Circular RNAs Could Encode Unique Proteins and Affect Cancer Pathways
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
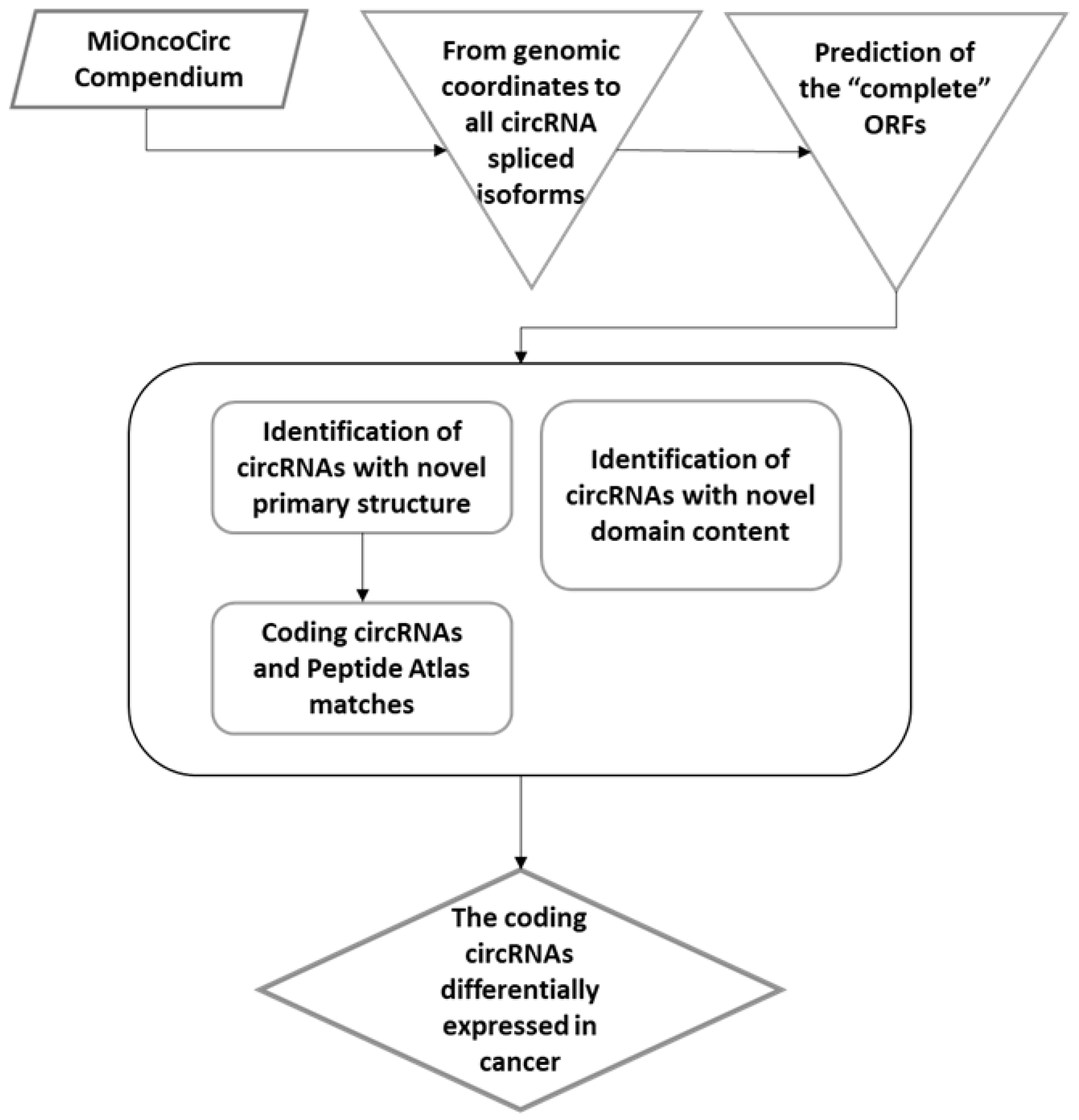
2.1. Cancer circRNA Selection
2.2. In Silico Characterization of Polypeptides Predicted from circRNAs
2.3. The Domain Structure of circRNA Encoded Proteins
2.4. Expression Profile of circRNAs with Unique Protein-Coding Potential in Cancer
2.5. Functional Characterization of the circRNAs Encoding Proteins
2.6. Mass Spectrometry Identification of Novel Peptides Derived from Coding circRNAs
2.7. Prediction of Internal Ribosome Entry Sites (IRES) in circRNAs with Coding Potential
2.8. Identification of m6A Sites in Coding-circRNA Sequences
3. Results
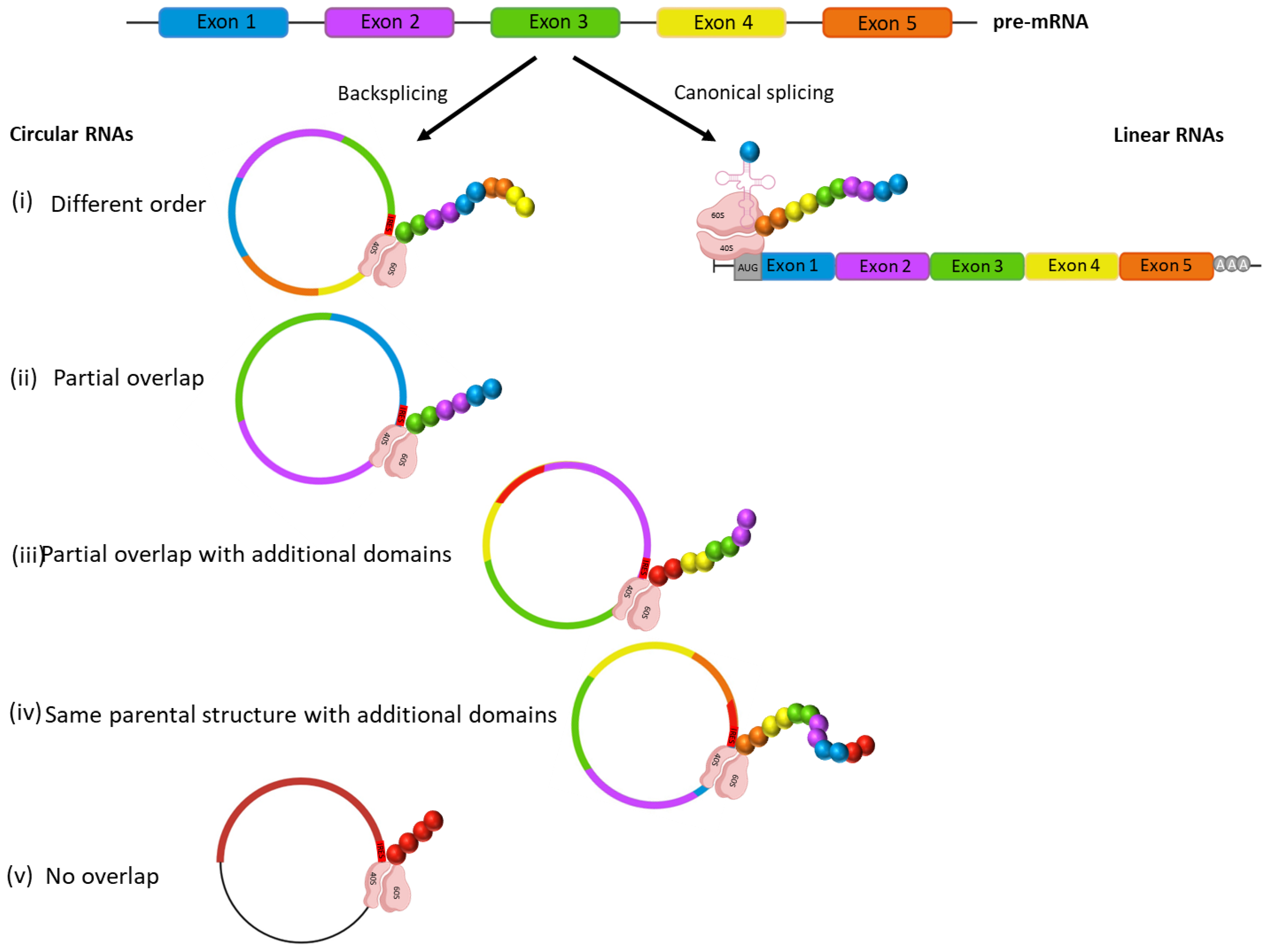
3.1. Cancer circRNAs Potentially Encode Novel Proteins
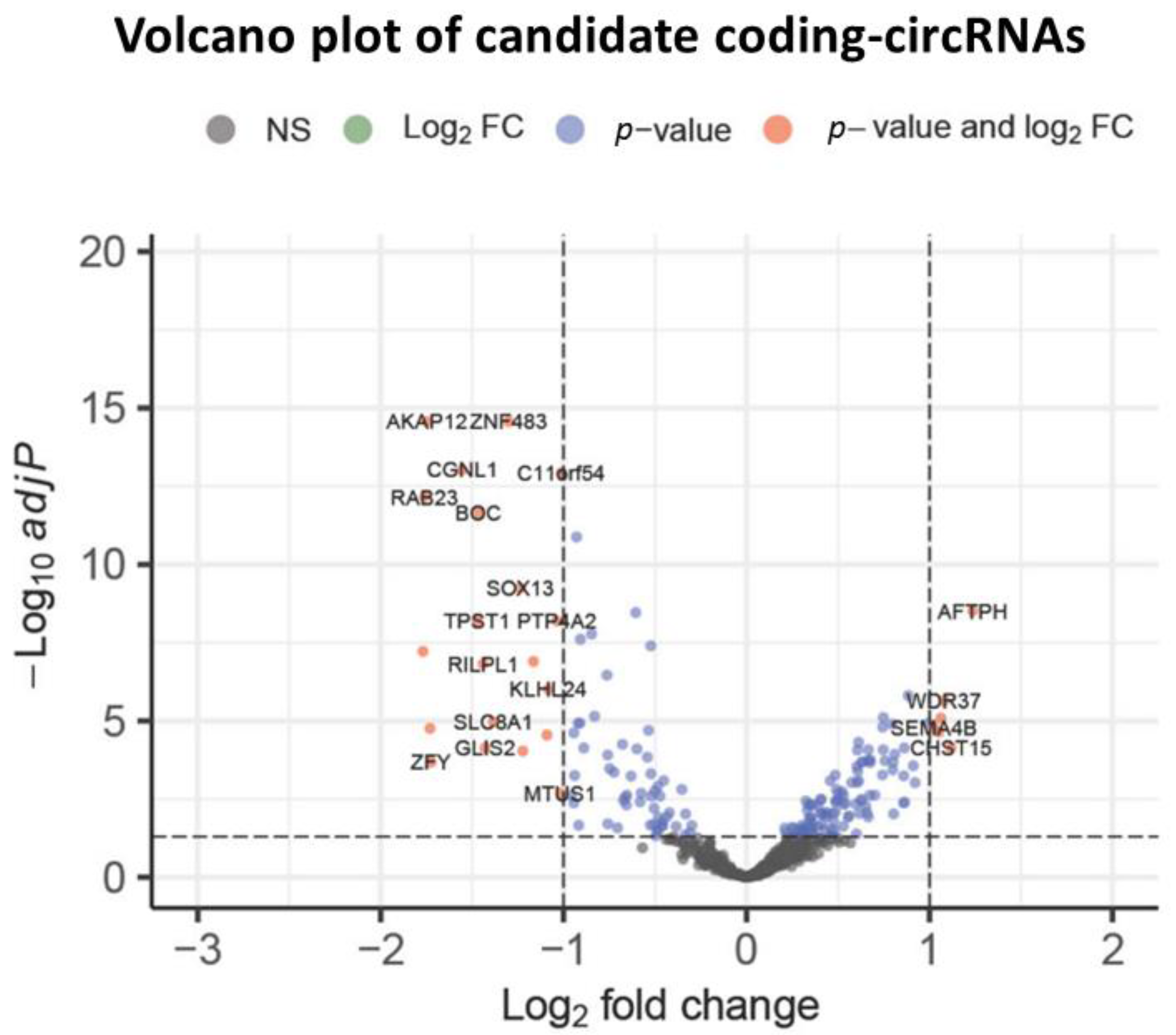
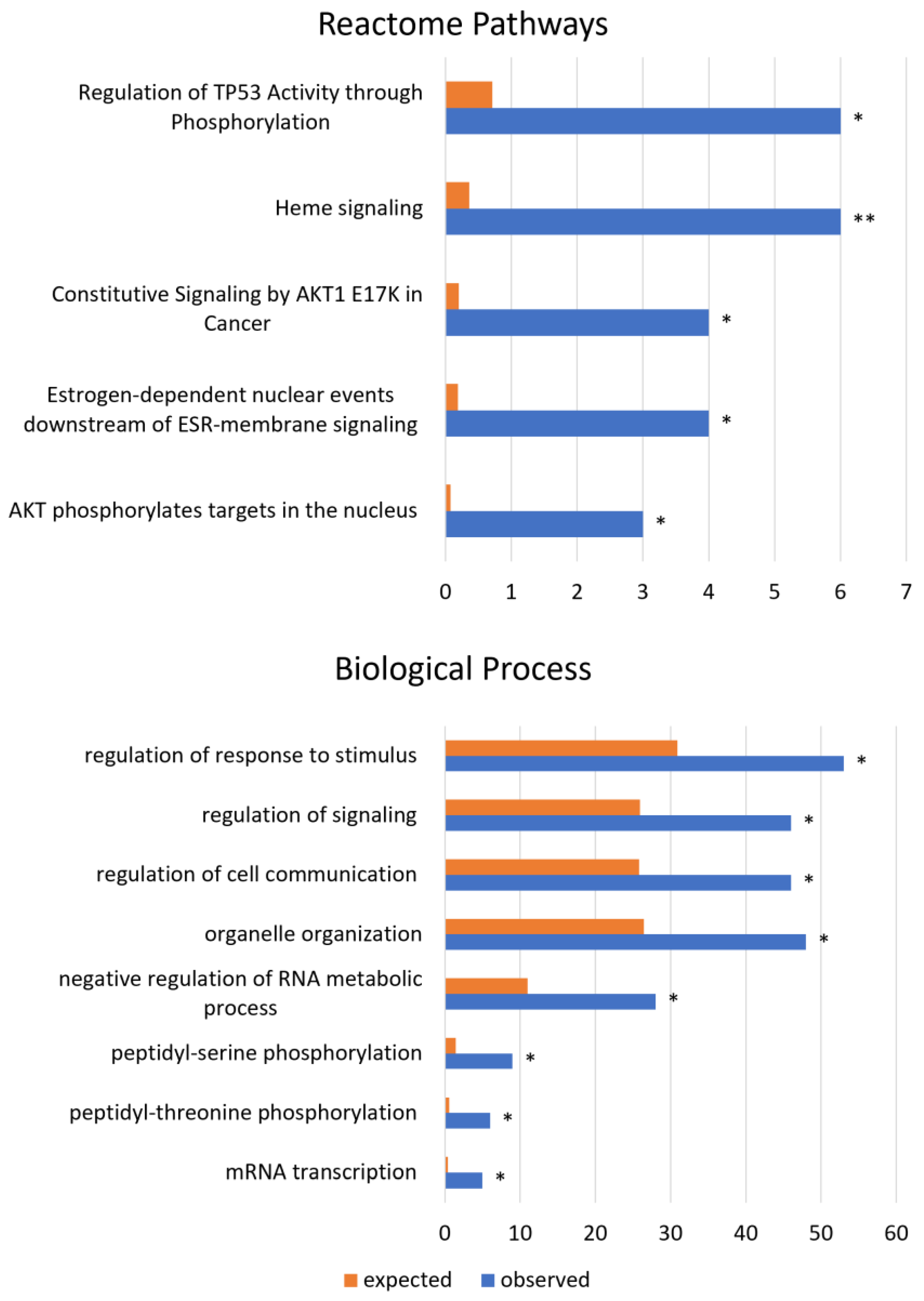
3.2. Expressed Coding circRNAs with Novel Coding Properties Are Involved in Cancer Pathways
3.3. The Coding circRNAs Differentially Expressed in Cancer Are Also Involved in AML
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409–423. [Google Scholar] [CrossRef] [PubMed]
- Smid, M.; Wilting, S.M.; Uhr, K.; Rodríguez-González, F.G.; de Weerd, V.; Prager-Van der Smissen, W.J.C.; van der Vlugt-Daane, M.; van Galen, A.; Nik-Zainal, S.; Butler, A.; et al. The circular RNome of primary breast cancer. Genome Res. 2019, 29, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The landscape of circular RNA in cancer. Cell 2019, 176, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, D.; Volinia, S.; Nicolet, D.; Świerniak, M.; Petri, A.; Mrózek, K.; Bill, M.; Pepe, F.; Walker, C.J.; Walker, A.E.; et al. Clinical and functional significance of circular RNAs in cytogenetically normal AML. Blood Adv. 2020, 4, 239–251. [Google Scholar] [CrossRef]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef]
- Abe, N.; Matsumoto, K.; Nishihara, M.; Nakano, Y.; Shibata, A.; Maruyama, H.; Shuto, S.; Matsuda, A.; Yoshida, M.; Ito, Y.; et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci. Rep. 2015, 5, 16435. [Google Scholar] [CrossRef]
- Chen, L.; Huang, C.; Wang, X.; Shan, G. Circular RNAs in eukaryotic cells. Curr. Genom. 2015, 16, 312–318. [Google Scholar] [CrossRef]
- Chen, X.; Han, P.; Zhou, T.; Guo, X.; Song, X.; Li, Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 2016, 6, 34985–34991. [Google Scholar] [CrossRef]
- Wawrzyniak, O.; Zarębska, Ż.; Kuczyński, K.; Gotz-Więckowska, A.; Rolle, K. Protein-related circular RNAs in human pathologies. Cells 2020, 9, 1841. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Gao, X.; Zhang, M.; Yan, S.; Sun, C.; Xiao, F.; Huang, N.; Yang, X.; Zhao, K.; Zhou, H.; et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. JNCI 2018, 110, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhao, K.; Xu, X.; Yang, Y.; Yan, S.; Wei, P.; Liu, H.; Xu, J.; Xiao, F.; Zhou, H.; et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018, 9, 4475–4492. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Huang, N.; Yang, X.; Luo, J.; Yan, S.; Xiao, F.; Chen, W.; Gao, X.; Zhao, K.; Zhou, H.; et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 2018, 37, 1805–1814. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.C.; Wong, C.W.; Liang, P.P.; Shi, M.; Cao, Y.; Rao, S.T.; Tsui, S.K.; Waye, M.M.; Zhang, Q.; Fu, W.M.; et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019, 20, 84–96. [Google Scholar] [CrossRef]
- Mo, D.; Li, X.; Raabe, C.A.; Rozhdestvensky, T.S.; Skryabin, B.V.; Brosius, J. Circular RNA encoded amyloid beta peptides-a novel putative player in Alzheimer’s Disease. Cells 2020, 9, 2196. [Google Scholar] [CrossRef]
- Cieslik, M.; Chugh, R.; Wu, Y.M.; Wu, M.; Brennan, C.; Lonigro, R.; Su, F.; Wang, R.; Siddiqui, J.; Mehra, R.; et al. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res. 2015, 25, 1372–1381. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.M.; Lonigro, R.J.; Vats, P.; Cobain, E.; Everett, J.; Cao, X.; Rabban, E.; Kumar-Sinha, C.; Raymond, V.; et al. Integrative clinical genomics of metastatic cancer. Nature 2017, 548, 297–303. [Google Scholar] [CrossRef]
- Mody, R.J.; Wu, Y.M.; Lonigro, R.J.; Cao, X.; Roychowdhury, S.; Vats, P.; Frank, K.M.; Prensner, J.R.; Asangani, I.; Palanisamy, N.; et al. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA 2015, 314, 913–925. [Google Scholar] [CrossRef]
- Desiere, F.; Deutsch, E.W.; King, N.L.; Nesvizhskii, A.I.; Mallick, P.; Eng, J.; Chen, S.; Eddes, J.; Loevenich, S.N.; Aebersold, R. The PeptideAtlas project. Nucleic Acids Res. 2006, 34, D655–D658. [Google Scholar] [CrossRef] [PubMed]
- Schwenk, J.M.; Omenn, G.S.; Sun, Z.; Campbell, D.S.; Baker, M.S.; Overall, C.M.; Aebersold, R.; Moritz, R.L.; Deutsch, E.W. The human plasma proteome draft of 2017: Building on the human plasma PeptideAtlas from mass spectrometry and complementary assays. J. Proteome Res. 2017, 16, 4299–4310. [Google Scholar] [CrossRef] [PubMed]
- Wesselhoeft, R.A.; Kowalski, P.S.; Anderson, D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018, 9, 2629–2639. [Google Scholar] [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017, 66, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gribskov, M. IRESpy: An XGBoost model for prediction of internal ribosome entry sites. BMC Bioinform. 2019, 20, 409–424. [Google Scholar] [CrossRef]
- Wen, S.Y.; Qadir, J.; Yang, B.B. Circular RNA translation: Novel protein isoforms and clinical significance. Trends Mol. Med. 2022, 28, 405–420. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Kulcheski, F.R.; Christoff, A.P.; Margis, R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 2016, 238, 42–51. [Google Scholar] [CrossRef]
- Li, J.; Sun, D.; Pu, W.; Wang, J.; Peng, Y. Circular RNAs in cancer: Biogenesis, function, and clinical significance. Trends Cancer 2020, 6, 319–336. [Google Scholar] [CrossRef]
- Fischer, J.W.; Leung, A.K. CircRNAs: A regulator of cellular stress. Crit. Rev. Biochem. Mol. 2017, 52, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell. 2017, 66, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lu, T.; Liu, D.; Zhang, C.; Zhang, Y.; Dong, F. Upregulated PPARG2 facilitates interaction with demethylated AKAP12 gene promoter and suppresses proliferation in prostate cancer. Cell Death Dis. 2021, 12, 528–544. [Google Scholar] [CrossRef]
- Schroeder, M.P.; Bastian, L.; Eckert, C.; Gökbuget, N.; James, A.R.; Tanchez, J.O.; Schlee, C.; Isaakidis, K.; Häupl, B.; Baum, K.; et al. Integrated analysis of relapsed B-cell precursor Acute Lymphoblastic Leukemia identifies subtype-specific cytokine and metabolic signatures. Sci. Rep. 2019, 9, 4188–4199. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Chen, Y.; Liu, Z.; Leng, Y.; Tian, Y. Expression profiles and prognostic significance of AFTPH in different tumors. FEBS Open Bio 2020, 10, 2666–2677. [Google Scholar] [CrossRef] [PubMed]
- Gui, C.P.; Liao, B.; Luo, C.G.; Chen, Y.H.; Tan, L.; Tang, Y.M.; Li, J.Y.; Hou, Y.; Song, H.D.; Lin, H.S.; et al. CircCHST15 is a novel prognostic biomarker that promotes clear cell renal cell carcinoma cell proliferation and metastasis through the miR-125a-5p/EIF4EBP1 axis. Mol. Cancer 2021, 20, 169–190. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Jia, Y.; Wang, B.; Yang, S.; Du, K.; Luo, Y.; Li, Y.; Zhu, B. Circular RNA CHST15 sponges miR-155-5p and miR-194-5p to promote the immune escape of lung cancer cells mediated by PD-L1. Front. Oncol. 2021, 11, 595609–595626. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, Y.H.; Xu, C.; Liang, Y.L.; Zhao, Y.; He, Q.M.; Li, J.Y.; Chen, K.L.; Qiao, H.; Liu, N.; et al. Chemotherapy-induced senescence reprogramming promotes nasopharyngeal carcinoma metastasis by circRNA-mediated PKR activation. Adv. Sci. 2023, 10, e2205668. [Google Scholar] [CrossRef]
- Zhang, L.; Peng, H.; Xu, Z.; Yang, Q.; Wang, Y.; Wang, H.; Bu, L. Circular RNA SOX13 promotes malignant behavior and cisplatin resistance in non-small cell lung cancer through targeting microRNA-3194-3p/microtubule-associated protein RP/EB family member 1. Bioengineered 2022, 13, 1814–1827. [Google Scholar] [CrossRef]
- Pedraz-Valdunciel, C.; Giannoukakos, S.; Potie, N.; Giménez-Capitán, A.; Huang, C.Y.; Hackenberg, M.; Fernandez-Hilario, A.; Bracht, J.; Filipska, M.; Aldeguer, E.; et al. Digital multiplexed analysis of circular RNAs in FFPE and fresh non-small cell lung cancer specimens. Mol. Oncol. 2022, 16, 2367–2383. [Google Scholar] [CrossRef]
- Ye, F.; Gao, G.; Zou, Y.; Zheng, S.; Zhang, L.; Ou, X.; Xie, X.; Tang, H. CircFBXW7 inhibits malignant progression by sponging miR-197-3p and encoding a 185-aa protein in triple-negative breast cancer. Mol. Ther. Nucleic Acids 2019, 18, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Qi, Z.; He, D.; Liu, J.; Xu, E.; Li, B.; Cai, S.; Sun, D.; Cheng, Y.; Shi, Q.; et al. Strategy for scanning peptide-coding circular RNAs in colorectal cancer based on bioinformatics analysis and experimental assays. Front. Cell Dev. Biol. 2022, 9, 815895–815906. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Li, X.; Li, F.; Wu, X.; Zhang, M.; Zhou, H.; Huang, N.; Yang, X.; Xiao, F.; Liu, D.; et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol. Cancer 2019, 18, 131–147. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, L.; Zhou, Y.; Wang, Q.; Zheng, Z.; Xu, B.; Wu, C.; Zhou, Q.; Hu, W.; Wu, C.; et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer 2019, 18, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jian, W.; Luo, Q.; Fang, L. CircSEMA4B inhibits the progression of breast cancer by encoding a novel protein SEMA4B-211aa and regulating AKT phosphorylation. Cell Death Dis. 2022, 13, 794–810. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, L.; Yang, M.; Wu, C.; Lan, R.; Wang, W.; Li, Y. Circ-SIRT1 inhibits cardiac hypertrophy via activating SIRT1 to promote autophagy. Cell Death Dis. 2021, 12, 1069. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Hou, X.T.; Guan, Y.X.; Chen, Y. CircRNA SCMH1 regulates the miR-200a-3p/ZEB1 signaling axis to promote diabetes-induced retinal epithelial-mesenchymal transition. Exp. Eye Res. 2022, 224, 109264–109275. [Google Scholar] [CrossRef]
- He, Y.; Qiu, F.; Qiao, B.; Zhang, N.; Fang, Z.; Feng, L.; Zhang, S.; Qiu, W. Blocking circ-SCMH1 (hsa_circ_0011946) suppresses acquired DDP resistance of oral squamous cell carcinoma (OSCC) cells both in vitro and in vivo by sponging miR-338-3p and regulating LIN28B. Cancer Cell Int. 2021, 21, 412–429. [Google Scholar] [CrossRef]
| GO Categories | GO | Obs | Exp | Fold | p Value | FDR |
|---|---|---|---|---|---|---|
| AKT phosphorylates targets in the nucleus | RP | 3 | 0.08 | 38.85 | 1.19 × 10−4 | 3.31 × 10−2 |
| Estrogen-dependent nuclear events downstream of ESR-membrane signaling | RP | 4 | 0.19 | 21.58 | 5.89 × 10−5 | 2.45 × 10−2 |
| Constitutive Signaling by AKT1 E17K in Cancer | RP | 4 | 0.2 | 19.92 | 7.79 × 10−5 | 2.77 × 10−2 |
| Heme signaling | RP | 6 | 0.36 | 16.53 | 3.14 × 10−6 | 2.61 × 10−3 |
| Regulation of TP53 Activity through Phosphorylation | RP | 6 | 0.71 | 8.45 | 1.08 × 10−4 | 3.38 × 10−2 |
| aryl hydrocarbon receptor binding | MF | 3 | 0.07 | 43.16 | 9.23 × 10−5 | 3.78 × 10−2 |
| protein serine kinase activity | MF | 14 | 2.8 | 4.99 | 1.30 × 10−6 | 2.13 × 10−3 |
| protein serine/threonine kinase activity | MF | 14 | 3.34 | 4.2 | 9.06 × 10−6 | 6.35 × 10−3 |
| protein serine/threonine/tyrosine kinase activity | MF | 14 | 3.46 | 4.05 | 1.34 × 10−5 | 7.33 × 10−3 |
| DNA binding | MF | 39 | 19.3 | 2.02 | 1.57 × 10−5 | 7.68 × 10−3 |
| rough endoplasmic reticulum | CC | 5 | 0.63 | 7.9 | 5.53 × 10−4 | 4.70 × 10−2 |
| transcription regulator complex | CC | 15 | 3.91 | 3.84 | 1.20 × 10−5 | 1.63 × 10−3 |
| nuclear speck | CC | 11 | 3.19 | 3.45 | 4.41 × 10−4 | 4.09 × 10−2 |
| centrosome | CC | 14 | 4.87 | 2.87 | 4.57 × 10−4 | 4.05 × 10−2 |
| cytosol | CC | 65 | 42.04 | 1.55 | 9.16 × 10−5 | 1.04 × 10−2 |
| mRNA transcription | BP | 5 | 0.38 | 13.21 | 5.80 × 10−5 | 3.49 × 10−2 |
| peptidyl-threonine phosphorylation | BP | 6 | 0.58 | 10.36 | 3.72 × 10−5 | 3.43 × 10−2 |
| peptidyl-serine phosphorylation | BP | 9 | 1.42 | 6.33 | 1.85 × 10−5 | 3.23 × 10−2 |
| negative regulation of RNA metabolic process | BP | 28 | 11.02 | 2.54 | 5.46 × 10−6 | 2.14 × 10−2 |
| organelle organization | BP | 48 | 26.44 | 1.82 | 2.44 × 10−5 | 3.19 × 10−2 |
| regulation of cell communication | BP | 46 | 25.81 | 1.78 | 5.71 × 10−5 | 3.58 × 10−2 |
| regulation of signaling | BP | 46 | 25.91 | 1.78 | 8.60 × 10−5 | 4.65 × 10−2 |
| regulation of response to stimulus | BP | 53 | 30.9 | 1.72 | 4.71 × 10−5 | 3.69 × 10−2 |
| Gene | chr | Start | End | Annotation |
|---|---|---|---|---|
| ABHD2 | chr15 | 89113724 | 89116521 | C-term|canonicalMet |
| ANKRD12 * | chr18 | 9182381 | 9221999 | C-term|canonicalMet|conservedStructure |
| ARAP2 * | chr4 | 36228581 | 36229645 | C-term|canonicalMet|lackingDomain |
| CLNS1A | chr11 | 77619605 | 77625818 | canonicalSTOP|conservedStructure|internalMet |
| CPSF6 | chr12 | 69251128 | 69262562 | canonicalSTOP|conservedStructure|internalMet |
| CSNK1G3 | chr5 | 123545416 | 123557564 | C-term|canonicalMet |
| FBXW7 | chr4 | 152411302 | 152412529 | C-term|canonicalMet |
| HIPK3 * | chr11 | 33286412 | 33287511 | C-term|canonicalMet|conservedStructure |
| KLHL8 | chr4 | 87195323 | 87195690 | C-term|canonicalMet |
| MGA | chr15 | 41668827 | 41669958 | C-term|canonicalMet|lackingDomain |
| NCOA2 * | chr8 | 70213902 | 70216764 | C-term|canonicalMet|novelDomainStructure |
| OMA1 | chr1 | 58506059 | 58539310 | C-term|canonicalMet|conservedStructure |
| PCMTD1 | chr8 | 51860844 | 51861246 | C-term|canonicalMet|conservedStructure |
| PDE3B | chr11 | 14771936 | 14789242 | C-term|internalMet |
| RELL1 | chr4 | 37631384 | 37638504 | canonicalSTOP|internalMet |
| RNF13 * | chr3 | 149846010 | 149921227 | C-term|canonicalMet|conservedStructure |
| RNF220 | chr1 | 44411980 | 44412722 | C-term|canonicalMet |
| RSRC1 | chr3 | 158122102 | 158123991 | C-term|canonicalMet |
| SATB1 * | chr3 | 18378169 | 18420991 | C-term|canonicalMet|lackingDomain |
| SHOC2 | chr10 | 110964124 | 110985765 | lackingDomain |
| SLC38A1 | chr12 | 46229152 | 46243314 | C-term|canonicalMet|conservedStructure |
| SLC8A1 * | chr2 | 40428472 | 40430304 | C-term|canonicalMet|conservedStructure |
| XPO1 * | chr2 | 61522610 | 61533903 | C-term|canonicalMet|conservedStructure |
| ZBTB44 | chr11 | 130260855 | 130261929 | C-term|canonicalMet|lackingDomain |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crudele, F.; Bianchi, N.; Terrazzan, A.; Ancona, P.; Frassoldati, A.; Gasparini, P.; D’Adamo, A.P.; Papaioannou, D.; Garzon, R.; Wójcicka, A.; et al. Circular RNAs Could Encode Unique Proteins and Affect Cancer Pathways. Biology 2023, 12, 493. https://doi.org/10.3390/biology12040493
Crudele F, Bianchi N, Terrazzan A, Ancona P, Frassoldati A, Gasparini P, D’Adamo AP, Papaioannou D, Garzon R, Wójcicka A, et al. Circular RNAs Could Encode Unique Proteins and Affect Cancer Pathways. Biology. 2023; 12(4):493. https://doi.org/10.3390/biology12040493
Chicago/Turabian StyleCrudele, Francesca, Nicoletta Bianchi, Anna Terrazzan, Pietro Ancona, Antonio Frassoldati, Paolo Gasparini, Adamo P. D’Adamo, Dimitrios Papaioannou, Ramiro Garzon, Anna Wójcicka, and et al. 2023. "Circular RNAs Could Encode Unique Proteins and Affect Cancer Pathways" Biology 12, no. 4: 493. https://doi.org/10.3390/biology12040493
APA StyleCrudele, F., Bianchi, N., Terrazzan, A., Ancona, P., Frassoldati, A., Gasparini, P., D’Adamo, A. P., Papaioannou, D., Garzon, R., Wójcicka, A., Gaj, P., Jażdżewski, K., Palatini, J., & Volinia, S. (2023). Circular RNAs Could Encode Unique Proteins and Affect Cancer Pathways. Biology, 12(4), 493. https://doi.org/10.3390/biology12040493








