Profiling the Hsp70 Chaperone Network in Heat-Induced Proteotoxic Stress Models of Human Neurons
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Differentiation
2.2. Induction of Heat Stress
2.3. Protein Aggregation Assay
2.4. Cell Proliferation Assays
2.5. RNA Isolation and cDNA Synthesis
2.6. Gene Arrays and RT-PCR
2.7. Protein Extraction and Concentration Determination
2.8. Immunoblotting
2.9. Expression Analysis
2.10. Expression Kinetics Analysis
2.11. Co-Expression Analysis
3. Results
3.1. Heat-Induced Proteotoxic Stress
3.2. Dose Responsive Heat-Stress Models
3.3. Chaperone Gene Expression during Heat-Shock Response
3.4. Temporal Analysis of Gene Expression
3.5. Protein Expression Profiling of Hsp70 Members
3.6. Extreme and Mild Heat Stress-Response Comparison
3.6.1. Gene Expression Analysis
3.6.2. Protein Expression Analysis
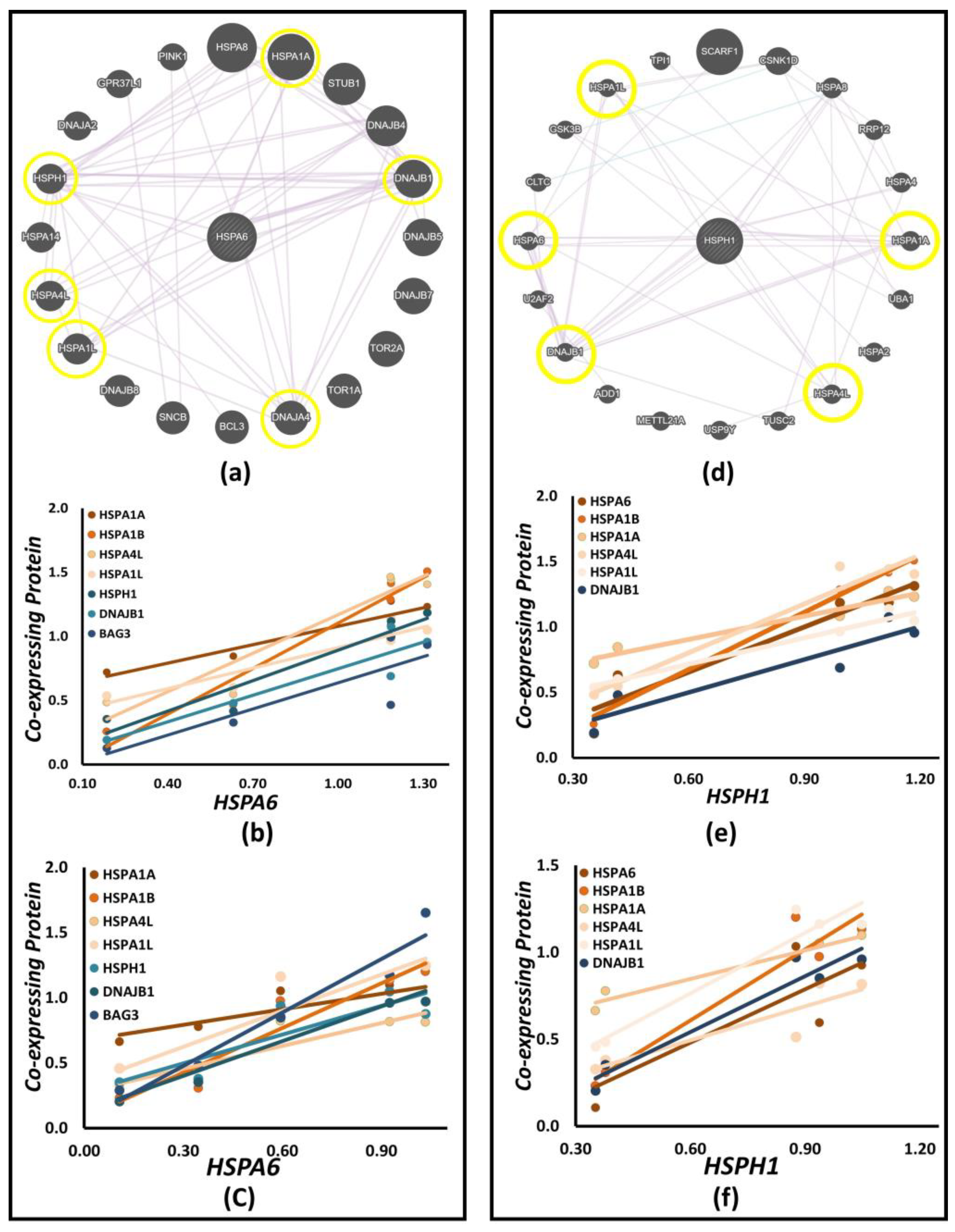
3.7. Expression Profiling of Hsp70 Co-Expressing Proteins
3.8. Co-Expression Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Watts, N.; Amann, M.; Arnell, N.; Ayeb-Karlsson, S.; Beagley, J.; Belesova, K.; Boykoff, M.; Byass, P.; Cai, W.; Campbell-Lendrum, D.; et al. The 2020 report of The Lancet Countdown on health and climate change: Responding to converging crises. Lancet 2021, 397, 129–170. [Google Scholar] [CrossRef]
- Sorensen, C.; Hess, J. Treatment and Prevention of Heat-Related Illness. N. Engl. J. Med. 2022, 387, 1404–1413. [Google Scholar] [CrossRef]
- Yoram Epstein, R.Y. Heatstroke. N. Engl. J. Med. 2019, 380, 2449–2459. [Google Scholar] [CrossRef]
- Bouchama, A.; Knochel, J.P. HEAT STROKE. N. Engl. J. Med. 2002, 346, 1978–1988. [Google Scholar] [CrossRef]
- Walter, E.J.; Carraretto, M. The neurological and cognitive consequences of hyperthermia. Crit. Care 2016, 20, 199. [Google Scholar] [CrossRef]
- Richter, K.; Haslbeck, M.; Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef]
- Lindberg, I.; Shorter, J.; Wiseman, R.L.; Chiti, F.; Dickey, C.A.; McLean, P.J. Chaperones in neurodegeneration. J. Neurosci. 2015, 35, 13853–13859. [Google Scholar] [CrossRef]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Ulrich Hartl, F. Molecular Chaperone Functions in Protein Folding and Proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- Hipp, M.S.; Kasturi, P.; Hartl, F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019, 20, 421–435. [Google Scholar] [CrossRef]
- Waudby, C.A.; Dobson, C.M.; Christodoulou, J. Nature and Regulation of Protein Folding on the Ribosome. Trends Biochem. Sci. 2019, 44, 914–926. [Google Scholar] [CrossRef]
- Saibil, H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef]
- Seo, J.H.; Park, J.H.; Lee, E.J.; Vo, T.T.L.; Choi, H.; Kim, J.Y.; Jang, J.K.; Wee, H.J.; Lee, H.S.; Jang, S.H.; et al. ARD1-mediated Hsp70 acetylation balances stress-induced protein refolding and degradation. Nat. Commun. 2016, 7, 12882. [Google Scholar] [CrossRef]
- White, M.G.; Luca, L.E.; Nonner, D.; Saleh, O.; Hu, B.; Barrett, E.F.; Barrett, J.N. Cellular mechanisms of neuronal damage from hyperthermia. Prog. Brain Res. 2007, 162, 347–371. [Google Scholar] [CrossRef]
- Yaqub, B.A. Neurologic manifestations of heatstroke at the Mecca pilgrimage. Neurology 1987, 37, 1004. [Google Scholar] [CrossRef]
- Jinwal, U.K.; Miyata, Y.; Iii, J.K.; Jones, J.R.; Trotter, J.H.; Chang, L.; Leary, J.O.; Morgan, D.; Lee, D.C.; Shults, C.L.; et al. Chemical Manipulation of Hsp70 ATPase Activity Regulates Tau Stability. Neurobiol. Dis. 2009, 29, 12079–12088. [Google Scholar] [CrossRef]
- Davis, A.K.; Pratt, W.B.; Lieberman, A.P.; Osawa, Y.; Arbor, A.; Arbor, A. Targeting Hsp70 facilitated protein quality control for treatment of polyglutamine diseases. Cell Mol. Life Sci. 2021, 77, 977–996. [Google Scholar] [CrossRef]
- Wang, A.M.; Miyata, Y.; Klinedinst, S.; Peng, H.; Jason, P.; Komiyama, T.; Li, X.; Morishima, Y.; Merry, D.E.; Pratt, W.B.; et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat. Chem. Biol. 2013, 9, 112–118. [Google Scholar] [CrossRef]
- Kovalevich, J.; Langford, D. Considerations for the Use of SH-SY5Y Neuroblastoma Cells in Neurobiology. Methods Mol. Biol. 2013, 1078, 9–21. [Google Scholar] [CrossRef]
- Deane, C.A.S.; Brown, I.R. Differential Targeting of Hsp70 Heat Shock Proteins HSPA6 and HSPA1A with Components of a Protein Disaggregation/Refolding Machine in Differentiated Human Neuronal Cells following Thermal Stress. Front. Neurosci. 2017, 11, 227. [Google Scholar] [CrossRef]
- Hamidi, H.; Lilja, J.; Ivaska, J. Using xCELLigence RTCA Instrument to Measure Cell Adhesion. Bio-Protoc. 2017, 7, e2646. [Google Scholar] [CrossRef]
- Spandidos, A.; Wang, X.; Wang, H.; Seed, B. PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2009, 38, 792–799. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, 214–220. [Google Scholar] [CrossRef]
- Mayer, M.P. Gymnastics of molecular chaperones. Mol. Cell 2010, 39, 321–331. [Google Scholar] [CrossRef]
- Schuermann, J.P.; Jiang, J.; Cuellar, J.; Llorca, O.; Wang, L.; Gimenez, L.E.; Jin, S.; Taylor, A.B.; Demeler, B.; Morano, K.A.; et al. Structure of the Hsp110:Hsc70 Nucleotide Exchange Machine. Mol. Cell 2008, 23, 232–243. [Google Scholar] [CrossRef]
- Saxena, A.; Banasavadi-Siddegowda, Y.K.; Fan, Y.; Bhattacharya, S.; Roy, G.; Giovannucci, D.R.; Frizzell, R.A.; Wang, X. Human heat shock protein 105/110 kDa (Hsp105/110) regulates biogenesis and quality control of misfolded cystic fibrosis transmembrane conductance regulator at multiple levels. J. Biol. Chem. 2012, 287, 19158–19170. [Google Scholar] [CrossRef]
- Wallace, E.W.J.; Kear-Scott, J.L.; Pilipenko, E.V.; Schwartz, M.H.; Laskowski, P.R.; Rojek, A.E.; Katanski, C.D.; Riback, J.A.; Dion, M.F.; Franks, A.M.; et al. Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell 2015, 162, 1286–1298. [Google Scholar] [CrossRef]
- Niforou, K.; Cheimonidou, C.; Trougakos, I.P. Molecular chaperones and proteostasis regulation during redox imbalance. Redox Biol. 2014, 2, 323–332. [Google Scholar] [CrossRef]
- Lamech, L.T.; Haynes, C.M. The unpredictability of prolonged activation of stress response pathways. J. Cell Biol. 2015, 209, 781–787. [Google Scholar] [CrossRef]
- Sørensen, J.G.; Nielsen, M.M.; Kruhøffer, M.; Justesen, J.; Loeschcke, V. Full genome gene expression analysis of the heat stress response in Drosophila melanogaster. Cell Stress Chaperones 2005, 10, 312–328. [Google Scholar] [CrossRef]
- Vihervaara, A.; Duarte, F.M.; Lis, J.T. Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 2018, 19, 385–397. [Google Scholar] [CrossRef]
- Mahat, D.B.; Salamanca, H.H.; Duarte, F.M.; Danko, C.G.; Lis, J.T. Mammalian Heat Shock Response and Mechanisms Underlying Its Genome-wide Transcriptional Regulation. Mol. Cell 2016, 62, 63–78. [Google Scholar] [CrossRef]
- Zheng, X.; Krakowiak, J.; Patel, N.; Beyzavi, A.; Ezike, J.; Khalil, A.S.; Pincus, D. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. eLife 2016, 90, e18638. [Google Scholar] [CrossRef]
- Zou, J.; Guo, Y.; Guettouche, T.; Smith, D.F.; Voellmy, R. Repression of Heat Shock Transcription Factor HSF1 Activation by HSP90 (HSP90 Complex) that Forms a Stress-Sensitive Complex with HSF1. Cell 1998, 94, 471–480. [Google Scholar] [CrossRef]
- Akerfelt, M.; Richard, I.; Morimoto, L.S. Heat shock factors: Integrators of cell stress, development and lifespan. Nat Rev Drug Discov 2011, 11, 545–555. [Google Scholar] [CrossRef]
- Anckar, J.; Sistonen, L. Regulation of HSF1 Function in the Heat Stress Response: Implications in Aging and Disease. Annu. Rev. Biochem. 2011, 80, 1089–1115. [Google Scholar] [CrossRef]
- Gomez-pastor, R.; Burchfiel, E.T.; Thiele, D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 4–19. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef]
- Zander, G.; Hackmann, A.; Bender, L.; Becker, D.; Lingner, T.; Salinas, G.; Krebber, H. MRNA quality control is bypassed for immediate export of stress-responsive transcripts. Nature 2016, 540, 593–596. [Google Scholar] [CrossRef]
- Rodrigues, D.C.; Silva, R.; Rondinelli, E.; Ürményi, T.P. Trypanosoma cruzi: Modulation of HSP70 mRNA stability by untranslated regions during heat shock. Exp. Parasitol. 2010, 126, 245–253. [Google Scholar] [CrossRef]
- Jovic, K.; Sterken, M.G.; Grilli, J.; Bevers, R.P.J.; Rodriguez, M.; Riksen, J.A.G.; Allesina, S.; Kammenga, J.E.; Snoek, L.B. Temporal dynamics of gene expression in heat-stressed Caenorhabditis elegans. PLoS ONE 2017, 12, e0189445. [Google Scholar] [CrossRef]
- Bouchama, A.; Aziz, M.A.; Mahri, S.A.L.; Gabere, M.N.; Dlamy, M.A.L.; Mohammad, S.; Abbad, M.A.L.; Hussein, M. A Model of Exposure to Extreme Environmental Heat Uncovers the Human Transcriptome to Heat Stress. Sci. Rep. 2017, 7, 9429. [Google Scholar] [CrossRef]
- Radons, J. The human HSP70 family of chaperones: Where do we stand? Cell Stress Chaperones 2016, 21, 379–404. [Google Scholar] [CrossRef]
- Maugeri, N.; Radhakrishnan, J.; Knight, J.C. Genetic determinants of HSP70 gene expression following heat shock. Hum. Mol. Genet. 2010, 19, 4939–4947. [Google Scholar] [CrossRef]
- Lee, J.H.; Han, Y.; Yoon, Y.M.; Yun, C.W.; Yun, S.P.; Kim, S.M.; Kwon, H.Y.; Jeong, D.; Baek, M.J.; Lee, H.J.; et al. Role of HSPA1L as a cellular prion protein stabilizer in tumor progression via HIF-1 α/GP78 axis. Oncogene 2017, 36, 6555–6567. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallstro, M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the Human Tissue-specific Expression by Genome-wide Integration of Transcriptomics and Antibody-based. Mol. Cell Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Deane, C.A.S.; Brown, I.R. Knockdown of Heat Shock Proteins HSPA6 (Hsp70B’) and HSPA1A (Hsp70-1) Sensitizes Differentiated Human Neuronal Cells to Cellular Stress. Neurochem. Res. 2018, 43, 340–350. [Google Scholar] [CrossRef]
- Zarouchlioti, C.; Parfitt, D.A.; Li, W.; Gittings, L.M.; Cheetham, M.E. DNAJ Proteins in neurodegeneration: Essential and protective factors. Philos. Trans. R. Soc. B Biol. Sci. 2017, 373, 20160534. [Google Scholar] [CrossRef]
- Rozales, K.; Younis, A.; Saida, N.; Meller, A.; Goldman, H.; Kellerman, L.; Heinrich, R.; Berlin, S.; Shalgi, R. Differential roles for DNAJ isoforms in HTT-polyQ and FUS aggregation modulation revealed by chaperone screens. Nat. Commun. 2022, 13, 516. [Google Scholar] [CrossRef]
- Park, S.; Arslan, F.; Kanneganti, V.; Barmada, S.J.; Purushothaman, P. Overexpression of a conserved HSP40 chaperone reduces toxicity of several neurodegenerative disease proteins. Prion 2018, 12, 16–22. [Google Scholar] [CrossRef]
- Stürner, E.; Behl, C. The role of the multifunctional bag3 protein in cellular protein quality control and in disease. Front. Mol. Neurosci. 2017, 10, 177. [Google Scholar] [CrossRef]
- Behl, C. Breaking BAG: The Co-Chaperone BAG3 in Health and Disease. Trends Pharmacol. Sci. 2016, 37, 672–688. [Google Scholar] [CrossRef]
- Rosati, A.; Graziano, V.; Laurenzi, V.D.; Pascale, M.; Turco, M.C. BAG3: A multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011, 2, e141. [Google Scholar] [CrossRef]
- Dragovic, Z.; Broadley, S.A.; Shomura, Y.; Bracher, A.; Hartl, F.U. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006, 25, 2519–2528. [Google Scholar] [CrossRef]
- Smith, H.L.; Li, W.; Cheetham, M.E. Molecular chaperones and neuronal proteostasis. Semin. Cell Dev. Biol. 2015, 40, 142–152. [Google Scholar] [CrossRef]
- Beretta, G.; Shala, A.L. Impact of Heat Shock Proteins in Neurodegeneration: Possible Therapeutical Targets. Ann. Neurosci. 2022, 29, 71–82. [Google Scholar] [CrossRef]
- Evans, C.G.; Wise, S.; Gestwicki, J.E. Heat Shock Proteins 70 and 90 Inhibit Early Stages of Amyloid β-(1–42) Aggregation In Vitro *. J. Biol. Chem. 2006, 281, 33182–33191. [Google Scholar] [CrossRef]



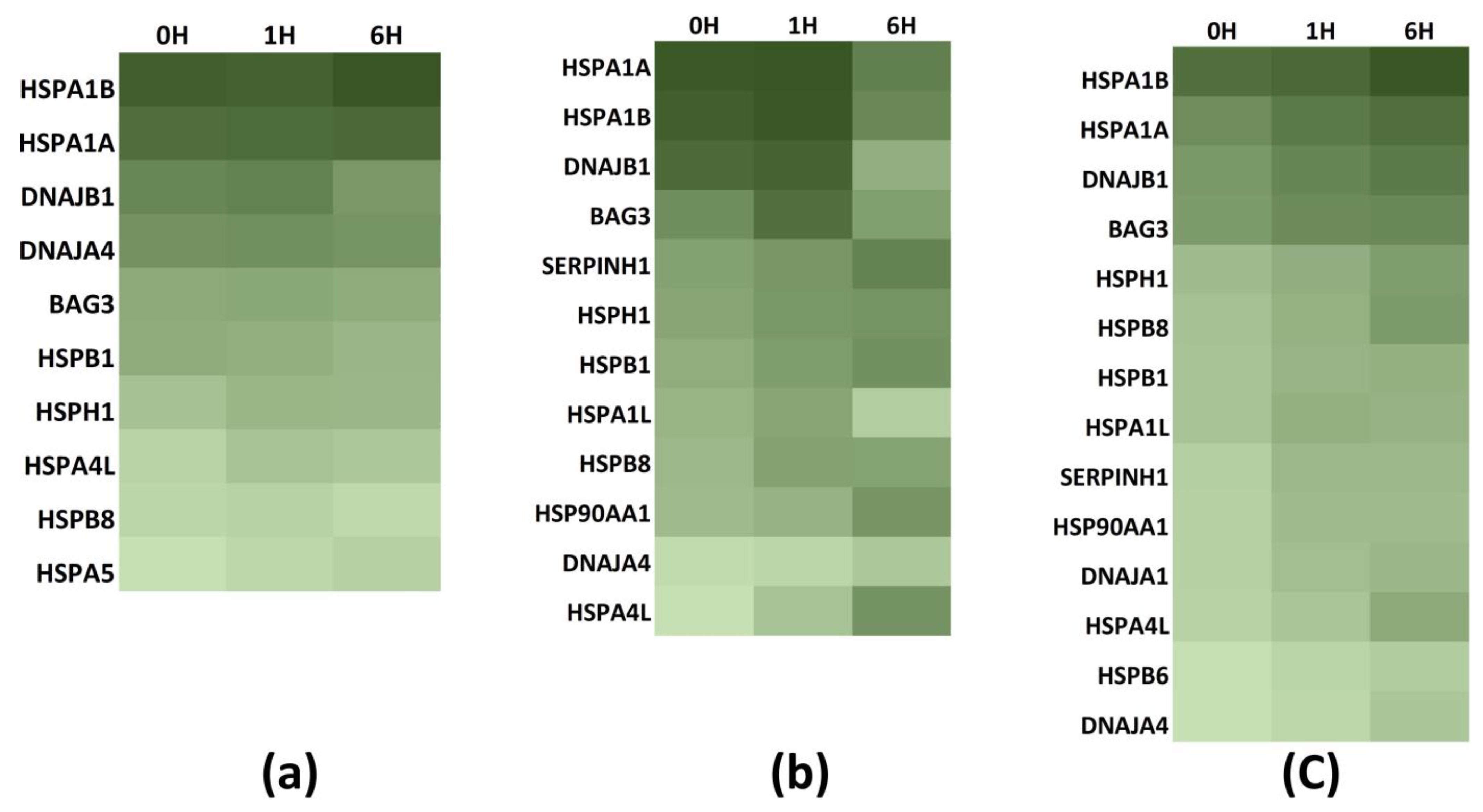


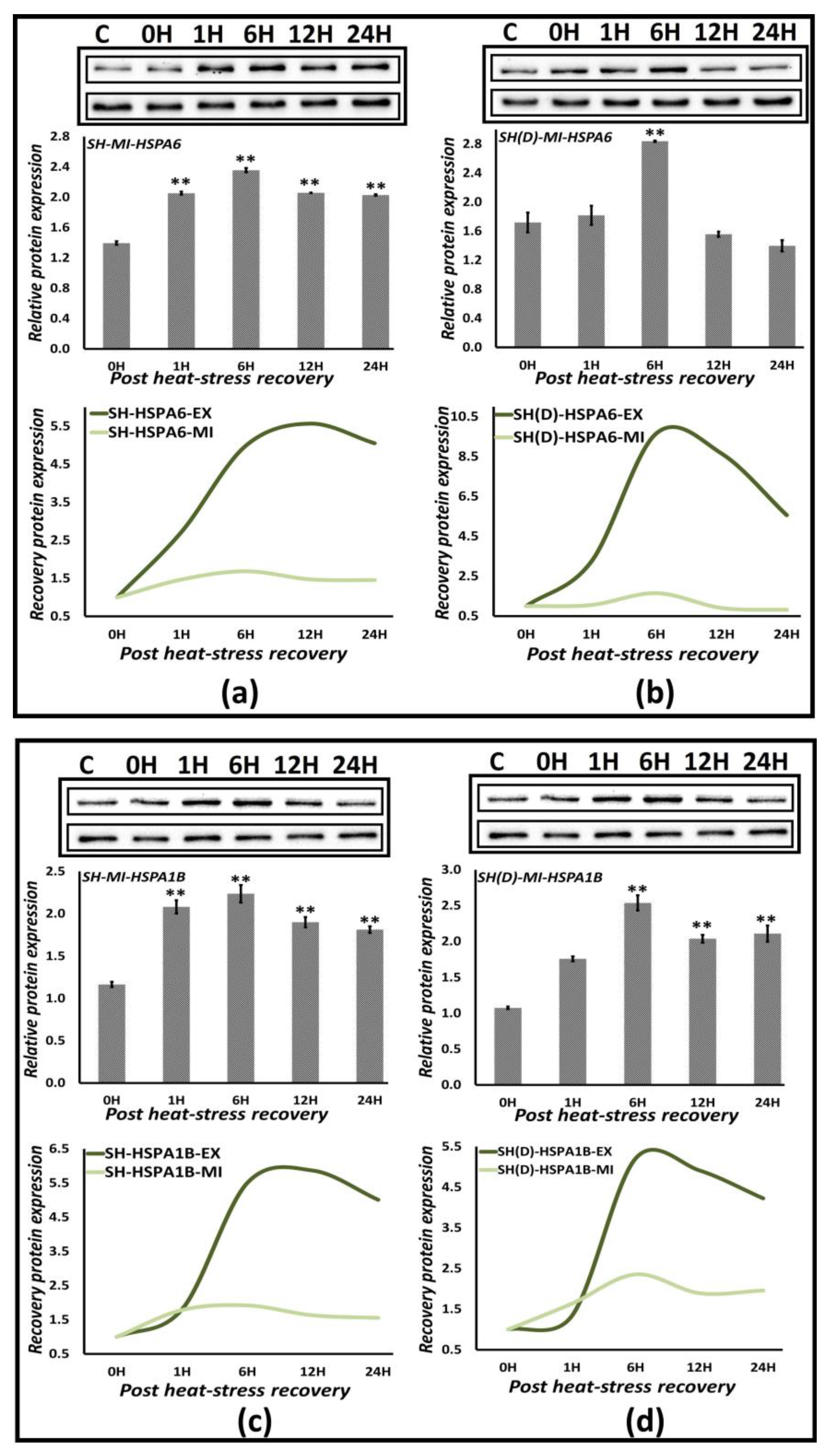


| Exposure Temperature [ET] | Exposure Duration [ED] | Hsp70 Induction |
|---|---|---|
| 39 °C | 1 h | 5.5 ± 0.7 |
| 2 h | 3.6 ± 1.1 | |
| 42 °C | 1 h | 25.1 ± 7.7 |
| 2 h | 96.6 ± 1.3 | |
| 44 °C | 1 h | 81.7 ± 7.4 |
| 2 h | 144.34 ± 4.38 |
| Chaperone Family | Overexpressed Genes | Proportion |
|---|---|---|
| Small Heat Shock Protein | HSPB1, HSPB8, CRYAB, HSPB6 | 4/8 |
| Heat shock 70 kDa protein | HSPA1A, HSPA1B, HSPA1L, HSPA4L, HSPA5, HSPA6 | 6/11 |
| Heat shock protein 90 kDa alpha (cytosolic), class A | HSP90AA1 | 1/1 |
| Heat shock protein 105 kDa | HSPH1 | 1/1 |
| DnaJ homolog subfamily A | DNAJA1, DNAJA4 | 2/4 |
| DnaJ homolog subfamily B | DNAJB1 | 1/11 |
| BAG family molecular chaperone regulator 3 | BAG3 | 1/1 |
| Serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) | SERPINH1 | 1/1 |
| Gene | Predicted Co-Expressing Interactors | Daoy | SH-SY5Y | SH-SY5Y(D) |
|---|---|---|---|---|
| HSPA1A | HSPB1, HSPA6, HSPA8, HSPH1, BAG3 | HSPA6, HSPH1, BAG3, HSPB1 | HSPA6, HSPH1, BAG3, HSPB1 | HSPA6, HSPH1, BAG3, HSPB1 |
| HSPA1B | HSPA1A, HSPH1, HSPA6, HSPA5, HYOU1 | HSPA1A, HSPA6, HSPH1 | HSPA1A, HSPA6, HSPH1 | HSPA1A, HSPA6, HSPH1 |
| HSPA6 | HSPA1A, DNAJB4, DNAJB1, DNDAJA4, DNAJB8, HSPA1L, HSPA4L, HSPH1 | HSPA1A, HSPA4L, HSPH1, DNAJB1, DNAJA4 | HSPA1A, HSPA1L, HSPA4L, HSPH1, DNAJB1, DNDAJA4 | HSPA1A, HSPA1L, HSPA4L, HSPH1, DNAJB1, DNDAJA4 |
| HSPA4L | HSPA8, RTN4, HSPH1, HSPA4, HSPA6, LUC7L2 | HSPA6, HSPH1 | HSPA6, HSPH1 | HSPA6, HSPH1 |
| HSPA1L | TOR2A, TPR1A, PRKN, SNCB, STUB1, HSPA6, HSPA4L, HSPH1 | HSPA6, HSPA4L, HSPH1 | HSPA6, HSPA4L, HSPH1 | HSPA6, HSPA4L, HSPH1 |
| HSPA4 | HSPH1, BAG3, HSPA4L | HSPA4L, HSPH1, BAG3 | HSPA4L, HSPH1, BAG3 | HSPA4L, HSPH1, BAG3 |
| BAG3 | HSPB8, BAG5, HSPA8, HSPA4, HSPA1A, SYNPO2, ETFA, HSP90AB1, GORASP2, CAPZA1, DNAJB4 | HSPA1A | HSPA1A, DNAJB4, HSPB8 | HSPA1A, DNAJB4, HSPB8 |
| DNAJA4 | HSPA6, HSPE, HSPA1A, DNAJB4, DNAJB1, HSPA8, DNAJB11 | HSPA1A, HSPA6, DNAJB1 | HSPA1A, HSPA6, DNAJB1 | HSPA1A, HSPA6, DNAJB1 |
| DNAJB1 | HSPA6, HSPA8, HSPH1, TARDBP, NUDC, DNAJB4, DNAJA1, HSPA1A | HSPA1A, HSPA6, HSPH1 | HSPA1A, HSPA6, HSPH1, DNAJA1 | HSPA1A, HSPA6, HSPH1, DNAJA1 |
| HSPH1 | HSPA8, HSPA4, HSPA1A, HSPA4L, DNAJB1, HSPA6, GSK3B, HSPA1L | HSPA1A, HSPA6, HSPA4L, DNAJB1 | HSPA1A, HSPA6, HSPA4L, HSPA1L, DNAJB1 | HSPA1A, HSPA6, HSPA4L, HSPA1L, DNAJB1 |
| DNAJA1 | TPP1, DNMA1, DNAJB1, HSPA9, HSPH1, HSPA6 | HSPA6, HSPH1, DNAJB1 | HSPA6, HSPH1, DNAJB1 | HSPA6, HSPH1, DNAJB1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alharbi, B.M.; Albinhassan, T.H.; Alzahrani, R.A.; Bouchama, A.; Mohammad, S.; Alomari, A.A.; Bin-Jumah, M.N.; AlSuhaibani, E.S.; Malik, S.S. Profiling the Hsp70 Chaperone Network in Heat-Induced Proteotoxic Stress Models of Human Neurons. Biology 2023, 12, 416. https://doi.org/10.3390/biology12030416
Alharbi BM, Albinhassan TH, Alzahrani RA, Bouchama A, Mohammad S, Alomari AA, Bin-Jumah MN, AlSuhaibani ES, Malik SS. Profiling the Hsp70 Chaperone Network in Heat-Induced Proteotoxic Stress Models of Human Neurons. Biology. 2023; 12(3):416. https://doi.org/10.3390/biology12030416
Chicago/Turabian StyleAlharbi, Bothina Mohammed, Tahani H. Albinhassan, Razan Ali Alzahrani, Abderrezak Bouchama, Sameer Mohammad, Awatif Abdulaziz Alomari, May Nasser Bin-Jumah, Entissar S. AlSuhaibani, and Shuja Shafi Malik. 2023. "Profiling the Hsp70 Chaperone Network in Heat-Induced Proteotoxic Stress Models of Human Neurons" Biology 12, no. 3: 416. https://doi.org/10.3390/biology12030416
APA StyleAlharbi, B. M., Albinhassan, T. H., Alzahrani, R. A., Bouchama, A., Mohammad, S., Alomari, A. A., Bin-Jumah, M. N., AlSuhaibani, E. S., & Malik, S. S. (2023). Profiling the Hsp70 Chaperone Network in Heat-Induced Proteotoxic Stress Models of Human Neurons. Biology, 12(3), 416. https://doi.org/10.3390/biology12030416






