Transcriptome Analysis on Hepatopancreas Reveals the Metabolic Dysregulation Caused by Vibrio parahaemolyticus Infection in Litopenaeus vannamei
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Shrimp Culture and VPAHPND Challenge
2.2. Molecular Detection of V. parahaemolyticus in the Hepatopancreas
2.3. Total RNA Extraction, Library Preparation, and Transcriptome Sequencing
2.4. Transcriptome Assembly and Gene Functional Annotation
2.5. Identification of Differentially Expressed Genes (DEGs)
2.6. Enrichment Analysis of DEGs
2.7. Validation of Differentially Expressed Genes by RT-qPCR
3. Results
3.1. The Loads of V. parahaemolyticus in Shrimp Hepatopancreas
3.2. Transcriptome Sequencing Data
3.3. Differentially Expressed Genes (DEGs) in Different Comparisons
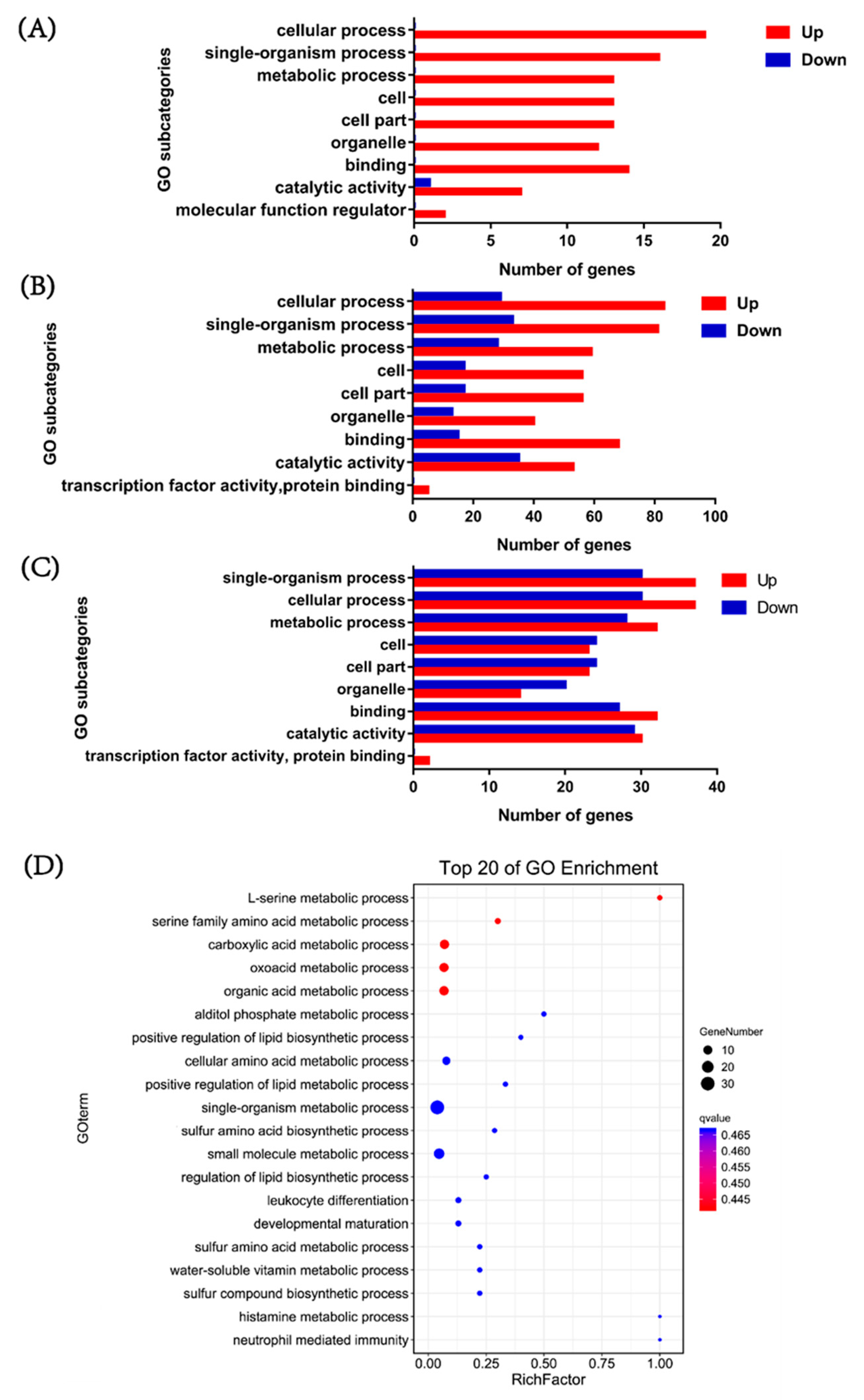
3.4. Functional Annotation of DEGs
3.5. Screening and Trend Analysis of Metabolism-Related DEGs
3.5.1. Glycolysis
3.5.2. Gluconeogenesis and Pentose Phosphate Pathway
3.5.3. Glycine, Serine and Threonine Metabolism
3.5.4. One Carbon Pool by Folate
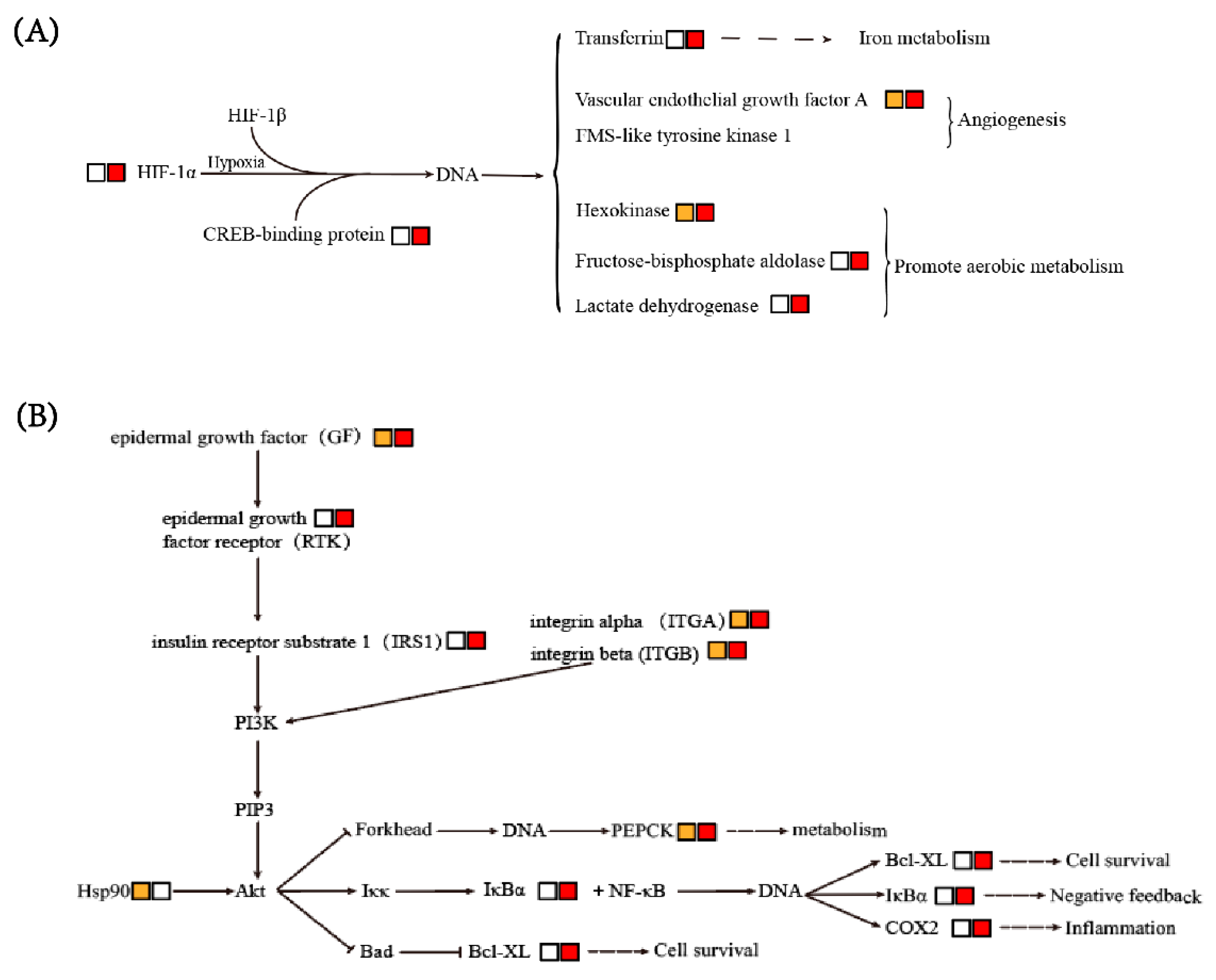
3.6. Enriched Signaling Pathways
3.6.1. HIF-1 Signaling Pathway
3.6.2. PI3K-Akt and NF-κB Signaling Pathways
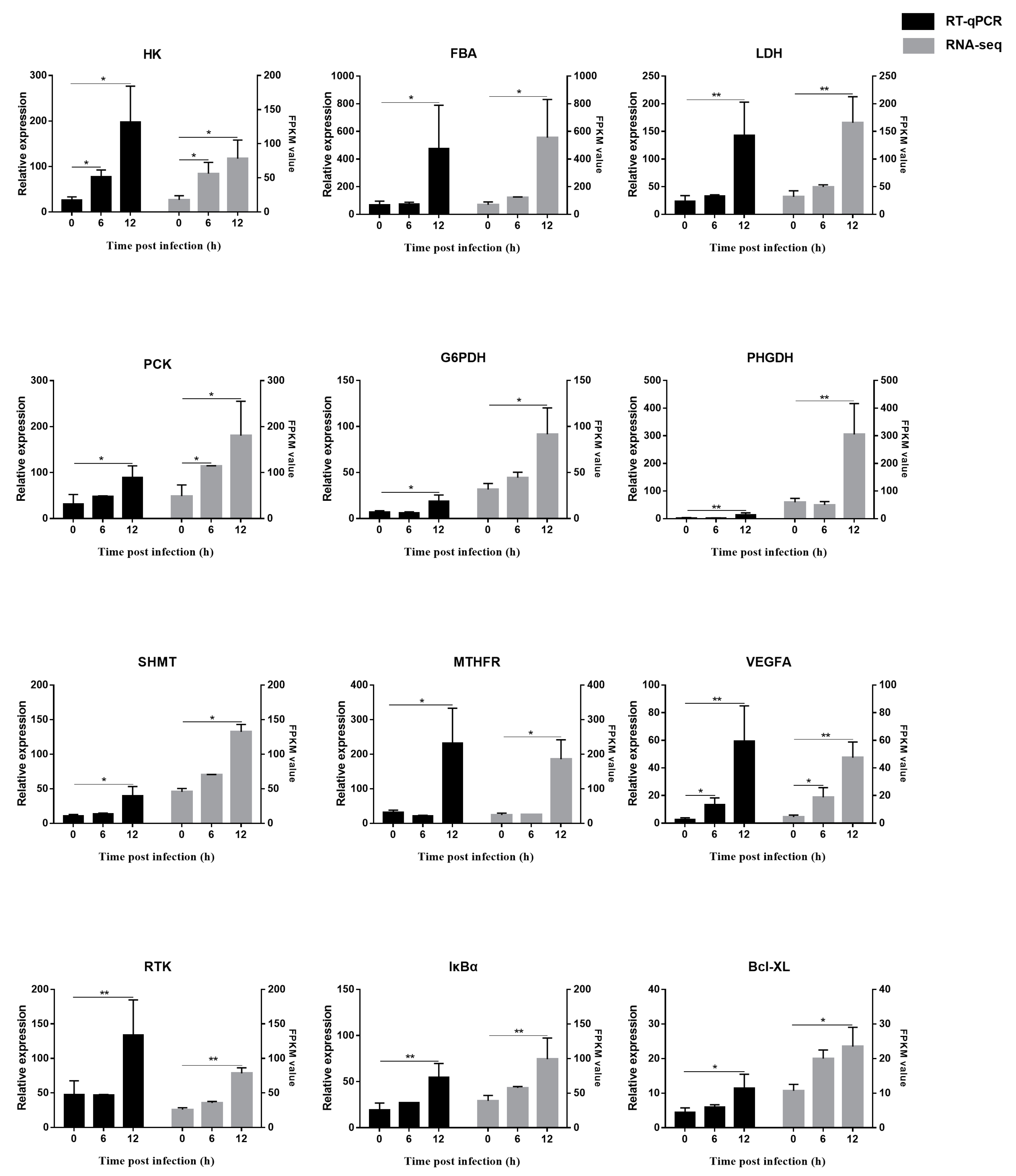
3.7. Verification of RNA-seq Results by RT-qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Colombo, G.M.; dos Santos Simião, C.; Ramírez, J.R.B.; de Sousa Araujo, A.C.; Gomes, R.M.M.; Buitagro, S.A.M.; Wasielesky, W.; Monserrat, J.M. Bioflocs enriched with lyophilized açaí (Euterpe oleracea) improved the survival and weight gain of Litopenaeus vannamei post-larvae cultivated in the BFT system. Aquaculture 2023, 566, 739230. [Google Scholar] [CrossRef]
- Thitamadee, S.; Prachumwat, A.; Srisala, J.; Jaroenlak, P.; Salachan, P.V.; Sritunyalucksana, K.; Flegel, T.W.; Itsathitphaisarn, O. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 2016, 452, 69–87. [Google Scholar] [CrossRef]
- Flegel, T.W. Historic emergence, impact and current status of shrimp pathogens in Asia. J. Invertebr. Pathol. 2012, 110, 166–173. [Google Scholar] [CrossRef]
- Hong, X.; Lu, L.; Xu, D. Progress in research on acute hepatopancreatic necrosis disease (AHPND). Aquac. Int. 2015, 24, 577–593. [Google Scholar] [CrossRef]
- Lee, C.T.; Chen, I.T.; Yang, Y.T.; Ko, T.P.; Huang, Y.T.; Huang, J.Y.; Huang, M.F.; Lin, S.J.; Chen, C.Y.; Lin, S.S.; et al. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. USA 2015, 112, 10798–10803. [Google Scholar] [CrossRef]
- Liu, L.; Xiao, J.; Xia, X.; Pan, Y.; Yan, S.; Wang, Y. Draft Genome Sequence of Vibrio owensii Strain SH-14, Which Causes Shrimp Acute Hepatopancreatic Necrosis Disease. Genome Announc. 2015, 3, e01395-15. [Google Scholar] [CrossRef]
- Kondo, H.; Van, P.T.; Dang, L.T.; Hirono, I. Draft Genome Sequence of Non-Vibrio parahaemolyticus Acute Hepatopancreatic Necrosis Disease Strain KC13.17.5, Isolated from Diseased Shrimp in Vietnam. Genome Announc. 2015, 3, e00978-15. [Google Scholar] [CrossRef]
- Dong, X.; Wang, H.; Xie, G.; Zou, P.; Guo, C.; Liang, Y.; Huang, J. An isolate of Vibrio campbellii carrying the pir(VP) gene causes acute hepatopancreatic necrosis disease. Emerg. Microbes Infect. 2017, 6, e2. [Google Scholar] [CrossRef]
- Restrepo, L.; Bayot, B.; Arciniegas, S.; Bajana, L.; Betancourt, I.; Panchana, F.; Reyes Munoz, A. PirVP genes causing AHPND identified in a new Vibrio species (Vibrio punensis) within the commensal Orientalis clade. Sci. Rep. 2018, 8, 13080. [Google Scholar] [CrossRef]
- Elgendy, M.; Soliman, W.S.; Hassan, H.A.; Kenawy, A.M.; Liala, A.M. Effect of Abrupt Environmental Deterioration on the Eruption of Vibriosis in Mari-Cultured Shrimp, Penaeus indicus, in Egypt. J. Fish. Aquat. Sci. 2005, 10, 146–158. [Google Scholar] [CrossRef]
- Zorriehzahra, M.J. Early Mortality Syndrome (EMS) as new Emerging Threat in Shrimp Industry. Adv. Anim. Vet. Sci. 2015, 3, 64–72. [Google Scholar] [CrossRef]
- Tran, L.; Nunan, L.; Redman, R.M.; Mohney, L.L.; Pantoja, C.R.; Fitzsimmons, K.; Lightner, D.V. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis. Aquat. Organ. 2013, 105, 45–55. [Google Scholar] [CrossRef]
- Roszer, T. The invertebrate midintestinal gland (“hepatopancreas”) is an evolutionary forerunner in the integration of immunity and metabolism. Cell Tissue Res. 2014, 358, 685–695. [Google Scholar] [CrossRef]
- Alday-Sanz, V.; Roque, A.; Turnbull, J.F. Clearing mechanisms of Vibrio vulnificus biotype I in the black tiger shrimp Penaeus monodon. Dis. Aquat. Organ. 2002, 48, 91–99. [Google Scholar] [CrossRef]
- Soo, T.C.C.; Devadas, S.; Mohamed Din, M.S.; Bhassu, S. Differential transcriptome analysis of the disease tolerant Madagascar-Malaysia crossbred black tiger shrimp, Penaeus monodon hepatopancreas in response to acute hepatopancreatic necrosis disease (AHPND) infection: Inference on immune gene response and interaction. Gut Pathog. 2019, 11, 39. [Google Scholar] [CrossRef]
- Qin, Z.; Babu, V.S.; Wan, Q.; Zhou, M.; Liang, R.; Muhammad, A.; Zhao, L.; Li, J.; Lan, J.; Lin, L. Transcriptome analysis of Pacific white shrimp (Litopenaeus vannamei) challenged by Vibrio parahaemolyticus reveals unique immune-related genes. Fish Shellfish Immunol. 2018, 77, 164–174. [Google Scholar] [CrossRef]
- Su, M.A.; Huang, Y.T.; Chen, I.T.; Lee, D.Y.; Hsieh, Y.C.; Li, C.Y.; Ng, T.H.; Liang, S.Y.; Lin, S.Y.; Huang, S.W.; et al. An invertebrate Warburg effect: A shrimp virus achieves successful replication by altering the host metabolome via the PI3K-Akt-mTOR pathway. PLoS Pathog. 2014, 10, e1004196. [Google Scholar] [CrossRef]
- Kumar, R.; Tung, T.C.; Ng, T.H.; Chang, C.C.; Chen, Y.L.; Chen, Y.M.; Lin, S.S.; Wang, H.C. Metabolic Alterations in Shrimp Stomach During Acute Hepatopancreatic Necrosis Disease and Effects of Taurocholate on Vibrio parahaemolyticus. Front. Microbiol. 2021, 12, 631468. [Google Scholar] [CrossRef]
- Yu, D.; Zhai, Y.; He, P.; Jia, R. Comprehensive Transcriptomic and Metabolomic Analysis of the Litopenaeus vannamei Hepatopancreas After WSSV Challenge. Front. Immunol. 2022, 13, 826794. [Google Scholar] [CrossRef]
- Liu, X.; Ji, C.; Zhao, J.; Wu, H. Differential metabolic responses of clam Ruditapes philippinarum to Vibrio anguillarum and Vibrio splendidus challenges. Fish Shellfish Immunol. 2013, 35, 2001–2007. [Google Scholar] [CrossRef]
- Liu, F.; Li, S.; Yu, Y.; Yuan, J.; Li, F. Pathogenicity of a Vibrio owensii strain isolated from Fenneropenaeus chinensis carrying pirAB genes and causing AHPND. Aquaculture 2020, 530, 735747. [Google Scholar] [CrossRef]
- Zhang, Q.; Yu, Y.; Wang, Q.; Liu, F.; Luo, Z.; Zhang, C.; Zhang, X.; Huang, H.; Xiang, J.; Li, F. Identification of Single Nucleotide Polymorphisms Related to the Resistance Against Acute Hepatopancreatic Necrosis Disease in the Pacific White Shrimp Litopenaeus vannamei by Target Sequencing Approach. Front. Genet. 2019, 10, 700. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, J.; Sun, Y.; Li, S.; Gao, Y.; Yu, Y.; Liu, C.; Wang, Q.; Lv, X.; Zhang, X.; et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Hong, S.S.; Lee, H.; Kim, K.W.J.C.R.; Association, T.O.J.o.K.C. HIF-1α: A Valid Therapeutic Target for Tumor Therapy. Cancer Res. Treat. 2004, 36, 343. [Google Scholar] [CrossRef]
- Velazquez-Lizarraga, A.E.; Juarez-Morales, J.L.; Racotta, I.S.; Villarreal-Colmenares, H.; Valdes-Lopez, O.; Luna-Gonzalez, A.; Rodriguez-Jaramillo, C.; Estrada, N.; Ascencio, F. Transcriptomic analysis of Pacific white shrimp (Litopenaeus vannamei, Boone 1931) in response to acute hepatopancreatic necrosis disease caused by Vibrio parahaemolyticus. PLoS ONE 2019, 14, e0220993. [Google Scholar] [CrossRef]
- Maralit, B.A.; Jaree, P.; Boonchuen, P.; Tassanakajon, A.; Somboonwiwat, K. Differentially expressed genes in hemocytes of Litopenaeus vannamei challenged with Vibrio parahaemolyticus AHPND (VPAHPND) and VPAHPND toxin. Fish Shellfish Immunol. 2018, 81, 284–296. [Google Scholar] [CrossRef]
- Ren, X.; Liu, P.; Li, J. Comparative transcriptomic analysis of Marsupenaeus japonicus hepatopancreas in response to Vibrio parahaemolyticus and white spot syndrome virus. Fish Shellfish Immunol. 2019, 87, 755–764. [Google Scholar] [CrossRef]
- Mai, H.N.; Caro, L.F.A.; Cruz-Flores, R.; White, B.N.; Dhar, A.K. Differentially Expressed Genes in Hepatopancreas of Acute Hepatopancreatic Necrosis Disease Tolerant and Susceptible Shrimp (Penaeus vannamei). Front. Immunol. 2021, 12, 634152. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, B.; Liu, M.; Jiang, K.; Wang, M.; Wang, L. Comparative transcriptome analysis reveals the potential influencing mechanism of dietary astaxanthin on growth and metabolism in Litopenaeus vannamei. Aquac. Rep. 2020, 16, 100259. [Google Scholar] [CrossRef]
- Sreedhar, A.; Zhao, Y. Dysregulated metabolic enzymes and metabolic reprogramming in cancer cells. Biomed. Rep. 2018, 8, 3–10. [Google Scholar] [CrossRef]
- Soto-Heredero, G.; Gomez de Las Heras, M.M.; Gabande-Rodriguez, E.; Oller, J.; Mittelbrunn, M. Glycolysis—A key player in the inflammatory response. FEBS J. 2020, 287, 3350–3369. [Google Scholar] [CrossRef]
- Warburg, O.A. On the Origin of Cancer Cell. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. Correction to: ‘The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 287. [Google Scholar] [CrossRef]
- Delgado, T.; Carroll, P.A.; Punjabi, A.S.; Margineantu, D.; Hockenbery, D.M.; Lagunoff, M. Induction of the Warburg effect by Kaposi’s sarcoma herpesvirus is required for the maintenance of latently infected endothelial cells. Proc. Natl. Acad. Sci. USA 2010, 107, 10696–10701. [Google Scholar] [CrossRef]
- Chen, I.T.; Aoki, T.; Huang, Y.T.; Hirono, I.; Chen, T.C.; Huang, J.Y.; Chang, G.D.; Lo, C.F.; Wang, H.C. White spot syndrome virus induces metabolic changes resembling the warburg effect in shrimp hemocytes in the early stage of infection. J. Virol. 2011, 85, 12919–12928. [Google Scholar] [CrossRef]
- Karniely, S.; Weekes, M.P.; Antrobus, R.; Rorbach, J.; van Haute, L.; Umrania, Y.; Smith, D.L.; Stanton, R.J.; Minczuk, M.; Lehner, P.J.; et al. Human Cytomegalovirus Infection Upregulates the Mitochondrial Transcription and Translation Machineries. mBio 2016, 7, e00029. [Google Scholar] [CrossRef]
- Kalezic, A.; Udicki, M.; Srdic Galic, B.; Aleksic, M.; Korac, A.; Jankovic, A.; Korac, B. Tissue-Specific Warburg Effect in Breast Cancer and Cancer-Associated Adipose Tissue-Relationship between AMPK and Glycolysis. Cancers 2021, 13, 2731. [Google Scholar] [CrossRef]
- Shim, H.; Dolde, C.; Lewis, B.C.; Wu, C.S.; Dang, G.; Jungmann, R.A.; Dalla-Favera, R.; Dang, C.V. c-Myc transactivation of LDH-A: Implications for tumor metabolismandgrowth. Proc. Natl. Acad. Sci. USA 1997, 94, 6658–6663. [Google Scholar] [CrossRef]
- Pedersen, B. High aerobic glycolysis of rat hepatoma cells in culture: Role of mitochondrial hexokinase. Proc. Natl. Acad. Sci. USA 1977, 74, 3735–3739. [Google Scholar]
- Rempel, A.; Mathupala, S.P.; Griffin, C.A.; Hawkins, A.L.; Pedersen, P.L. Glucose catabolism in cancer cells: Amplification of the gene encoding type II hexokinase. Cancer Res. 1996, 56, 2468–2471. [Google Scholar]
- Miao, P.; Sheng, S.; Sun, X.; Liu, J.; Huang, G. Lactate dehydrogenase A in cancer: A promising target for diagnosis and therapy. IUBMB Life 2013, 65, 904–910. [Google Scholar] [CrossRef]
- Valvona, C.J.; Fillmore, H.L.; Nunn, P.B.; Pilkington, G.J. The Regulation and Function of Lactate Dehydrogenase A: Therapeutic Potential in Brain Tumor. Brain Pathol. 2016, 26, 3–17. [Google Scholar] [CrossRef]
- Lai, H.C.; Ng, T.H.; Ando, M.; Lee, C.T.; Chen, I.T.; Chuang, J.C.; Mavichak, R.; Chang, S.H.; Yeh, M.D.; Chiang, Y.A.; et al. Pathogenesis of acute hepatopancreatic necrosis disease (AHPND) in shrimp. Fish Shellfish. Immunol. 2015, 47, 1006–1014. [Google Scholar] [CrossRef]
- Soonthornchai, W.; Chaiyapechara, S.; Jarayabhand, P.; Soderhall, K.; Jiravanichpaisal, P. Interaction of Vibrio spp. with the Inner Surface of the Digestive Tract of Penaeus monodon. PLoS ONE 2015, 10, e0135783. [Google Scholar] [CrossRef]
- O’Neill, L.A.; Kishton, R.J.; Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016, 16, 553–565. [Google Scholar] [CrossRef]
- Ramos-Montoya, A.; Lee, W.N.; Bassilian, S.; Lim, S.; Trebukhina, R.V.; Kazhyna, M.V.; Ciudad, C.J.; Noe, V.; Centelles, J.J.; Cascante, M. Pentose phosphate cycle oxidative and nonoxidative balance: A new vulnerable target for overcoming drug resistance in cancer. Int. J. Cancer 2006, 119, 2733–2741. [Google Scholar] [CrossRef]
- Puleston, D.J.; Villa, M.; Pearce, E.L. Ancillary Activity: Beyond Core Metabolism in Immune Cells. Cell Metab. 2017, 26, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Inada, M.; Sudhakaran, R.; Kihara, K.; Nishi, J.; Yoshimine, M.; Mekata, T.; Kono, T.; Sakai, M.; Yoshida, T.; Itami, T. Molecular cloning and characterization of the NADPH oxidase from the kuruma shrimp, Marsupenaeus japonicus: Early gene up-regulation after Vibrio penaeicida and poly(I:C) stimulations in vitro. Mol. Cell. Probes 2012, 26, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Anduro, G.A.; Barillas-Mury, C.V.; Peregrino-Uriarte, A.B.; Gupta, L.; Gollas-Galvan, T.; Hernandez-Lopez, J.; Yepiz-Plascencia, G. The cytosolic manganese superoxide dismutase from the shrimp Litopenaeus vannamei: Molecular cloning and expression. Dev. Comp. Immunol. 2006, 30, 893–900. [Google Scholar] [CrossRef]
- Muñoz, M.; Cedeño, R.; Rodrı, J.; van der Knaap, W.P.; Mialhe, E.; Bachère, E. Measurement of reactive oxygen intermediate production in haemocytes of the penaeid shrimp, Penaeus vannamei. Aquaculture 2000, 191, 89–107. [Google Scholar] [CrossRef]
- Newman, A.C.; Maddocks, O.D.K. Serine and Functional Metabolites in Cancer. Trends Cell Biol. 2017, 27, 645–657. [Google Scholar] [CrossRef]
- Kikuchi, G.; Motokawa, Y.; Yoshida, T.; Hiraga, K. Glycine cleavage system: Reaction mechanism, physiological significance, and hyperglycinemia. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2008, 84, 246–263. [Google Scholar] [CrossRef]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, M.; Wang, L.; Liu, M.; Jiang, K.; Xia, S.; Qi, C.; Wang, B. Analysis of the expression of metabolism-related genes and histopathology of the hepatopancreas of white shrimp Litopenaeus vannamei fed with aflatoxin B1. Aquaculture 2018, 485, 191–196. [Google Scholar] [CrossRef]
- Pan, S.; Fan, M.; Liu, Z.; Li, X.; Wang, H. Serine, glycine and onecarbon metabolism in cancer (Review). Int. J. Oncol. 2021, 58, 158–170. [Google Scholar] [CrossRef]
- Soñanez-Organis, J.G.; Racotta, I.S.; Yepiz-Plascencia, G. Silencing of the hypoxia inducible factor 1 –HIF-1- obliterates the effects of hypoxia on glucose and lactate concentrations in a tissue-specific manner in the shrimp Litopenaeus vannamei. J. Exp. Mar. Biol. Ecol. 2010, 393, 51–58. [Google Scholar] [CrossRef]
- Semenza, G.L.J.G.D. HIF-1 and human disease: One highly involved factor. Genes Dev. 2000, 14, 1983–1991. [Google Scholar] [CrossRef]
- Kilic-Eren, M.; Boylu, T.; Tabor, V. Targeting PI3K/Akt represses Hypoxia inducible factor-1alpha activation and sensitizes Rhabdomyosarcoma and Ewing’s sarcoma cells for apoptosis. Cancer Cell Int. 2013, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Katso, R.; Okkenhaug, K.; Ahmadi, K.; White, S.; Timms, J.; Biol, M.W. Cellular function of phosphoinositide 3-kinases: Implications for development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 2001, 17, 615–675. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, X.; Yuan, Z.; Wang, H. Metabolic Reprogramming in Immune Response and Tissue Inflammation. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 1990–2001. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, M.; Li, S.; Liu, Y.; Yu, Y.; Li, F. Transcriptome Analysis on Hepatopancreas Reveals the Metabolic Dysregulation Caused by Vibrio parahaemolyticus Infection in Litopenaeus vannamei. Biology 2023, 12, 417. https://doi.org/10.3390/biology12030417
Miao M, Li S, Liu Y, Yu Y, Li F. Transcriptome Analysis on Hepatopancreas Reveals the Metabolic Dysregulation Caused by Vibrio parahaemolyticus Infection in Litopenaeus vannamei. Biology. 2023; 12(3):417. https://doi.org/10.3390/biology12030417
Chicago/Turabian StyleMiao, Miao, Shihao Li, Yuan Liu, Yang Yu, and Fuhua Li. 2023. "Transcriptome Analysis on Hepatopancreas Reveals the Metabolic Dysregulation Caused by Vibrio parahaemolyticus Infection in Litopenaeus vannamei" Biology 12, no. 3: 417. https://doi.org/10.3390/biology12030417
APA StyleMiao, M., Li, S., Liu, Y., Yu, Y., & Li, F. (2023). Transcriptome Analysis on Hepatopancreas Reveals the Metabolic Dysregulation Caused by Vibrio parahaemolyticus Infection in Litopenaeus vannamei. Biology, 12(3), 417. https://doi.org/10.3390/biology12030417




