Identification of CNGCs in Glycine max and Screening of Related Resistance Genes after Fusarium solani Infection
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
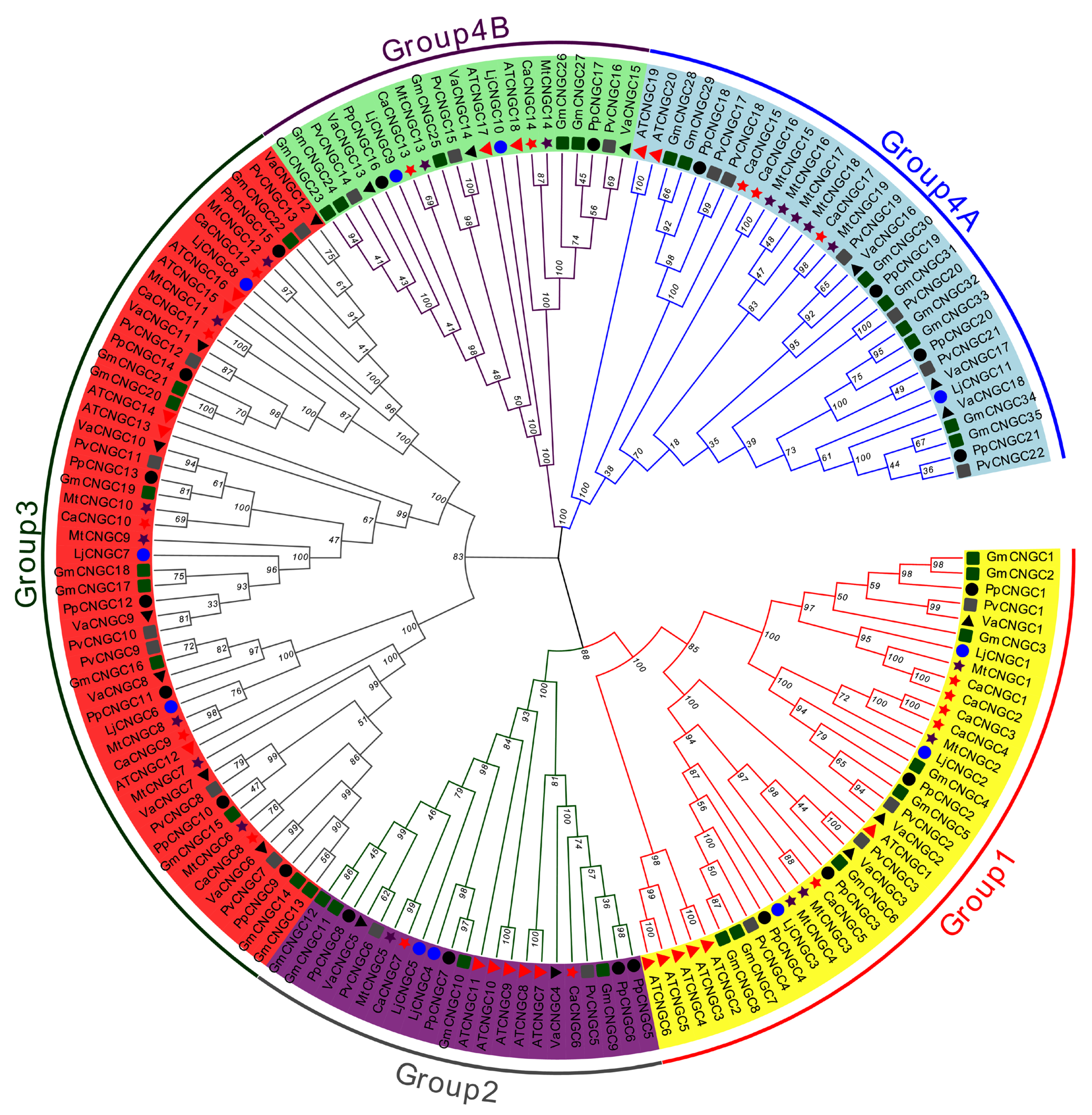
2.1. Identification and Phylogenetic Analysis
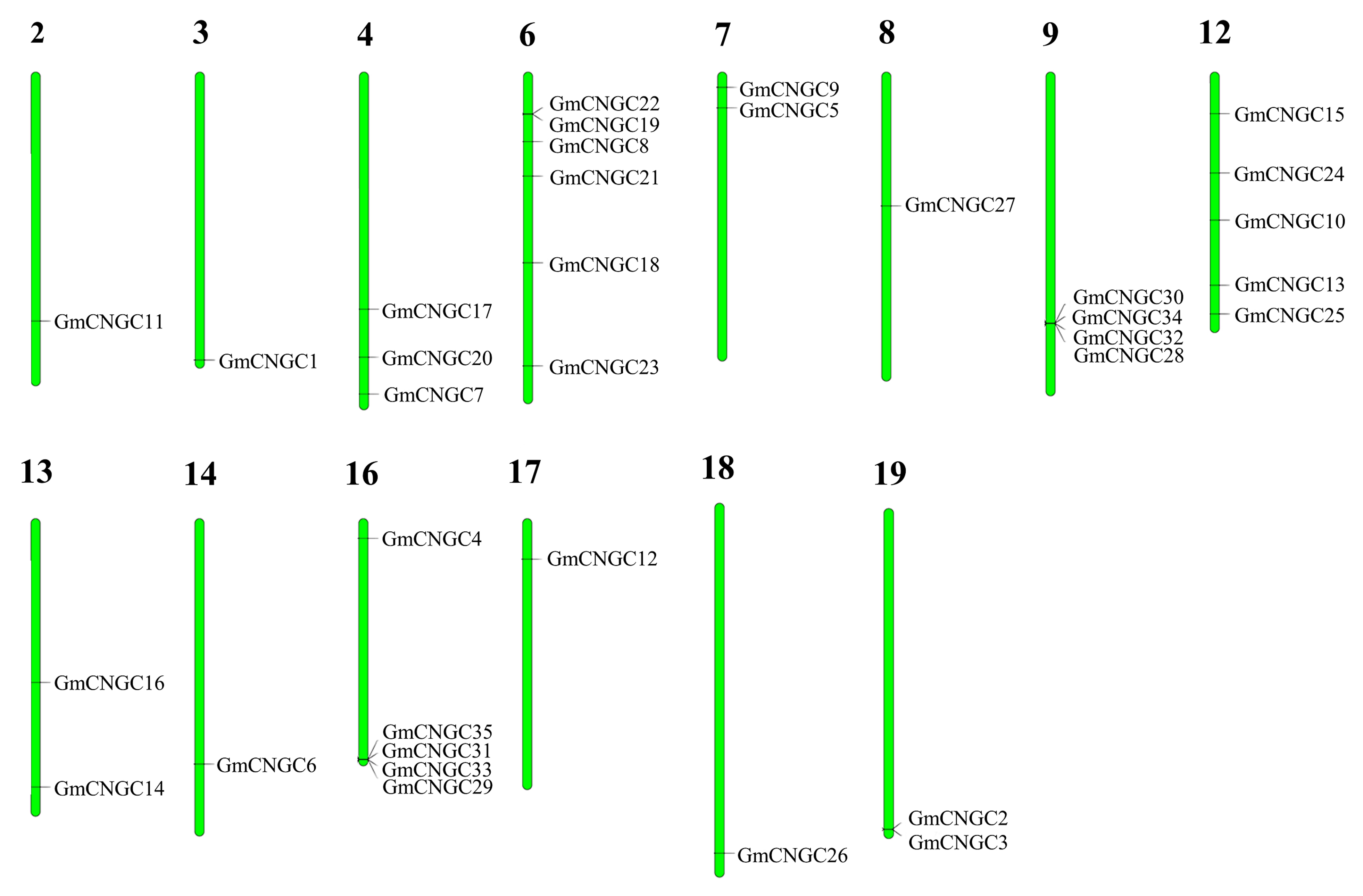
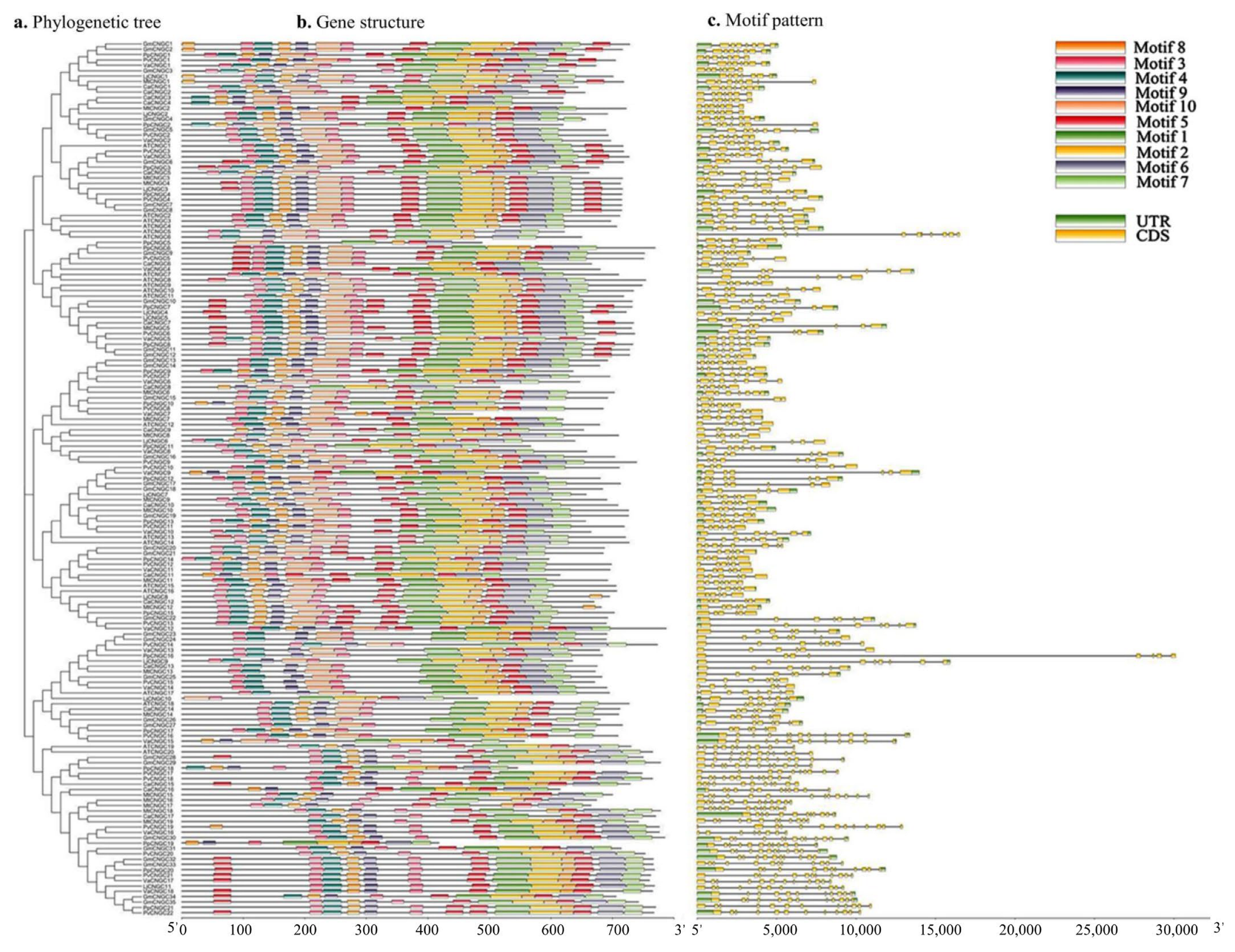
2.2. Location of CNGC Genes on Chromosomes and Conserved Motifs
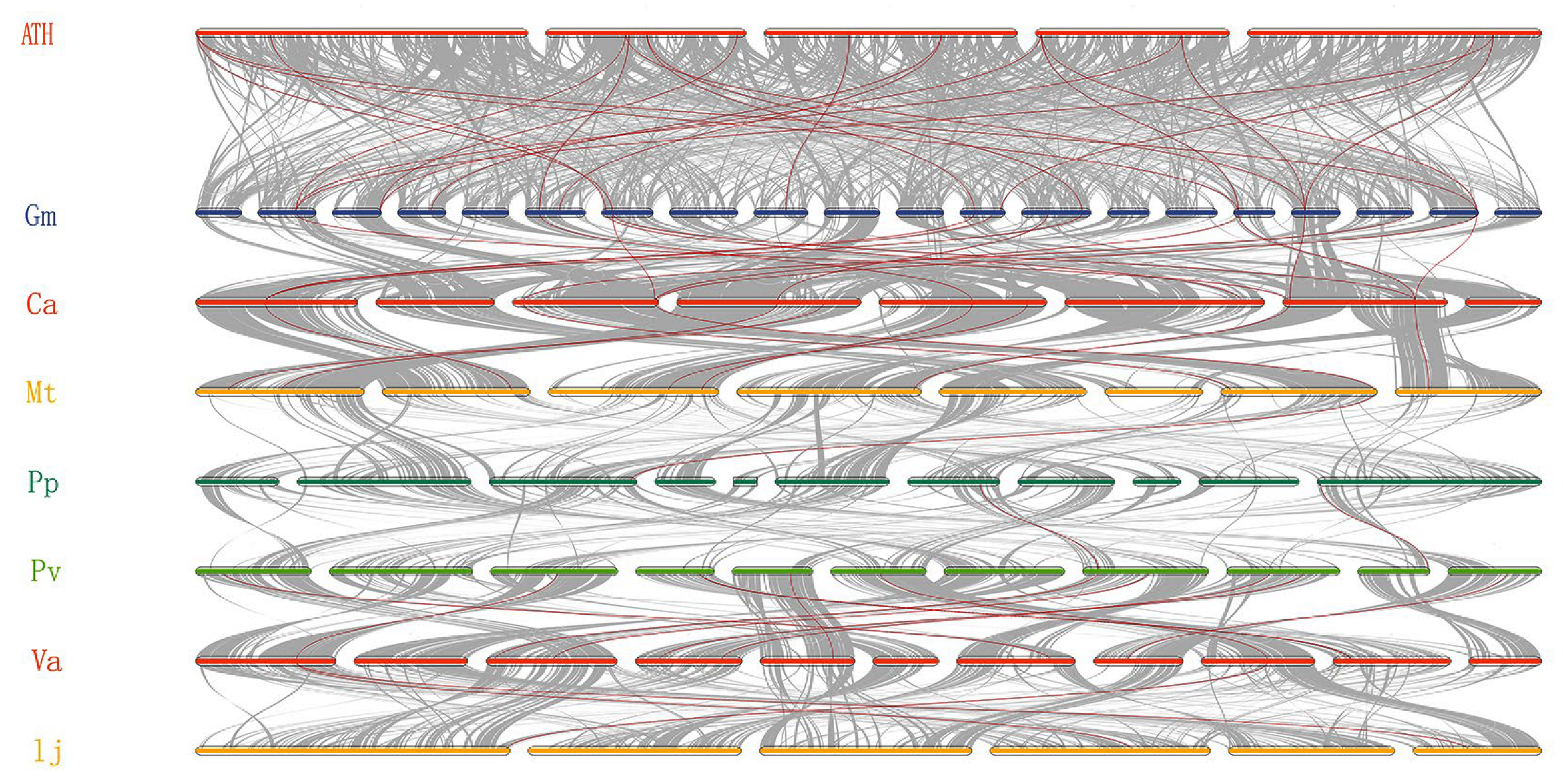
2.3. Collinear Analysis of CNGC Genes
2.4. Analysis of Cis-Acting Element
2.5. Pathogen Isolation, Purification, and Morphological Identification
2.6. Pathogenicity Detection
2.7. Molecular Identification of Pathogenic Fungi
2.8. Pathogen Inoculation and Sampling Time
2.9. Screening of Differentially Expressed Genes (DEGs)
2.10. Analysis of Enrichment
2.11. Protein–protein Interaction (PPI) Prediction
3. Results
3.1. Identification and Phylogenetic Analysis of the CNGC Gene Family
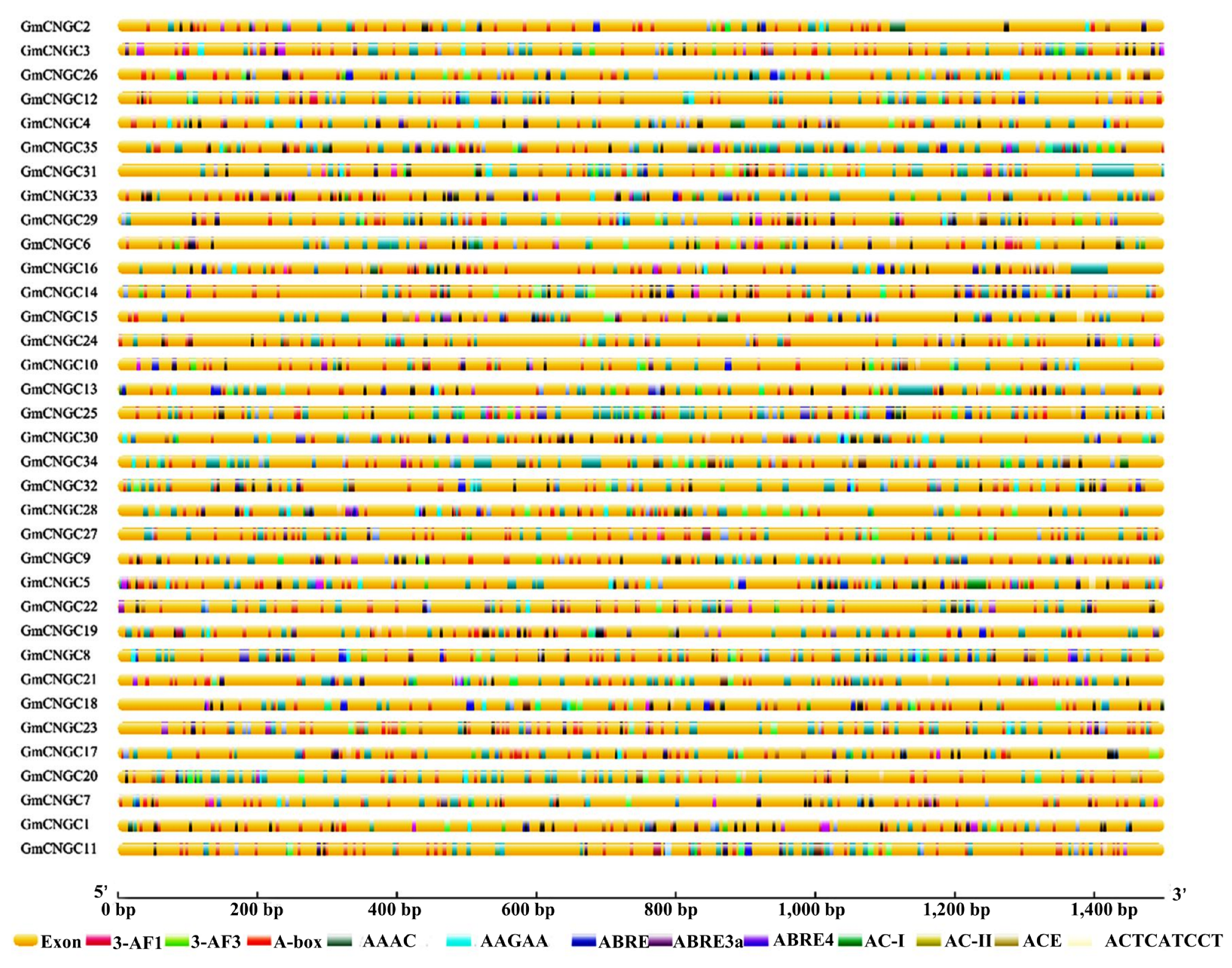
3.2. Distribution, Gene Structure, and Motif Analysis of GmCNGC Genes on Chromosomes
3.3. Collinearity and Evolutionary Analysis of CNGC Genes
3.4. The Cis-Acting Element in the CNGC Gene Promoter Sequence
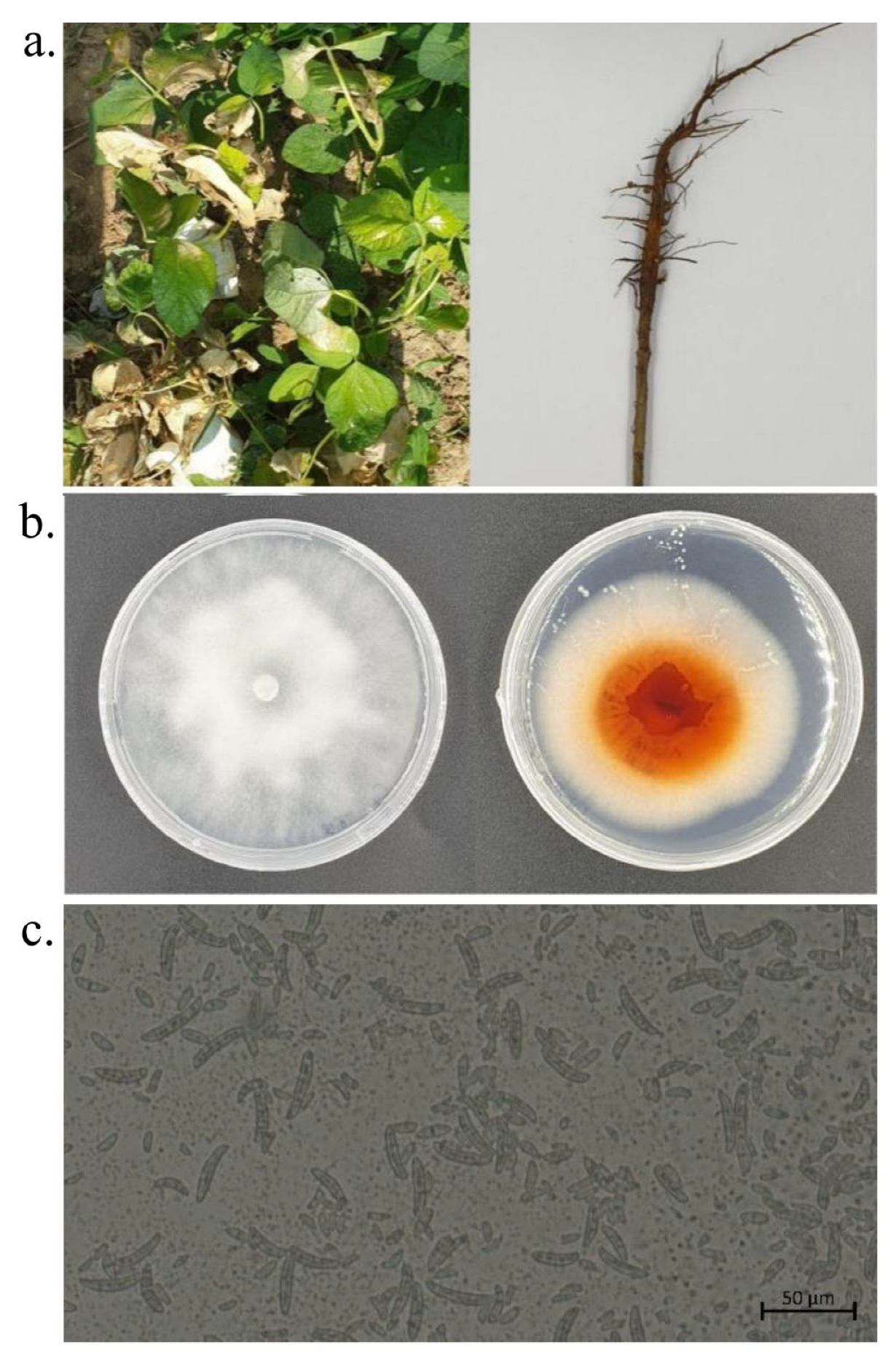
3.5. Symptoms of Field Diseases
3.6. Pathogen Isolation, Purification, and Morphological Identification
3.7. Pathogenicity Detection
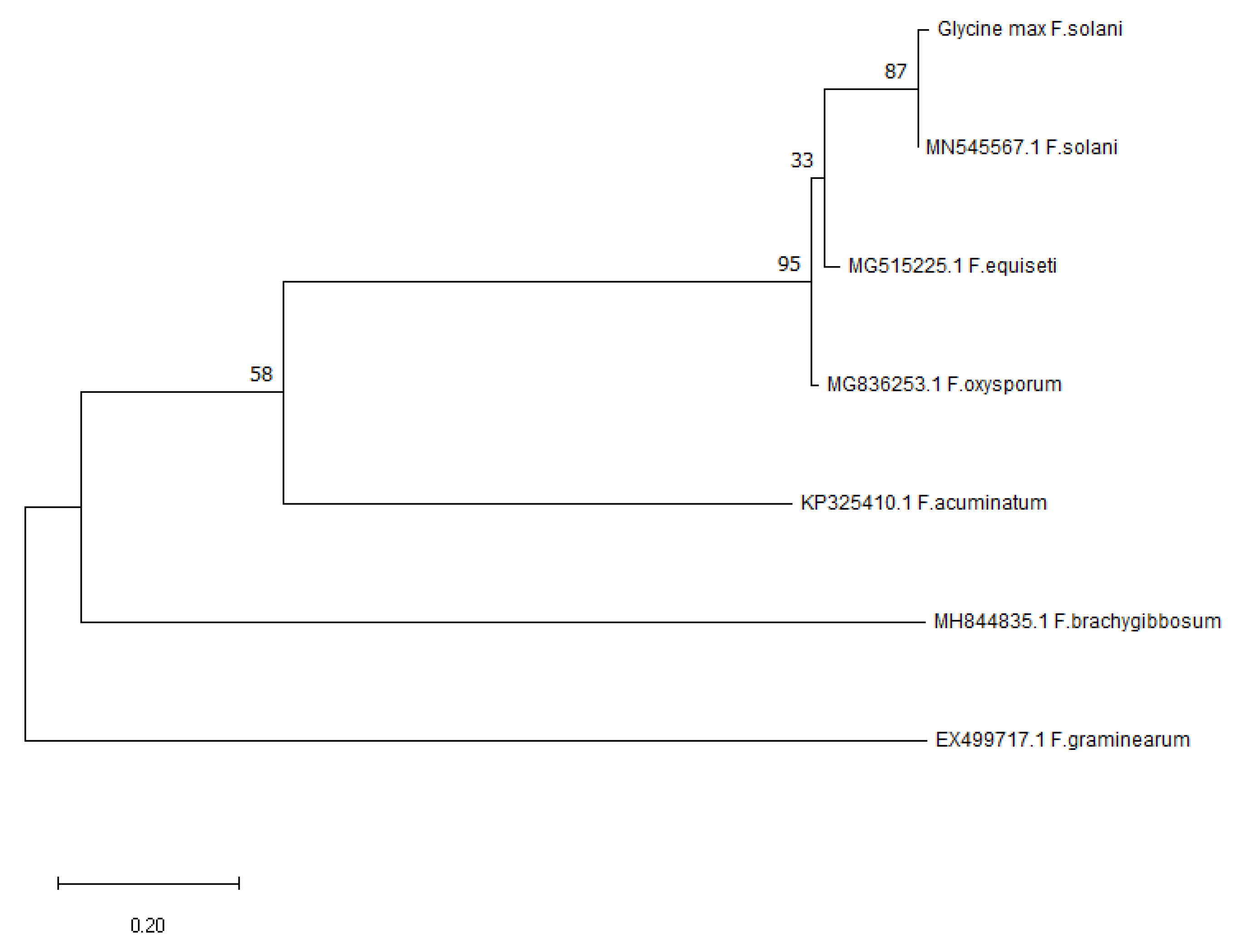
3.8. Molecular Identification and Analysis of Pathogen
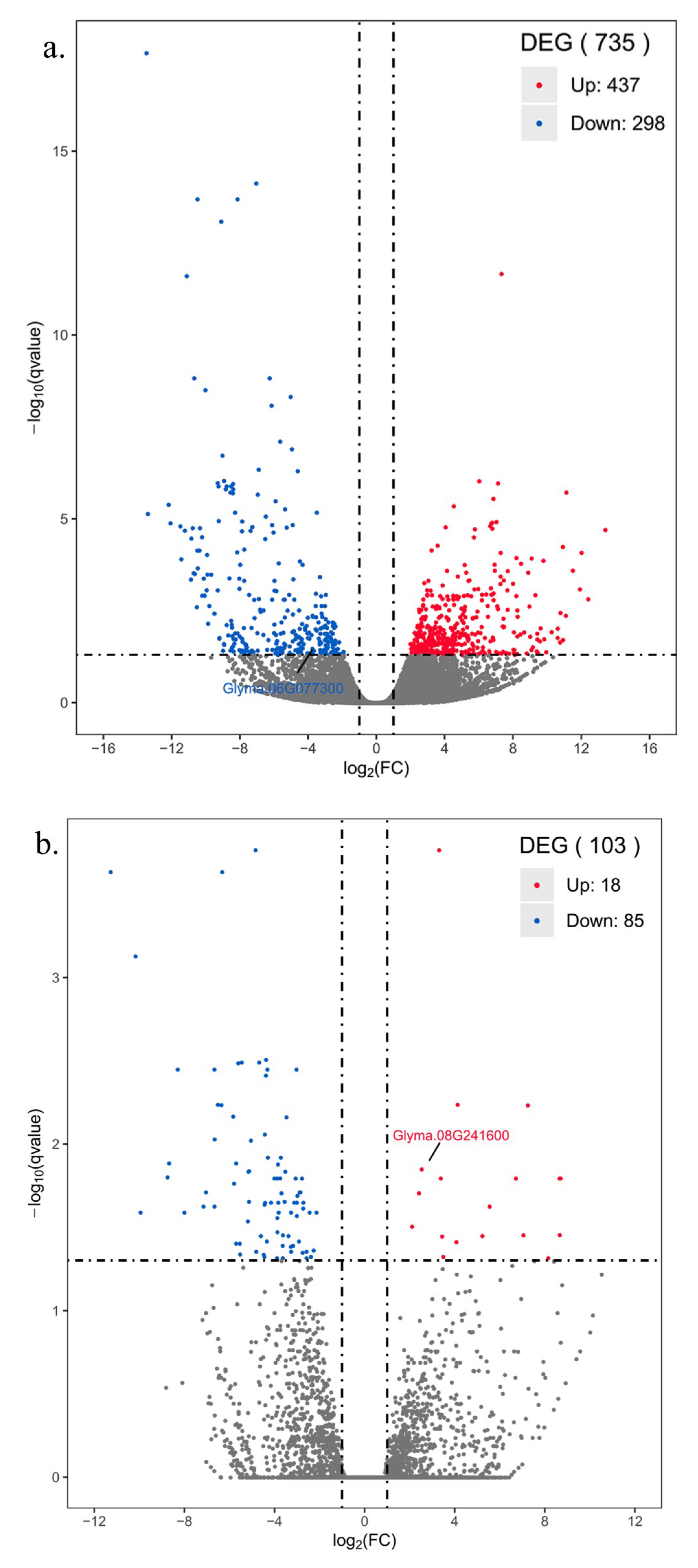
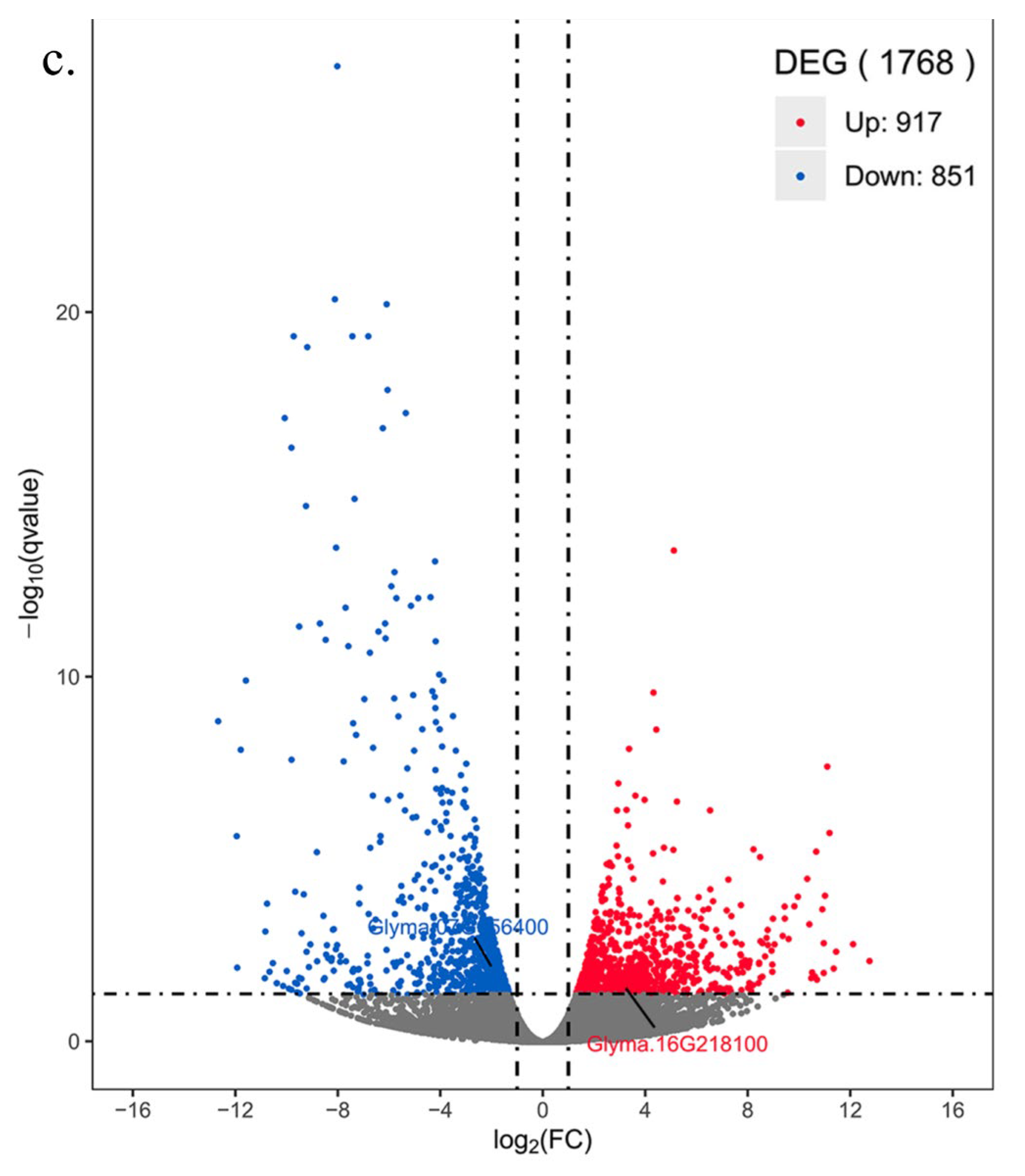
3.9. Screening of DEGs after Disease Stress
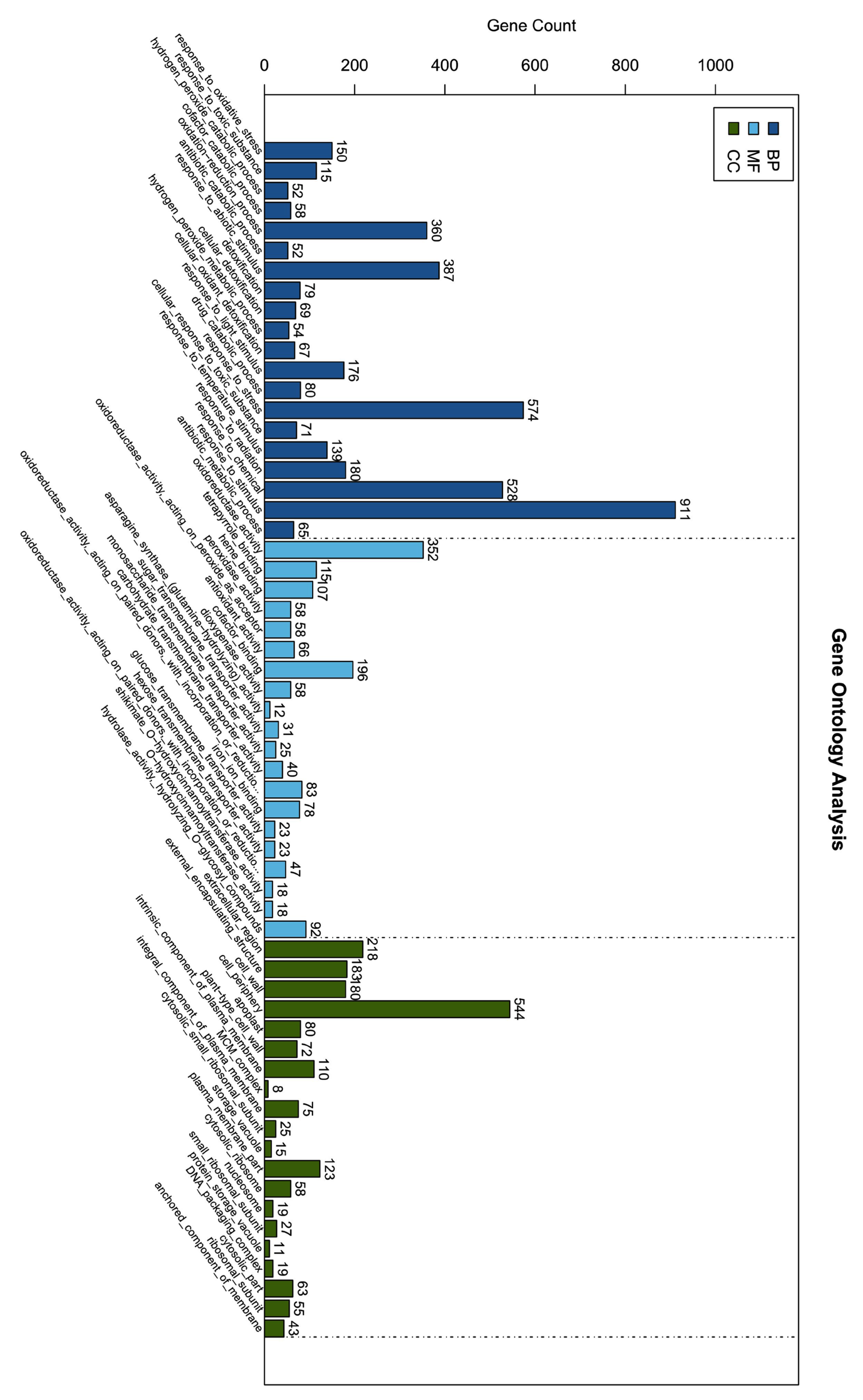
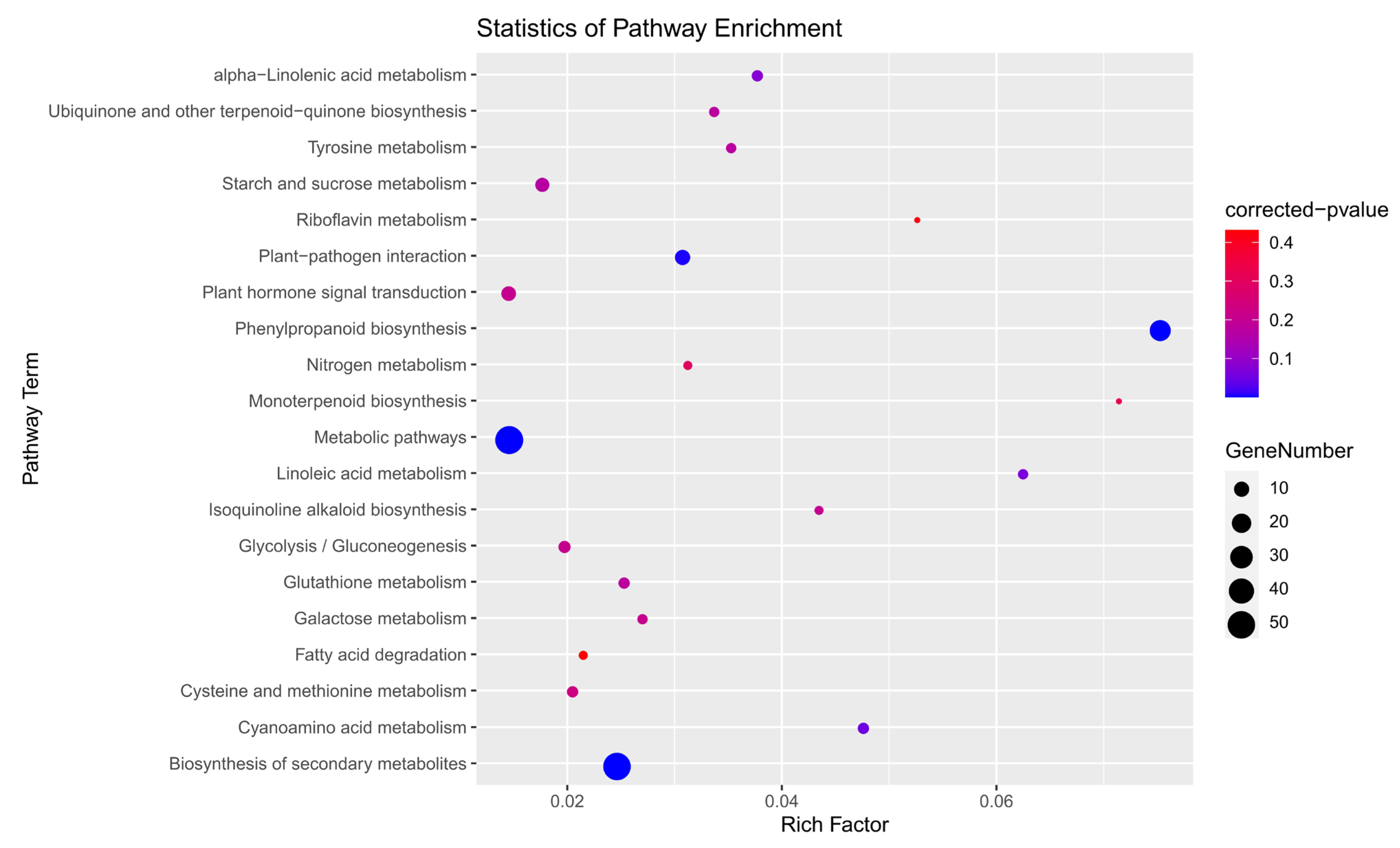
3.10. GO and KEGG Enrichment Analysis
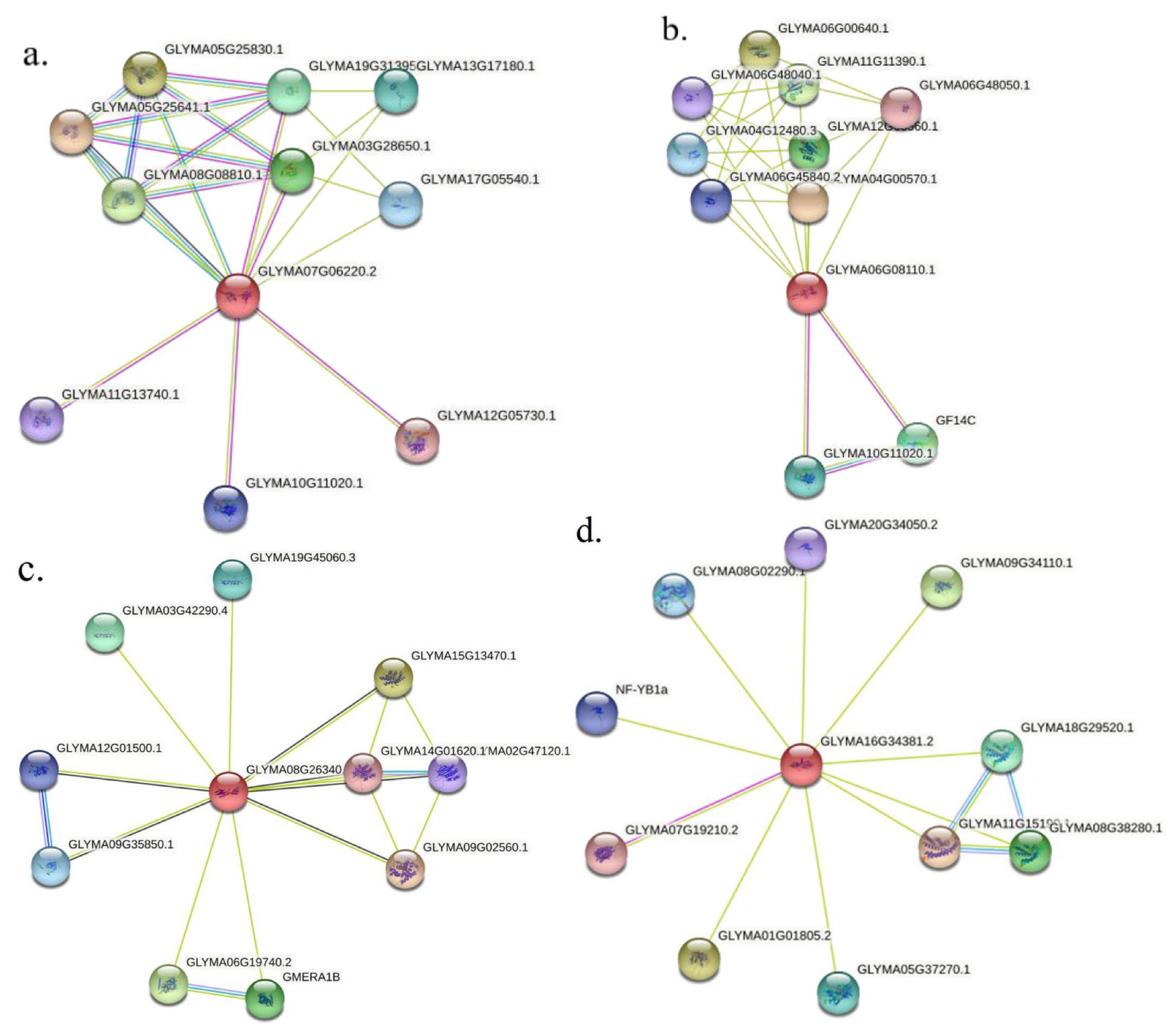
3.11. Protein Interaction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feng, Z.J.; Xu, S.C.; Liu, N.; Zhang, G.W.; Hu, Q.Z.; Gong, Y.M. Soybean TCP transcription factors: Evolution, classification, protein interaction and stress and hormone responsiveness. Plant Physiol. Biochem. 2018, 127, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Dhaubhadel, S.; Marsolais, F. Transcriptomics of Legume Seed: Soybean a Model Grain Legume; Springer Science+Business Media: New York, NY, USA, 2012. [Google Scholar]
- Li, M.H. Investigation and Identification of Soybean Field Diseases in Northeast China; Shenyang Agricultural University: Shenyang, China, 2016. [Google Scholar]
- Talke, I.N.; Damien, B.; Maathuis, F.; Sanders, D. CNGCs: Prime targets of plant cyclic nucleotide signalling? Trends Plant Sci. 2003, 8, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Zelman, A.K.; Dawe, A.; Gehring, C.; Berkowitz, G.A. Evolutionary and Structural Perspectives of Plant Cyclic Nucleotide-Gated Cation Channels. Front. Plant Sci. 2012, 3, 95. [Google Scholar] [CrossRef] [PubMed]
- Cukkemane, A.; Seifert, R.; Kaupp, U.B. Cooperative and uncooperative cyclic-nucleotide-gated ion channels. Trends Biochem. Sci. 2011, 36, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Young, E.C.; Krougliak, N. Distinct Structural Determinants of Efficacy and Sensitivity in the Ligand-binding Domain of Cyclic Nucleotide-gated Channels. J. Biol. Chem. 2003, 279, 3553–3562. [Google Scholar] [CrossRef]
- Schuurink, R.C.; Shartzer, S.F.; Fath, A.; Jones, R.L. Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone. Proc. Natl. Acad. Sci. USA 1998, 95, 1944–1949. [Google Scholar] [CrossRef]
- Arazi, T.; Kaplan, B.; Fromm, H. A high-affinity calmodulin-binding site in a tobacco plasma-membrane channel protein coincides with a characteristic element of cyclic nucleotide-binding domains. Plant Mol. Biol. 2000, 42, 591–601. [Google Scholar] [CrossRef]
- Köhler, C.; Neuhaus, G. Characterisation of calmodulin binding to cyclic nucleotide-gated ion channels from Arabidopsis thaliana. FEBS Lett. 2000, 471, 133–136. [Google Scholar] [CrossRef]
- Reddy, V.S.; Ali, G.S.; Reddy, A. Genes Encoding Calmodulin-binding Proteins in the Arabidopsis Genome. J. Biol. Chem. 2002, 277, 9840–9852. [Google Scholar] [CrossRef]
- Frietsch, S.; Wang, Y.F.; Sladek, C.; Poulsen, L.R.; Romanowsky, S.M.; Schroeder, J.L.; Harper, J.F. A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proc. Natl. Acad. Sci. USA 2007, 104, 14531–14536. [Google Scholar] [CrossRef]
- Zhang, S.; Pan, Y.; Tian, W.; Dong, M.; Zhu, H.; Luan, S.; Li, L. Arabidopsis CNGC14 Mediates Calcium Influx Required for Tip Growth in Root Hairs. Mol. Plant 2017, 10, 1004–1006. [Google Scholar] [CrossRef]
- Ma, W.; Smigel, A.; Walker, R.K.; Moeder, W.; Yoshioka, K.; Berkowitz, J.A. Leaf Senescence Signaling: The Ca2+-Conducting Arabidopsis Cyclic Nucleotide Gated Channel2 Acts through Nitric Oxide to Repress Senescence Programming. Plant Physiol. 2010, 154, 733–743. [Google Scholar] [CrossRef]
- Charpentier, M.; Sun, J.; Martins, T.V.; Radhakrishnan, G.V.; Findlay, K.; Soumpourou, E.; Thouin, J.; Véry, A.; Sanders, D.; Morris, R.J.; et al. Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 2016, 352, 1102–1105. [Google Scholar] [CrossRef]
- Jin, Y.; Jing, W.; Zhang, Q.; Zhang, W. Cyclic nucleotide gated channel 10 negatively regulates salt tolerance by mediating Na+ transport in Arabidopsis. J. Plant. Res. 2015, 128, 211–220. [Google Scholar] [CrossRef]
- Shao, H.B.; Chu, L.Y.; Shao, M.A.; Li, S.Q.; Yao, J.C. Bioengineering plant resistance to abiotic stresses by the global calcium signal system. Biotechnol. Adv. 2008, 26, 503–510. [Google Scholar]
- Tunc-Ozdemir, M.; Tang, C.; Ishka, M.R.; Brown, E.; Groves, N.R.; Myers, C.T.; Rato, C.; Poulsen, L.R.; McDowell, S.; Miller, G.; et al. A cyclic nucleotide-gated channel (CNGC16) in pollen is critical for stress tolerance in pollen reproductive development. Plant Physiol. 2013, 161, 1010–1020. [Google Scholar] [CrossRef]
- Nawaz, Z.; Kakar, K.U.; Ullah, R.; Yu, S.; Zhang, J.; Shu, Q.Y.; Ren, X.L. Genome-wide identification, evolution and expression analysis of cyclic nucleotide-gated channels in tobacco (Nicotiana tabacum L.). Genomics 2019, 111, 142–158. [Google Scholar] [CrossRef]
- Clough, S.J.; Fengler, K.A.; Yu, I.C.; Lippok, B.; Smith Jr, R.K.; Bent, A.F. The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 2000, 97, 9323–9328. [Google Scholar] [CrossRef]
- Ali, R.; Ma, W.; Lemtiri-Chlieh, F.; Tsaltas, D.; Leng, Q.; Bodman, S.; Berkowitz, G.A. Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 2007, 19, 1081–1095. [Google Scholar] [CrossRef]
- Li, Q.Q.; Yang, S.Q.; Ren, J.; Ye, X.; Jiang, X.; Liu, Z. Genome-wide identification and functional analysis of the cyclic nucleotide-gated channel gene family in Chinese cabbage. Biotech 2019, 9, 114. [Google Scholar] [CrossRef]
- Tao, X.; Chengjie, C.; Chuhao, L.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar]
- Kakar, K.U.; Zarqa, N.; Khadija, K.; Ali, E.; Almoneafy, A.A.; Ullah, R.; Ren, X.L.; Shu, Q.Y. Comprehensive genomic analysis of the CNGC gene family in Brassica oleracea: Novel insights into synteny, structures, and transcript profiles. BMC Genom. 2017, 18, 869. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, 458–460. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar]
- Kim, M.Y.; Kang, Y.J.; Lee, T.; Lee, S.H. Divergence of Flowering-Related Genes in Three Legume Species. Plant Genome 2013, 6, 1–12. [Google Scholar] [CrossRef]
- Xu, L.; Qiao, X.; Zhang, M.; Zhang, S. Genome-Wide analysis of aluminum-activated malate transporter family genes in six rosaceae species, and expression analysis and functional characterization on malate accumulation in Chinese white pear. Plant Sci. 2018, 24, 431–442. [Google Scholar]
- Ling, L.; Zhang, W.; An, Y.; Du, B.; Wang, D.; Guo, C. Genome-wide analysis of the TCP transcription factor genes in five legume genomes and their response to salt and drought stresses. Funct. Integr. Genom. 2020, 20, 537–550. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, 49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.V.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Fang, Z.D. Research Methods of Plant Diseases; China Agriculture Press: Beijing, China, 1998. [Google Scholar]
- Sohrabi, M.; Samsampour, D.; Bagheri, A. Molecular Identification of Fungal Endophytes of Medicinal Plant Citrullus colocynthis (L.) Schrad as a Medicinal Plant: Role of Tissue Type and Sampling Location on the Diversity. Mol. Biotechnol. 2023, 1–8, published online ahead of print. [Google Scholar] [CrossRef]
- Cong, L.L.; Kang, J.M.; Zhang, T.J.; Long, R.; Yang, Q. Identification and pathogenicity test of pathogenic Fusarium of alfafa root rot. Acta Agrestia Sin. 2017, 25, 857–865. [Google Scholar]
- Liu, M.; Wang, L.J.; Cao, S.; Wu, X.; Huang, J. Study on Identification of Cabbage Seedborne Fusarium Species and Their Pathogenicity. Seed 2012, 31, 26–29. [Google Scholar]
- Wang, J.; Ren, J.D.; Li, W.Y.; Shang, Q.X. Pathogen identification of soybean black spot. J. Beijing Agric. Coll. 2021, 36, 92–96. [Google Scholar]
- Tiwari, P.; Srivastava, D.; Chauhan, A.S.; Indoliya, Y.; Singh, P.K.; Tiwari, S.; Fatima, T.; Mishra, S.K.; Dwivedi, S.; Agarwal, L.; et al. Root system architecture, physiological analysis and dynamic transcriptomics unravel the drought-responsive traits in rice genotypes. Ecotoxicol. Environ. Saf. 2021, 207, 111252. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Carbon, S.; Douglass, E.; Good, B.M.; Unni, D.R.; Harris, N.L.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; Hartline, E.; et al. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, 325–334. [Google Scholar]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, 317–325. [Google Scholar] [CrossRef]
- Wang, J.T.; Ramage, D.; Amin, N.; Baliga, N.S.; Wang, J.Y.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar]
- Song, W.; Song, S.; Lan, J.; Zheng, D.; Xu, X.; Huang, L.; Yan, C.; Liu, Y.; Tang, W. First Report of root rot Caused by Fusarium solani and F. breve on kiwifruit in China. Plant Dis. 2022. (published online ahead of print, 2022 Nov 10). [Google Scholar]
- Zhang, G.; Yan, X.; Zhang, S.; Zheng, D.; Xu, X.; Huang, L.; Yan, C.; Liu, Y.; Tang, W. The jasmonate-ZIM domain gene VqJAZ4 from the Chinese wild grape Vitis quinquangularis improves resistance to powdery mildew in Arabidopsis thaliana. Plant Physiol. Bioch. 2019, 143, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, Q.; Sun, L.X.; He, Z. The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res. 2006, 13, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ying, J.; Kuzma, M.; Chalifoux, M.; Sample, A.; McArthur, C.; Uchacz, T.; Sarvas, C.; Wan, J.; Dennis, D.T.; et al. Molecular tailoring of farnesylation for plant drought tolerance and yield protection. Plant J. 2005, 43, 413–424. [Google Scholar] [CrossRef]
- Nawaz, Z.; Kakar, K.U.; Saand, M.A.; Shu, Q.Y. Cyclic nucleotide-gated ion channel gene family in rice, identification, characterization and experimental analysis of expression response to plant hormones, biotic and abiotic stresses. BMC Genomics 2014, 15, 853. [Google Scholar] [CrossRef]
- Saand, M.A.; Xu, Y.P.; Li, W.; Wang, J.P.; Cai, X.Z. Cyclic nucleotide gated channel gene family in tomato, genome-wide identification and functional analyses in disease resistance. Front. Plant Sci. 2015, 6, 303. [Google Scholar] [CrossRef]
- Chen, J.Q.; Hao, Y.; Gu, J.P.; Li, L.; Liu, Z.; Jiang, X.; Zou, H.; Wei, S.; Zhang, S.; Wu, J. Genomic characterization, phylogenetic comparison and differential expression of the cyclic nucleotide-gated channels gene family in pear (Pyrus bretchneideri Rehd). Genomics 2015, 105, 39–52. [Google Scholar] [CrossRef]
- Guo, J.; Islam, M.A.; Lin, H.; Ji, C.; Duan, Y.; Liu, P.; Zeng, Q.; Day, B.; Kang, Z.; Guo, J. Genome-Wide Identification of Cyclic Nucleotide-Gated Ion Channel Gene Family in Wheat and Functional Analyses of TaCNGC14 and TaCNGC16. Front. Plant Sci. 2018, 9, 18. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Khaldi, N.; Shields, D.C. Shift in the isoelectric-point of milk proteins as a consequence of adaptive divergence between the milks of mammalian species. Biol. Direct. 2011, 6, 40. [Google Scholar] [CrossRef]
- Wang, X.G.; Yang, B.; Guo, C.J.; Zhang, L.R.; Shen, Q. Identification and pathogenicity test of pathogens causing potato Fusarium root rot in Ningxia Hui Autonomous Region. Acta Phytophy Sin. 2020, 47, 609–619. [Google Scholar]
- Fang, X.C.; Qu, H.L.; Zhao, S.F.; Guo, K.; Ma, M. Pathogen identification of root rot of processed pepper in Xinjiang. Northern. Hort. 2016, 15, 108–112. [Google Scholar]
- Tian, F.M.; Chen, Q.; Wang, H.; Zhuo, P.; Sun, J. Isolation, Identification and Biological Characteristics of the Pathogen Bacte-ria of Zanthoxylum bungeanum Root Rot in Longnan. J. Northeast. Agric. Sci. 2022, 47, 95–107. [Google Scholar]
- Han, F.Y.; Wang, Y.; Yu, C.Z.; Gao, J.; Yang, S.; Jia, R.; Hu, S.; Zhu, C. Isolation and identification of carrot root rot caused by Fusarium solani. J. Northern Agric. 2020, 48, 70–75. [Google Scholar]
- Yan, L.Y.; Zhang, Y.Y.; Ma, K.H.; Song, H.; Wang, S. Multiple PCR Detection of Fusarium oxysporum and Fusarium solani in Cucurbitaceous Plants. Acta Agric. Boreali-Sin. 2014, 29, 44–47. [Google Scholar]
- Wang, J.; Liang, Z.S.; Kang, B.; Luo, M.J. Identification of Root Rot pathogen of Panax notoginseng from Wenshan. J. Northwest Univ. 2015, 30, 158–163. [Google Scholar]
- Shi, J.L.; Li, Y.Q.; Hu, K.M.; Ren, J.G.; Liu, H.M. Isolation and identification of pathogens from rotted root of Pinellia ternata in Guizhou province. Microbiol China. 2015, 42, 289–299. [Google Scholar]
- Moon, J.Y.; Belloeil, C.; Ianna, M.L.; Shin, R. Arabidopsis CNGC Family Members Contribute to Heavy Metal Ion Uptake in Plants. Int. J. Mol. Sci. 2019, 20, 413. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Zhang, A.; Ren, Y.; Wu, F.; Wng, G.; Xu, Y.; Lei, C.; Zhu, S.; Pan, T.; et al. A cyclic nucleotide-gated channel mediates cytoplasmic calcium elevation and disease resistance in rice. Cell Res. 2019, 29, 820–831. [Google Scholar] [CrossRef]
- Zhang, W.; Dong, C.; Zhang, Y.; Zhu, J.; Dai, H.; Bai, S. An apple cyclic nucleotide-gated ion channel gene highly responsive to Botryosphaeria dothidea infection enhances the susceptibility of Nicotiana benthamiana to bacterial and fungal pathogens. Plant Sci. 2018, 269, 94–105. [Google Scholar] [CrossRef]
- Zhang, X.R.; Xu, Y.P.; Cai, X.Z. SlCNGC1 and SlCNGC14 Suppress Xanthomonas oryzae pv. oryzicola-Induced Hypersensitive Response and Non-host Resistance in Tomato. Front. Plant Sci. 2018, 9, 285. [Google Scholar] [CrossRef] [PubMed]
- Ladwig, F.; Dahlke, R.I.; Stührwohldt, N.; Hartmann, J.; Harter, K.; Sauter, M. Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell 2015, 27, 1718–1729. [Google Scholar] [CrossRef] [PubMed]
- Meena, M.K.; Prajapati, R.; Krishna, D.; Divakaran, K.; Pandey, Y.; Reichelt, M.; Mathew, M.K.; Boland, W.; Mithöfer, A.; Vadassery, J. The Ca2+ channel CNGC19 regulates Arabidopsis defense against spodoptera herbivory. Plant Cell 2019, 31, 1539–1562. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Verma, R.; Gehring, C.; Yamaguchi, Y.; Zhao, Y.; Ryan, C.A.; Berkowitz, G.A. Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc. Natl. Acad. Sci. USA 2010, 107, s21193. [Google Scholar] [CrossRef]

| Species | Number of CNGC | Group | ||||
|---|---|---|---|---|---|---|
| I | II | III | IV-A | IV-B | ||
| Glycine max | 35 | 8 | 4 | 10 | 8 | 5 |
| Cicer arietinum | 17 | 5 | 2 | 5 | 3 | 2 |
| Lotus japonicus | 11 | 3 | 2 | 3 | 1 | 2 |
| Medicago truncatula | 19 | 4 | 1 | 7 | 5 | 2 |
| Phaseolus vulgaris | 22 | 4 | 2 | 7 | 6 | 3 |
| Cajanus cajan (Pigeon Pea) | 21 | 4 | 4 | 7 | 4 | 2 |
| Vigna angularis | 18 | 3 | 2 | 7 | 3 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Y.; Wang, J.; Bai, Y.; Ban, L.; Ren, J.; Shang, Q.; Li, W. Identification of CNGCs in Glycine max and Screening of Related Resistance Genes after Fusarium solani Infection. Biology 2023, 12, 439. https://doi.org/10.3390/biology12030439
Cui Y, Wang J, Bai Y, Ban L, Ren J, Shang Q, Li W. Identification of CNGCs in Glycine max and Screening of Related Resistance Genes after Fusarium solani Infection. Biology. 2023; 12(3):439. https://doi.org/10.3390/biology12030439
Chicago/Turabian StyleCui, Yuxing, Jingxuan Wang, Yingxue Bai, Liping Ban, Junda Ren, Qiaoxia Shang, and Weiyu Li. 2023. "Identification of CNGCs in Glycine max and Screening of Related Resistance Genes after Fusarium solani Infection" Biology 12, no. 3: 439. https://doi.org/10.3390/biology12030439
APA StyleCui, Y., Wang, J., Bai, Y., Ban, L., Ren, J., Shang, Q., & Li, W. (2023). Identification of CNGCs in Glycine max and Screening of Related Resistance Genes after Fusarium solani Infection. Biology, 12(3), 439. https://doi.org/10.3390/biology12030439






