annelida, Onuphidae). Zool. Scr. 2020, 49, 516–534. [Google Scholar] [CrossRef]
- Hektoen, M.M.; Willassen, E.; Budaeva, N. Phylogeny and Cryptic Diversity of Diopatra (Onuphidae, Annelida) in the East Atlantic. Biology 2022, 11, 327. [Google Scholar] [CrossRef]
- Paxton, H. The Diopatra chiliensis confusion—Redescription of D. chiliensis (Polychaeta, Onuphidae) and implicated species. Zool. Scr. 1998, 27, 31–48. [Google Scholar] [CrossRef]
- Paxton, H. Diopatra nishii, a new brooding species of Onuphidae (Annelida) from Japan. Mar. Biodivers. 2015, 45, 711–717. [Google Scholar] [CrossRef]
- Paxton, H. A new species of Diopatra (Annelida: Onuphidae) from Namibia, southwestern Africa. Mar. Biodivers. 2016, 46, 889–895. [Google Scholar] [CrossRef]
- Paxton, H.; Arias, A. Unveiling a surprising diversity of the genus Diopatra Audouin & Milne Edwards, 1833 (Annelida: Onuphidae) in the Macaronesian region (eastern North Atlantic) with the description of four new species. Zootaxa 2017, 4300, 505. [Google Scholar]
- Paxton, H.; Fadlaoui, S.; Lechapt, J.P. Diopatra marocensis, a New Brooding Species of Onuphidae (Annelida, Polychaeta). J. Mar. Biol. Assoc. United Kingd. 1995, 75, 949–955. [Google Scholar] [CrossRef]
- Pires, A.; Paxton, H.; Quintino, V.; Rodrigues, A.M. Diopatra (Annelida: Onuphidae) diversity in European waters with the description of Diopatra micrura, new species. Zootaxa 2010, 2395, 17–33. [Google Scholar] [CrossRef]
- World Polychaeta Database. Diopatra cuprea (Bosc, 1802). Read, G., Fauchald, K., Eds.; Accessed through: World Register of Marine Species. 2023. Available online: https://www.marinespecies.org/aphia.php?p=taxdetails&id=157339 (accessed on 15 March 2023).
- Seixas, V.C.; Steiner, T.M.; Solé-Cava, A.M.; Amaral, A.C.Z.; Paiva, P.C. Hidden diversity within the Diopatra cuprea complex (Annelida: Onuphidae): Morphological and genetics analyses reveal four new species in the south-west Atlantic. Zool. J. Linn. Soc. 2021, 191, 637–671. [Google Scholar] [CrossRef]
- Berke, S.K. Biogeographic variability in ecosystem engineering: Patterns in the abundance and behavior of the tube-building polychaete Diopatra cuprea. Mar. Ecol. Prog. Ser. 2012, 447, 1–13. [Google Scholar] [CrossRef]
- Berke, S. A Review of Diopatra Ecology: Current Knowledge, Open Questions, and Future Threats for an Ecosystem Engineering Polychaete. Biology 2022, 11, 1485. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, M.S.; Gurgel, C.F.D.; Fredericq, S.; McGlathery, K.J. Gracilaria vermiculophylla (Rhodophyta, Gracilariales) in Hog Island Bay, Virginia: A cryptic alien and invasive macroalga and taxonomic correction. J. Phycol. 2005, 42, 139–141. [Google Scholar] [CrossRef]
- Keller, E.L.; Berke, S.K.; Needham, C.N.; Salerno, C.R. A double-edged sword: Infaunal responses to Agarophyton vermiculophyllum in the mid-Atlantic United States. Estuaries Coasts J. ERF 2019, 42, 1924–1937. [Google Scholar] [CrossRef]
- Kollars, N.M.; Byers, J.E.; Sotka, E.E. Invasive décor: An association between a native decorator worm and a non-native seaweed can be mutualistic. Mar. Ecol. Prog. Ser. 2016, 545, 135–145. [Google Scholar] [CrossRef]
- Mott, A.W.; Krueger-Hadfield, S.A.; Blakeslee, A.M.H.; Fowler, A.E. Native tube-building polychaete prefers to anchor non-native alga over other macrophytes. Oecologia 2022, 198, 967–980. [Google Scholar] [CrossRef]
- Thomsen, M.S.; McGlathery, K. Facilitation of macroalgae by the sedimentary tube forming polychaete Diopatra cuprea. Estuar. Coast. Shelf Sci. 2005, 62, 63–73. [Google Scholar] [CrossRef]
- Pollock, L.W. A Practical Guide to the Marine Animals of Northeastern North America; Rutgers University Press: New Brunswick, NJ, USA; London, UK, 1998; p. 367. Available online: http://books.google.com/books?id=i1AmT31cuR4C (accessed on 15 March 2023).
- Berke, S.K.; Mahon, A.R.; Lima, F.P.; Halanych, K.M.; Wethey, D.S.; Woodin, S.A. Range Shifts and Species Diversity in Marine Ecosystem Engineers: Patterns and Predictions for European Sedimentary Habitats. Glob. Ecol. Biogeogr. 2010, 19, 223–232. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. ape 5.0, an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 15 March 2023).
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Archer, F.I.; Adams, P.E.; Schneiders, B.B. strataG: An R package for manipulating, summarizing and analysing population genetic data. Mol. Ecol. Resour. 2016, 17, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Leray, M.; Paulay, G.; Mulcahy, D.G. Diopatra Ornata Voucher BMBM0561 Cytochrome Oxidase Subunit 1 (COI) Gene, Partial cds; Mitochondrial. 2019. Available online: https://www.ncbi.nlm.nih.gov/nuccore/MN138386 (accessed on 15 March 2023).
- Harper, K.E.; Scheinberg, L.A.; Boyer, K.E.; Sotka, E.E. Global distribution of cryptic native, introduced and hybrid lineages in the widespread estuarine amphipod Ampithoe valida. Conserv. Genet. 2022, 23, 791–806. [Google Scholar] [CrossRef]
- Bors, E.K.; Rowden, A.A.; Maas, E.W.; Clark, M.R.; Shank, T.M. Patterns of Deep-Sea Genetic Connectivity in the New Zealand Region: Implications for Management of Benthic Ecosystems. PLoS ONE 2012, 7, e49474. [Google Scholar] [CrossRef]
- Avise, J.C. Molecular Population Structure and the Biogeographic History of a Regional Fauna: A Case History with Lessons for Conservation Biology. Oikos 1992, 63, 62–76. [Google Scholar] [CrossRef]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Soltis, D.E.; Morris, A.B.; McLachlan, J.S.; Manos, P.S.; Soltis, P.S. Comparative phylogeography of unglaciated eastern North America. Mol. Ecol. 2006, 15, 4261–4293. [Google Scholar] [CrossRef] [PubMed]
- Wares, J.P. Community genetics in the Northwestern Atlantic intertidal. Mol. Ecol. 2002, 11, 1131–1144. [Google Scholar] [CrossRef]
- Hilbish, T.J. Population genetics of marine species: The interaction of natural selection and historically differentiated populations. J. Exp. Mar. Biol. Ecol. 1996, 200, 67–83. [Google Scholar] [CrossRef]
- Zerebecki, R.A.; Sotka, E.E.; Hanley, T.C.; Bell, K.L.; Gehring, C.; Nice, C.C.; Richards, C.L.; Hughes, A.R. Repeated Genetic and Adaptive Phenotypic Divergence across Tidal Elevation in a Foundation Plant Species. Am. Nat. 2021, 198, E152–E169. [Google Scholar] [CrossRef]
- Galaska, M.P.; Wethey, D.S.; Arias, A.; Dubois, S.F.; Halanych, K.M.; Woodin, S.A. The Impact of Aquaculture on the Genetics and Distribution of the Onuphid Annelid Diopatra Biscayensis. Ecol. Evol. 2021, 11, 6184–6194. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).