miR-4432 Targets FGFBP1 in Human Endothelial Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
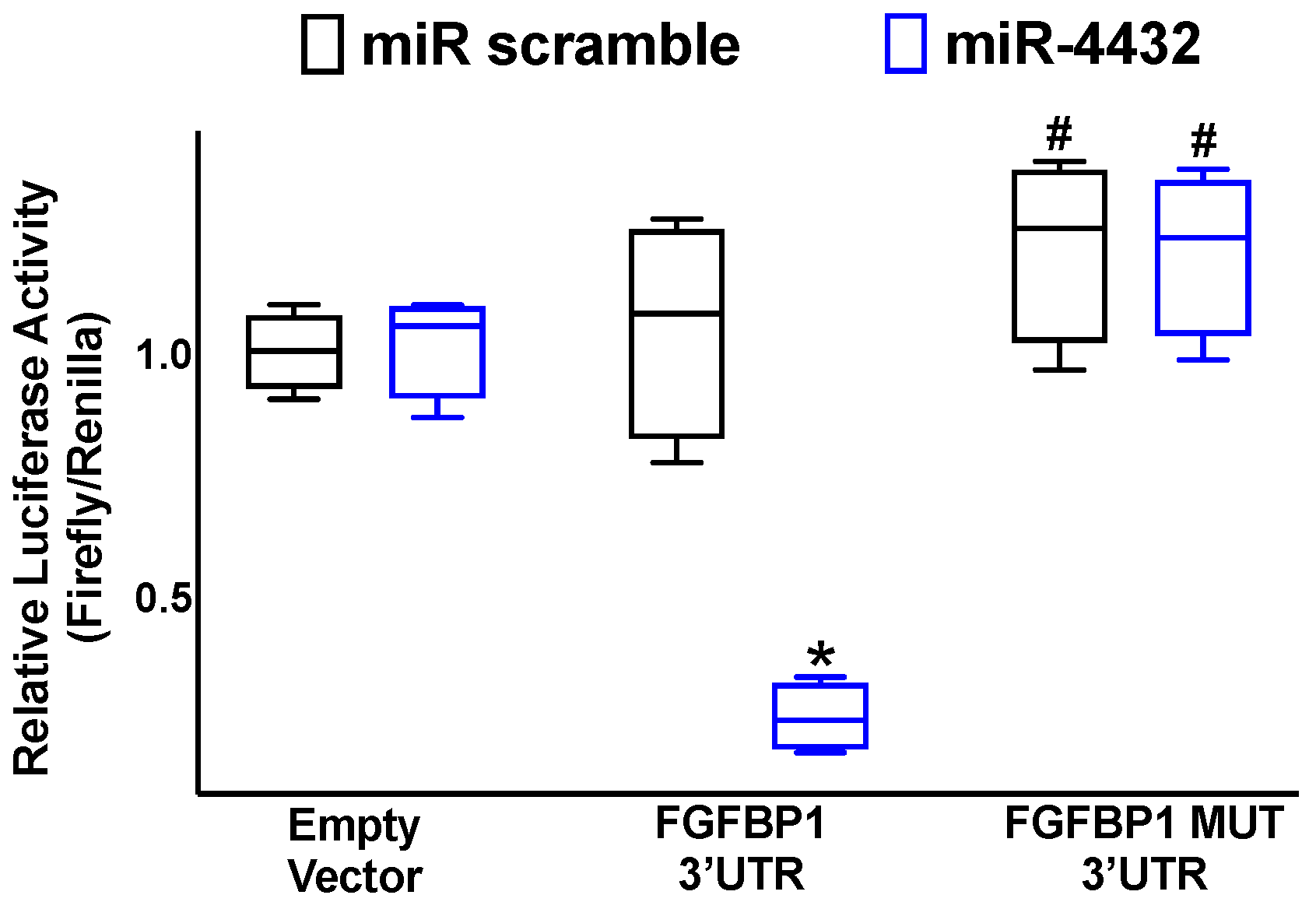
2.1. miR-4432 Targets FGFBP1 in a Conservative Manner
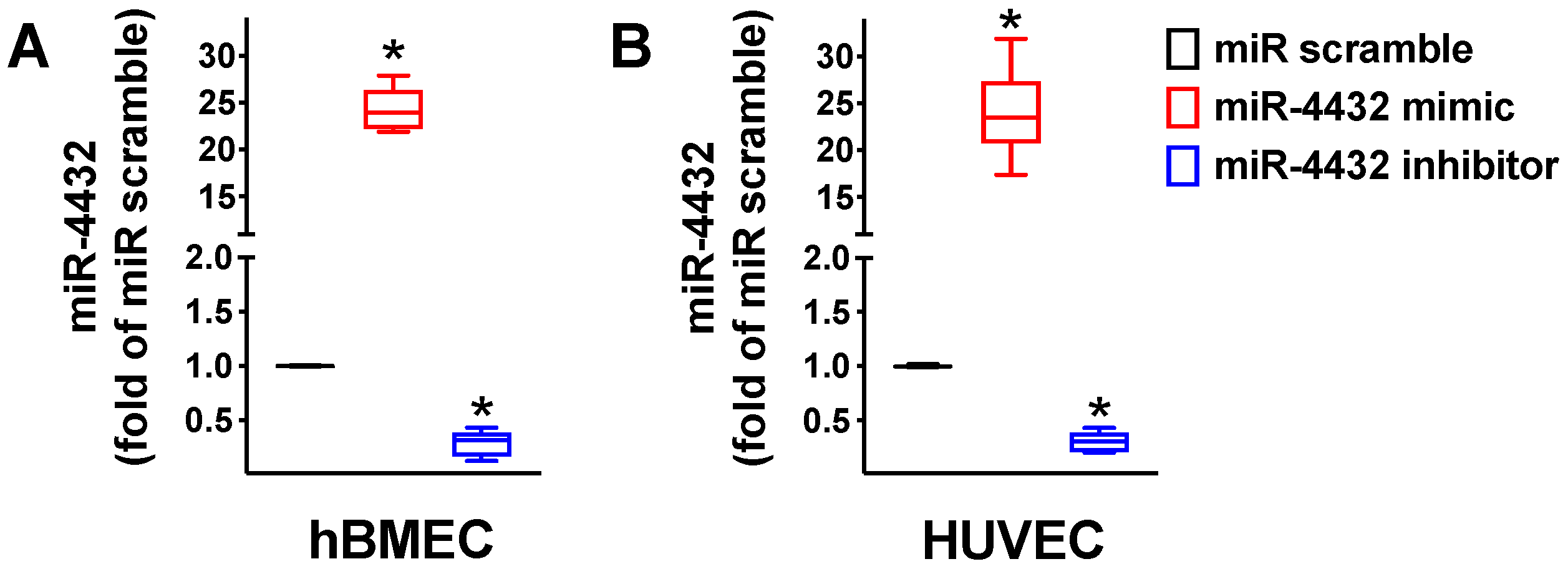
2.2. miR-4432 Regulates FGFBP1 Transcription in Endothelial Cells
2.3. FGFBP1 Expression Is Controlled by miR-4432
2.4. miR-4432 Regulates Mitochondrial Oxidative Stress in Human ECs
3. Discussion
4. Methods
4.1. Cells and Other Reagents
4.2. Identification of miR-4432 as a Modulator of FGFBP1
4.3. Immunoblotting
4.4. Mitochondrial ROS
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, B.; Perel, P.; Mensah, G.A.; Ezzati, M. Global epidemiology, health burden and effective interventions for elevated blood pressure and hypertension. Nat. Rev. Cardiol. 2021, 18, 785–802. [Google Scholar] [CrossRef]
- Bruno, R.M.; Masi, S.; Taddei, M.; Taddei, S.; Virdis, A. Essential Hypertension and Functional Microvascular Ageing. High Blood Press. Cardiovasc. Prev. 2018, 25, 35–40. [Google Scholar] [CrossRef]
- Saxena, T.; Ali, A.O.; Saxena, M. Pathophysiology of essential hypertension: An update. Expert Rev. Cardiovasc. Ther. 2018, 16, 879–887. [Google Scholar] [CrossRef]
- Vanhoutte, P.M.; Shimokawa, H.; Feletou, M.; Tang, E.H. Endothelial dysfunction and vascular disease—A 30th anniversary update. Acta Physiol. 2017, 219, 22–96. [Google Scholar] [CrossRef]
- Daiber, A.; Steven, S.; Weber, A.; Shuvaev, V.V.; Muzykantov, V.R.; Laher, I.; Li, H.; Lamas, S.; Munzel, T. Targeting vascular (endothelial) dysfunction. Br. J. Pharmacol. 2017, 174, 1591–1619. [Google Scholar] [CrossRef] [PubMed]
- Ruppert, V.; Maisch, B. Genetics of human hypertension. Herz 2003, 28, 655–662. [Google Scholar] [CrossRef]
- Kolifarhood, G.; Sabour, S.; Akbarzadeh, M.; Sedaghati-Khayat, B.; Guity, K.; Rasekhi Dehkordi, S.; Amiri Roudbar, M.; Hadaegh, F.; Azizi, F.; Daneshpour, M.S. Genome-wide association study on blood pressure traits in the Iranian population suggests ZBED9 as a new locus for hypertension. Sci. Rep. 2021, 11, 11699. [Google Scholar] [CrossRef]
- Mompeo, O.; Freidin, M.B.; Gibson, R.; Hysi, P.G.; Christofidou, P.; Segal, E.; Valdes, A.M.; Spector, T.D.; Menni, C.; Mangino, M. Genome-Wide Association Analysis of Over 170,000 Individuals from the UK Biobank Identifies Seven Loci Associated with Dietary Approaches to Stop Hypertension (DASH) Diet. Nutrients 2022, 14, 4431. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Warren, H.R.; Hiltunen, T.P.; McDonough, C.W.; El Rouby, N.; Salvi, E.; Wang, Z.; Garofalidou, T.; Fyhrquist, F.; Kontula, K.K.; et al. Genome-Wide Meta-Analysis of Blood Pressure Response to beta(1)-Blockers: Results From ICAPS (International Consortium of Antihypertensive Pharmacogenomics Studies). J. Am. Heart Assoc. 2019, 8, e013115. [Google Scholar] [CrossRef]
- Cottarelli, A.; Corada, M.; Beznoussenko, G.V.; Mironov, A.A.; Globisch, M.A.; Biswas, S.; Huang, H.; Dimberg, A.; Magnusson, P.U.; Agalliu, D.; et al. Fgfbp1 promotes blood-brain barrier development by regulating collagen IV deposition and maintaining Wnt/beta-catenin signaling. Development 2020, 147, dev185140. [Google Scholar] [CrossRef]
- Propson, N.E.; Roy, E.R.; Litvinchuk, A.; Kohl, J.; Zheng, H. Endothelial C3a receptor mediates vascular inflammation and blood-brain barrier permeability during aging. J. Clin. Investig. 2021, 131, e140966. [Google Scholar] [CrossRef]
- Kadry, H.; Noorani, B.; Cucullo, L. A blood-brain barrier overview on structure, function, impairment, and biomarkers of integrity. Fluids Barriers CNS 2020, 17, 69. [Google Scholar] [CrossRef] [PubMed]
- Ben-Zvi, A.; Liebner, S. Developmental regulation of barrier- and non-barrier blood vessels in the CNS. J. Intern. Med. 2022, 292, 31–46. [Google Scholar] [CrossRef]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.T.B.; Clark, I.M.; Le, L.T.T. MicroRNA-Based Diagnosis and Therapy. Int. J. Mol. Sci. 2022, 23, 7167. [Google Scholar] [CrossRef]
- Santulli, G.; Wronska, A.; Uryu, K.; Diacovo, T.G.; Gao, M.; Marx, S.O.; Kitajewski, J.; Chilton, J.M.; Akat, K.M.; Tuschl, T.; et al. A selective microRNA-based strategy inhibits restenosis while preserving endothelial function. J. Clin. Investig. 2014, 124, 4102–4114. [Google Scholar] [CrossRef] [PubMed]
- Rafehi, H.; El-Osta, A. HDAC Inhibition in Vascular Endothelial Cells Regulates the Expression of ncRNAs. Noncoding RNA 2016, 2, 4. [Google Scholar] [CrossRef] [PubMed]
- Naik, P.; Cucullo, L. In vitro blood-brain barrier models: Current and perspective technologies. J. Pharm. Sci. 2012, 101, 1337–1354. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Chen, S.; Liu, X.; Lin, L.; Huang, X.; Guo, Z.; Liu, J.; Wang, Y.; Yuan, W.; et al. Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis 2011, 215, 286–293. [Google Scholar] [CrossRef]
- Taniyama, Y.; Griendling, K.K. Reactive oxygen species in the vasculature: Molecular and cellular mechanisms. Hypertension 2003, 42, 1075–1081. [Google Scholar] [CrossRef]
- De Giusti, V.C.; Caldiz, C.I.; Ennis, I.L.; Perez, N.G.; Cingolani, H.E.; Aiello, E.A. Mitochondrial reactive oxygen species (ROS) as signaling molecules of intracellular pathways triggered by the cardiac renin-angiotensin II-aldosterone system (RAAS). Front. Physiol. 2013, 4, 126. [Google Scholar] [CrossRef] [PubMed]
- Welch, W.J. Angiotensin II-dependent superoxide: Effects on hypertension and vascular dysfunction. Hypertension 2008, 52, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Tassi, E.; Lai, E.Y.; Li, L.; Solis, G.; Chen, Y.; Kietzman, W.E.; Ray, P.E.; Riegel, A.T.; Welch, W.J.; Wilcox, C.S.; et al. Blood Pressure Control by a Secreted FGFBP1 (Fibroblast Growth Factor-Binding Protein). Hypertension 2018, 71, 160–167. [Google Scholar] [CrossRef]
- Piotti, A.; Novelli, D.; Meessen, J.; Ferlicca, D.; Coppolecchia, S.; Marino, A.; Salati, G.; Savioli, M.; Grasselli, G.; Bellani, G.; et al. Endothelial damage in septic shock patients as evidenced by circulating syndecan-1, sphingosine-1-phosphate and soluble VE-cadherin: A substudy of ALBIOS. Crit. Care 2021, 25, 113. [Google Scholar] [CrossRef] [PubMed]
- Mauricio, M.D.; Aldasoro, M.; Ortega, J.; Vila, J.M. Endothelial dysfunction in morbid obesity. Curr. Pharm. Des. 2013, 19, 5718–5729. [Google Scholar] [CrossRef]
- Bordy, R.; Totoson, P.; Prati, C.; Marie, C.; Wendling, D.; Demougeot, C. Microvascular endothelial dysfunction in rheumatoid arthritis. Nat. Rev. Rheumatol. 2018, 14, 404–420. [Google Scholar] [CrossRef]
- Bijl, M. Endothelial activation, endothelial dysfunction and premature atherosclerosis in systemic autoimmune diseases. Neth. J. Med. 2003, 61, 273–277. [Google Scholar]
- Budhiraja, R.; Parthasarathy, S.; Quan, S.F. Endothelial dysfunction in obstructive sleep apnea. J. Clin. Sleep Med. 2007, 3, 409–415. [Google Scholar] [CrossRef]
- Kim, J.A.; Montagnani, M.; Koh, K.K.; Quon, M.J. Reciprocal relationships between insulin resistance and endothelial dysfunction: Molecular and pathophysiological mechanisms. Circulation 2006, 113, 1888–1904. [Google Scholar] [CrossRef]
- Tomaszewski, M.; Charchar, F.J.; Nelson, C.P.; Barnes, T.; Denniff, M.; Kaiser, M.; Debiec, R.; Christofidou, P.; Rafelt, S.; van der Harst, P.; et al. Pathway analysis shows association between FGFBP1 and hypertension. J. Am. Soc. Nephrol. 2011, 22, 947–955. [Google Scholar] [CrossRef]
- Braun, M.C.; Herring, S.M.; Gokul, N.; Monita, M.; Bell, R.; Hicks, M.J.; Wenderfer, S.E.; Doris, P.A. Hypertensive renal disease: Susceptibility and resistance in inbred hypertensive rat lines. J. Hypertens 2013, 31, 2050–2059. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Cao, X.; Li, L.; Wang, X.; Wang, Q.; Jiang, S.; Tang, C.; Zhou, S.; Xu, N.; Cui, Y.; et al. Acute Kidney Injury Sensitizes the Brain Vasculature to Ang II (Angiotensin II) Constriction via FGFBP1 (Fibroblast Growth Factor Binding Protein 1). Hypertension 2020, 76, 1924–1934. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.Y.; Bai, W.D.; Liu, J.Q.; Zheng, Z.; Guan, H.; Zhou, Q.; Su, L.L.; Xie, S.T.; Wang, Y.C.; Li, J.; et al. Up-regulation of FGFBP1 signaling contributes to miR-146a-induced angiogenesis in human umbilical vein endothelial cells. Sci. Rep. 2016, 6, 25272. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.E.; Tassi, E.; Liu, X.H.; Wellstein, A. Role of fibroblast growth factor-binding protein in the pathogenesis of HIV-associated hemolytic uremic syndrome. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 290, R105–R113. [Google Scholar] [CrossRef]
- Liu, X.H.; Aigner, A.; Wellstein, A.; Ray, P.E. Up-regulation of a fibroblast growth factor binding protein in children with renal diseases. Kidney Int. 2001, 59, 1717–1728. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.E.; Al-Attar, A.; Liu, X.H.; Das, J.R.; Tassi, E.; Wellstein, A. Expression of a Secreted Fibroblast Growth Factor Binding Protein-1 (FGFBP1) in Angioproliferative Kaposi Sarcoma. J. AIDS Clin. Res. 2014, 5, 309. [Google Scholar] [CrossRef] [PubMed]
- Ueyama, T.; Sakuma, M.; Nakatsuji, M.; Uebi, T.; Hamada, T.; Aiba, A.; Saito, N. Rac-Dependent Signaling from Keratinocytes Promotes Differentiation of Intradermal White Adipocytes. J. Investig. Dermatol. 2020, 140, 75–84.e6. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Chen, Z.; Shang, X.; Tian, D.; Wang, D.; Wu, K.; Fan, D.; Xia, L. Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology 2015, 61, 1920–1933. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, M.; Hu, Q.; Xu, W.; Liu, W.; Sun, Q.; Ye, Z.; Fan, G.; Xu, X.; Yu, X.; et al. FGFBP1, a downstream target of the FBW7/c-Myc axis, promotes cell proliferation and migration in pancreatic cancer. Am. J. Cancer Res. 2019, 9, 2650–2664. [Google Scholar]
- Tassi, E.; Wellstein, A. The angiogenic switch molecule, secreted FGF-binding protein, an indicator of early stages of pancreatic and colorectal adenocarcinoma. Semin Oncol. 2006, 33, S50–S56. [Google Scholar] [CrossRef]
- Li, F.; Zhang, H.; Wang, Y.; Yao, Z.; Xie, K.; Mo, Q.; Fan, Q.; Hou, L.; Deng, F.; Tan, W. FGFBP1 as a potential biomarker predicting bacillus Calmette-Guerin response in bladder cancer. Front. Immunol. 2022, 13, 954836. [Google Scholar] [CrossRef] [PubMed]
- Ray, R.; Cabal-Manzano, R.; Moser, A.R.; Waldman, T.; Zipper, L.M.; Aigner, A.; Byers, S.W.; Riegel, A.T.; Wellstein, A. Up-regulation of fibroblast growth factor-binding protein, by beta-catenin during colon carcinogenesis. Cancer Res. 2003, 63, 8085–8089. [Google Scholar]
- Schulze, D.; Plohmann, P.; Hobel, S.; Aigner, A. Anti-tumor effects of fibroblast growth factor-binding protein (FGF-BP) knockdown in colon carcinoma. Mol. Cancer 2011, 10, 144. [Google Scholar] [CrossRef]
- Czubayko, F.; Liaudet-Coopman, E.D.; Aigner, A.; Tuveson, A.T.; Berchem, G.J.; Wellstein, A. A secreted FGF-binding protein can serve as the angiogenic switch in human cancer. Nat. Med. 1997, 3, 1137–1140. [Google Scholar] [CrossRef]
- Zhou, M.; Sutliff, R.L.; Paul, R.J.; Lorenz, J.N.; Hoying, J.B.; Haudenschild, C.C.; Yin, M.; Coffin, J.D.; Kong, L.; Kranias, E.G.; et al. Fibroblast growth factor 2 control of vascular tone. Nat. Med. 1998, 4, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Folkman, J.; Klagsbrun, M. Vascular physiology. A family of angiogenic peptides. Nature 1987, 329, 671–672. [Google Scholar] [CrossRef] [PubMed]
- Doetschman, T.; Shull, M.; Kier, A.; Coffin, J.D. Embryonic stem cell model systems for vascular morphogenesis and cardiac disorders. Hypertension 1993, 22, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Mone, P.; Gambardella, J.; Wang, X.; Jankauskas, S.S.; Matarese, A.; Santulli, G. miR-24 Targets the Transmembrane Glycoprotein Neuropilin-1 in Human Brain Microvascular Endothelial Cells. Noncoding RNA 2021, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, J.; Jankauskas, S.S.; Kansakar, U.; Varzideh, F.; Avvisato, R.; Prevete, N.; Sidoli, S.; Mone, P.; Wang, X.; Lombardi, A.; et al. Ketone bodies rescue mitochondrial dysfunction via epigenetic remodeling. JACC Basic Transl. Sci. 2023; in press. [Google Scholar]
- Mone, P.; Varzideh, F.; Jankauskas, S.S.; Pansini, A.; Lombardi, A.; Frullone, S.; Santulli, G. SGLT2 Inhibition via Empagliflozin Improves Endothelial Function and Reduces Mitochondrial Oxidative Stress: Insights From Frail Hypertensive and Diabetic Patients. Hypertension 2022, 79, 1633–1643. [Google Scholar] [CrossRef]






| Genes | F/R | Sequence (5′-to-3′) | bp |
|---|---|---|---|
| FGFBP1 | Forward | GG AGG AGC TGT GAG TAA CGT | 113 |
| Reverse | TG TCA GGT AGA GTG CAA GGG | ||
| GAPDH | Forward | GG CTC CCT TGG GTA TAT GGT | 94 |
| Reverse | TT GAT TTT GGA GGG ATC TCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avvisato, R.; Mone, P.; Jankauskas, S.S.; Varzideh, F.; Kansakar, U.; Gambardella, J.; De Luca, A.; Matarese, A.; Santulli, G. miR-4432 Targets FGFBP1 in Human Endothelial Cells. Biology 2023, 12, 459. https://doi.org/10.3390/biology12030459
Avvisato R, Mone P, Jankauskas SS, Varzideh F, Kansakar U, Gambardella J, De Luca A, Matarese A, Santulli G. miR-4432 Targets FGFBP1 in Human Endothelial Cells. Biology. 2023; 12(3):459. https://doi.org/10.3390/biology12030459
Chicago/Turabian StyleAvvisato, Roberta, Pasquale Mone, Stanislovas S. Jankauskas, Fahimeh Varzideh, Urna Kansakar, Jessica Gambardella, Antonio De Luca, Alessandro Matarese, and Gaetano Santulli. 2023. "miR-4432 Targets FGFBP1 in Human Endothelial Cells" Biology 12, no. 3: 459. https://doi.org/10.3390/biology12030459
APA StyleAvvisato, R., Mone, P., Jankauskas, S. S., Varzideh, F., Kansakar, U., Gambardella, J., De Luca, A., Matarese, A., & Santulli, G. (2023). miR-4432 Targets FGFBP1 in Human Endothelial Cells. Biology, 12(3), 459. https://doi.org/10.3390/biology12030459








