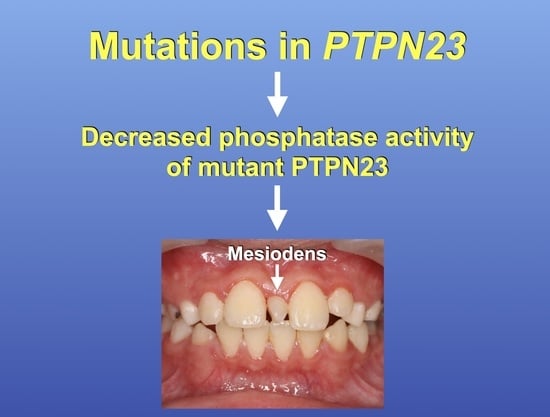
Genetic Variants in Protein Tyrosine Phosphatase Non-Receptor Type 23 Are Responsible for Mesiodens Formation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Examination, Sample Collection, and DNA Extraction
2.2. Whole Exome Sequencing and Sanger Direct Sequencing
2.3. Gene and Protein Expression
2.4. Computational Structural Analysis of Mutants
2.5. PTPN23 Mutant Protein Stability
2.6. PTPN23 Mutant Phosphatase Activity
3. Results
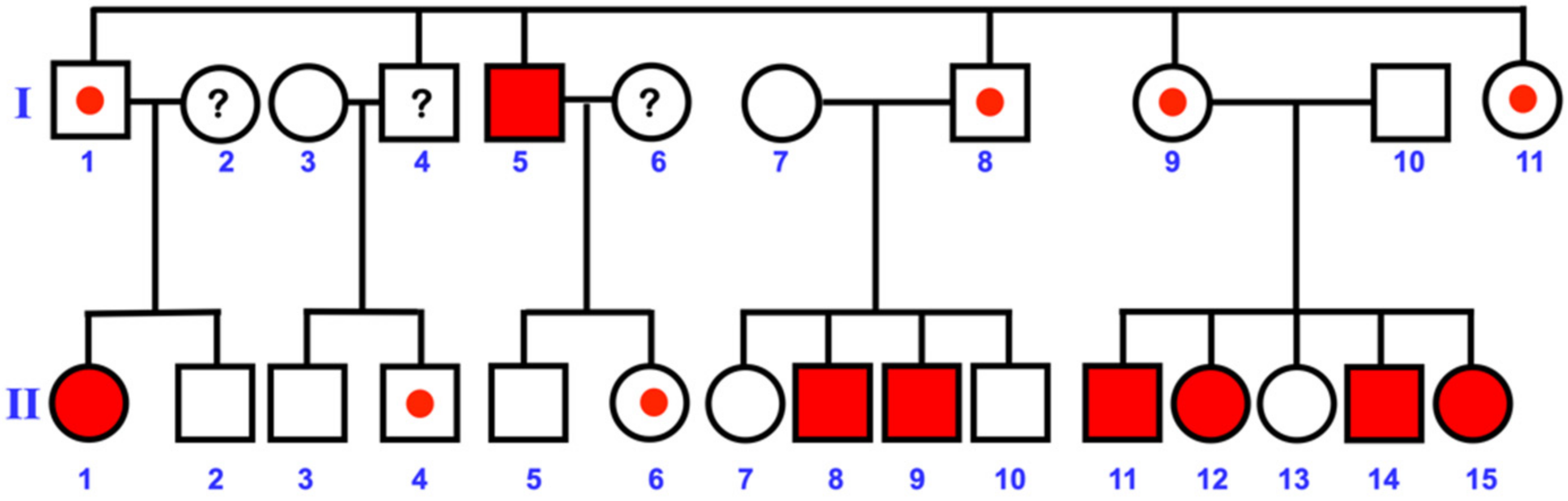
3.1. Varied Mesiodens Phenotypes within the Hmong Family (Family 1)
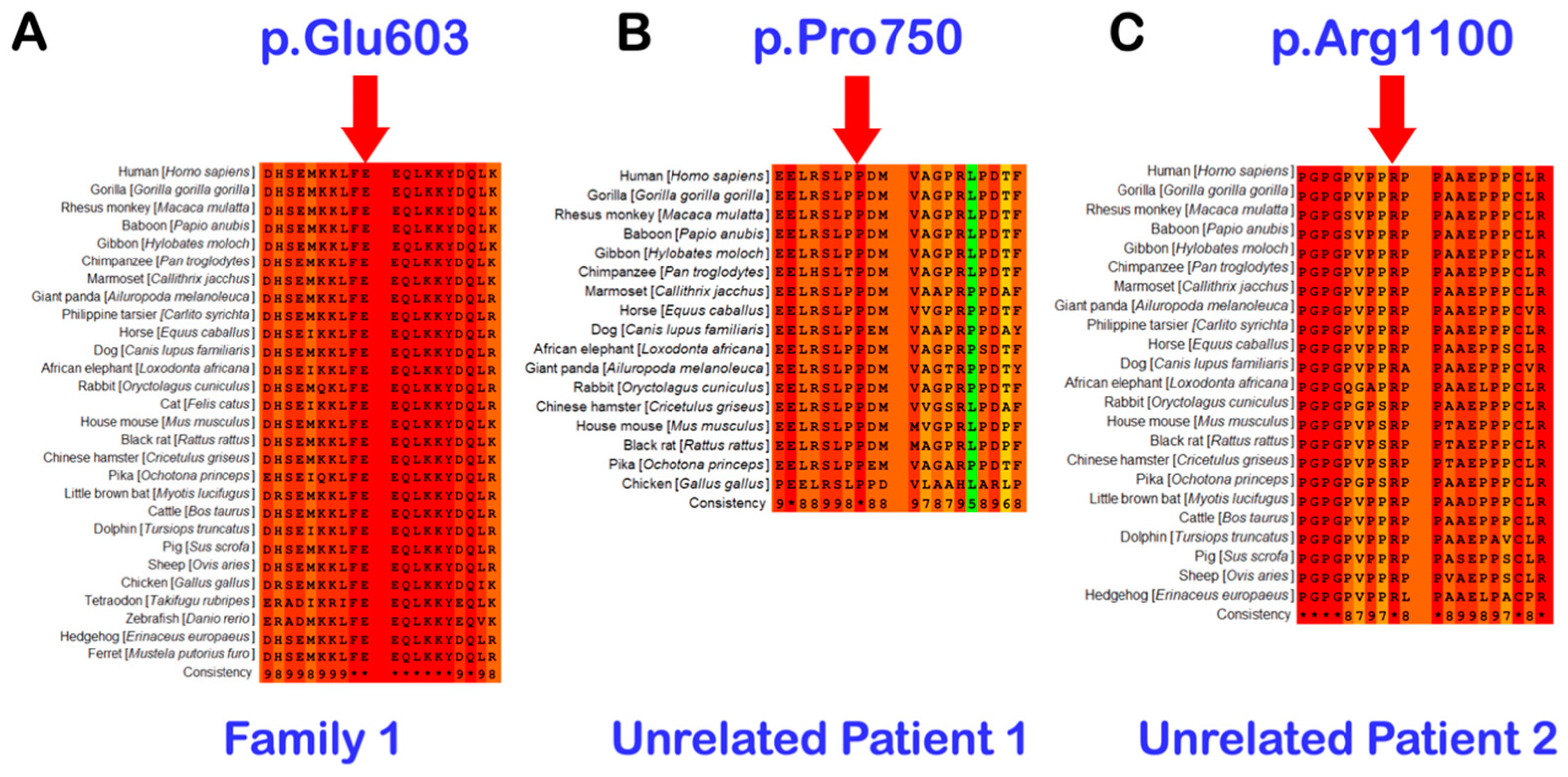
3.2. Whole Exome Sequencing, Sanger Direct Sequencing, and Bioinformatic Analysis
3.3. Unrelated Mesiodens Patients Identified with Rare PTPN23 Variants
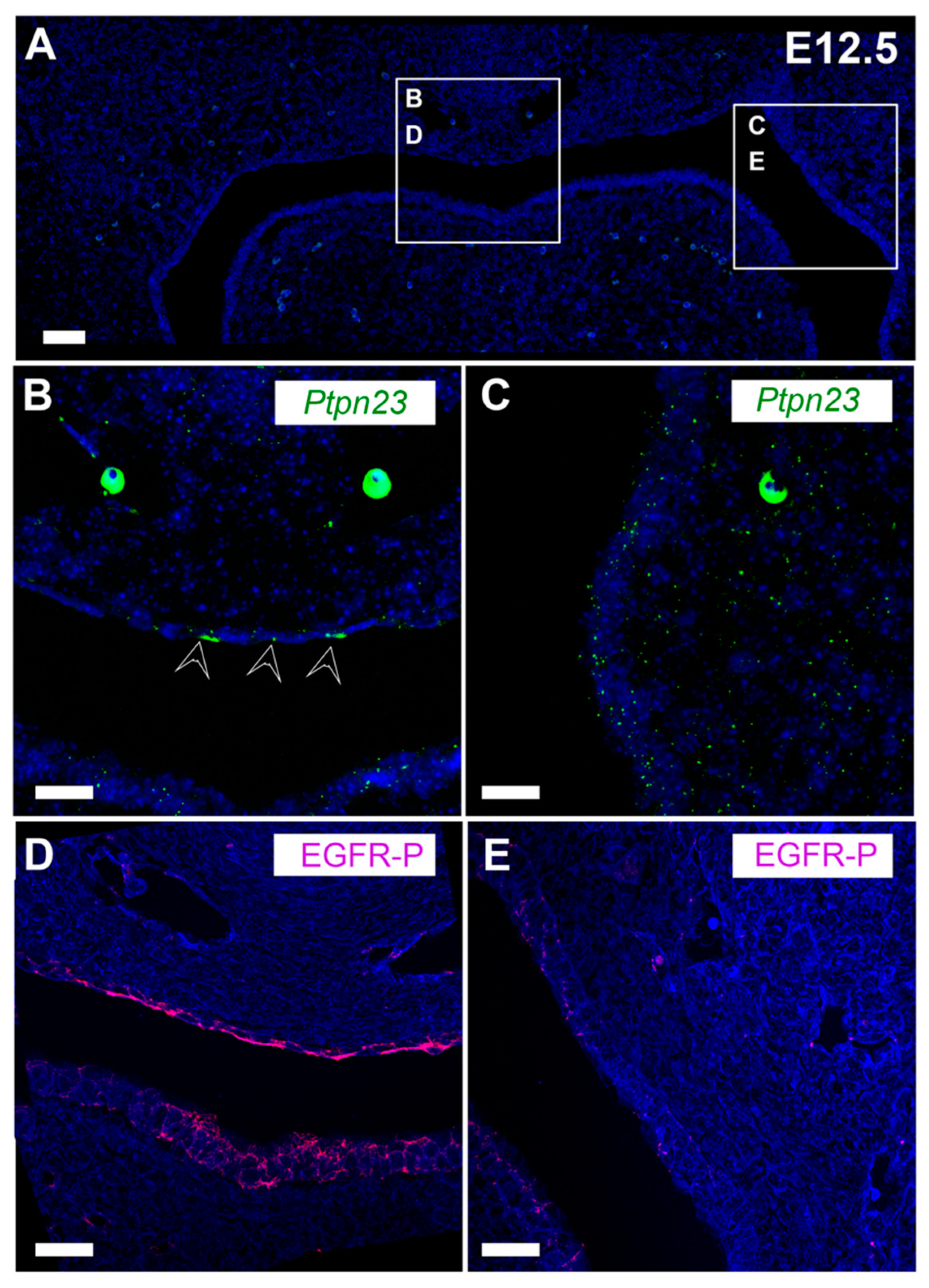
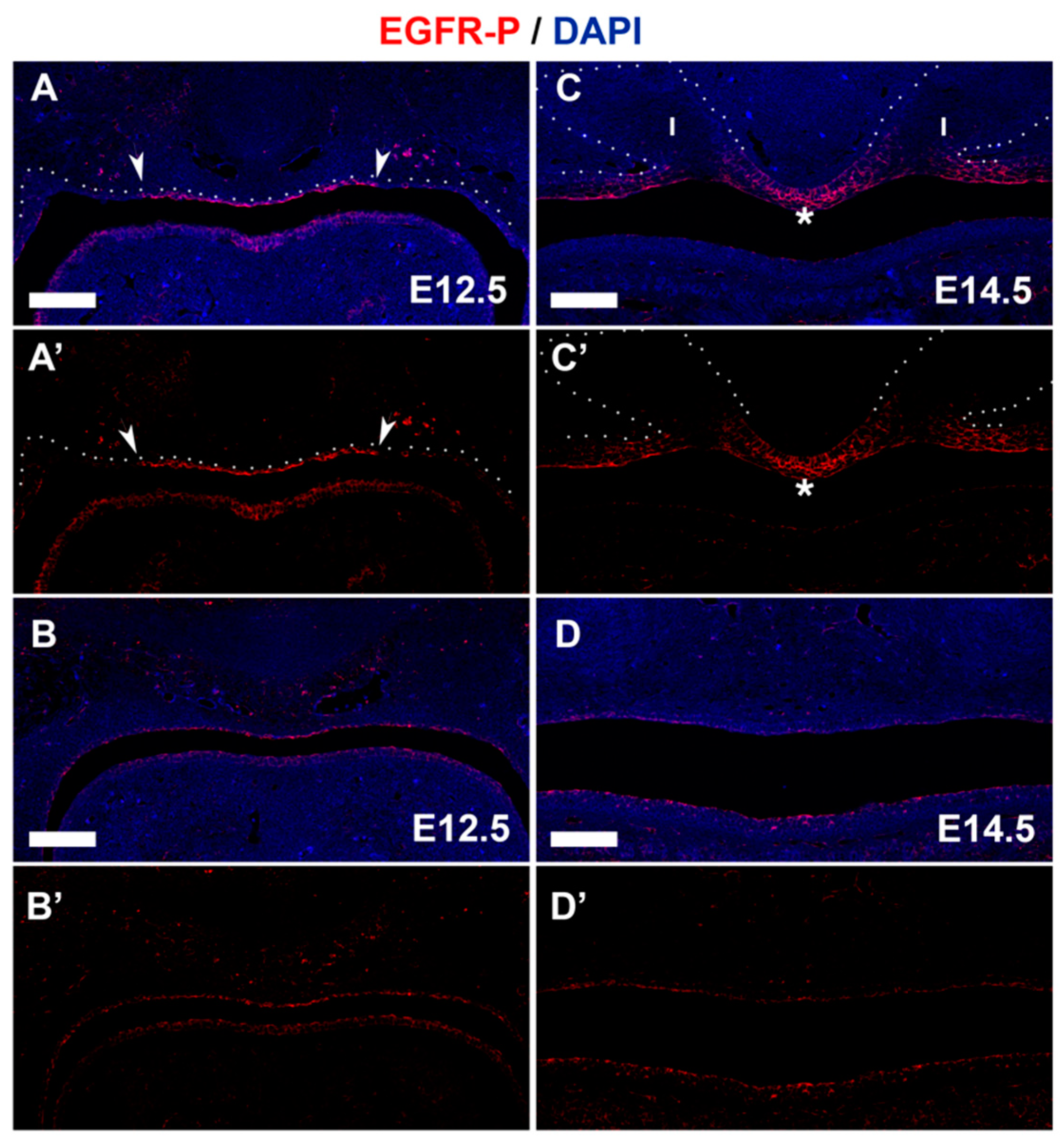
3.4. PTPN23 and Its Relationship to EGFR during Early Murine Tooth Development
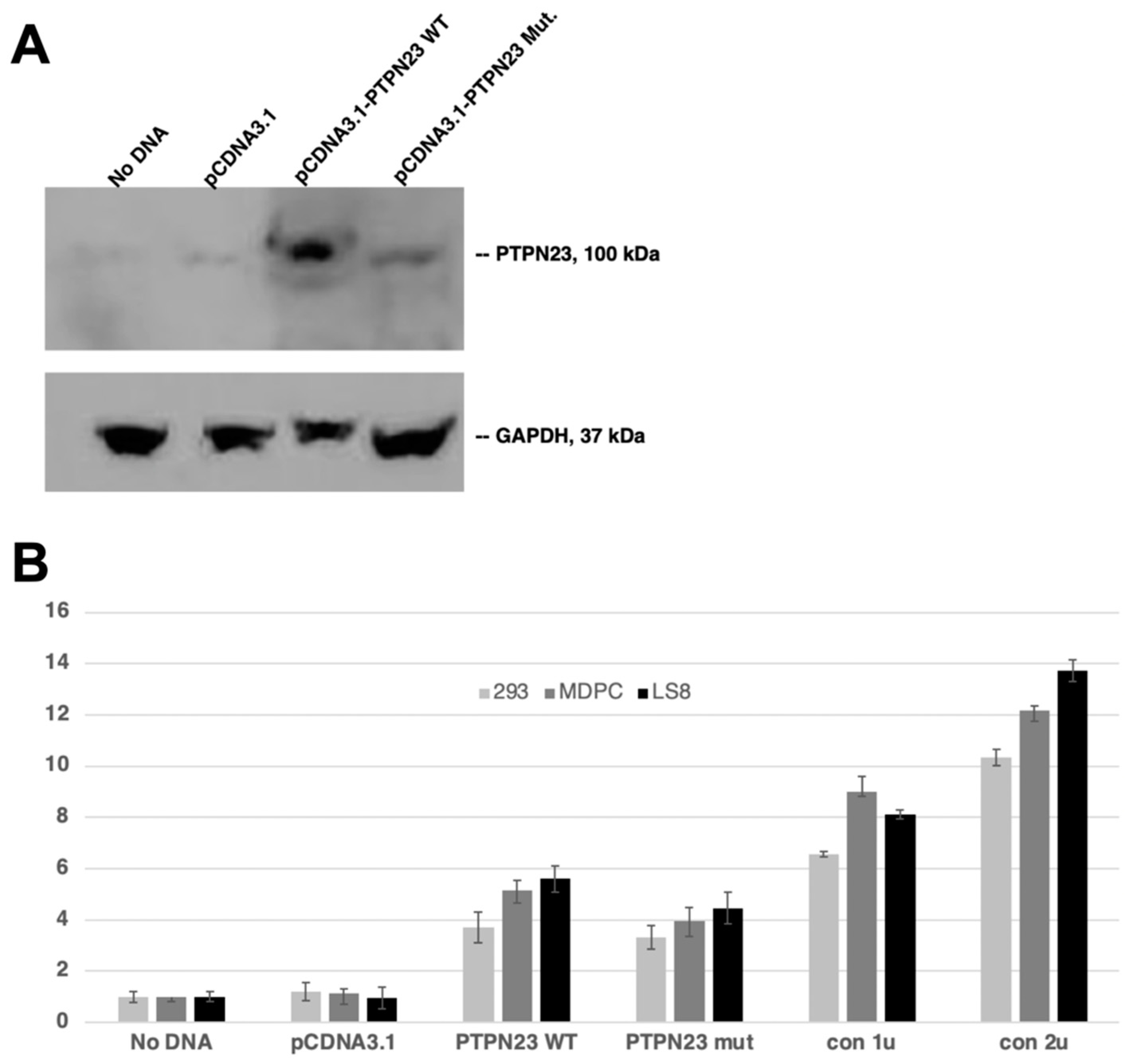
3.5. Decreased Phosphatase Activity of Mutant PTPN23
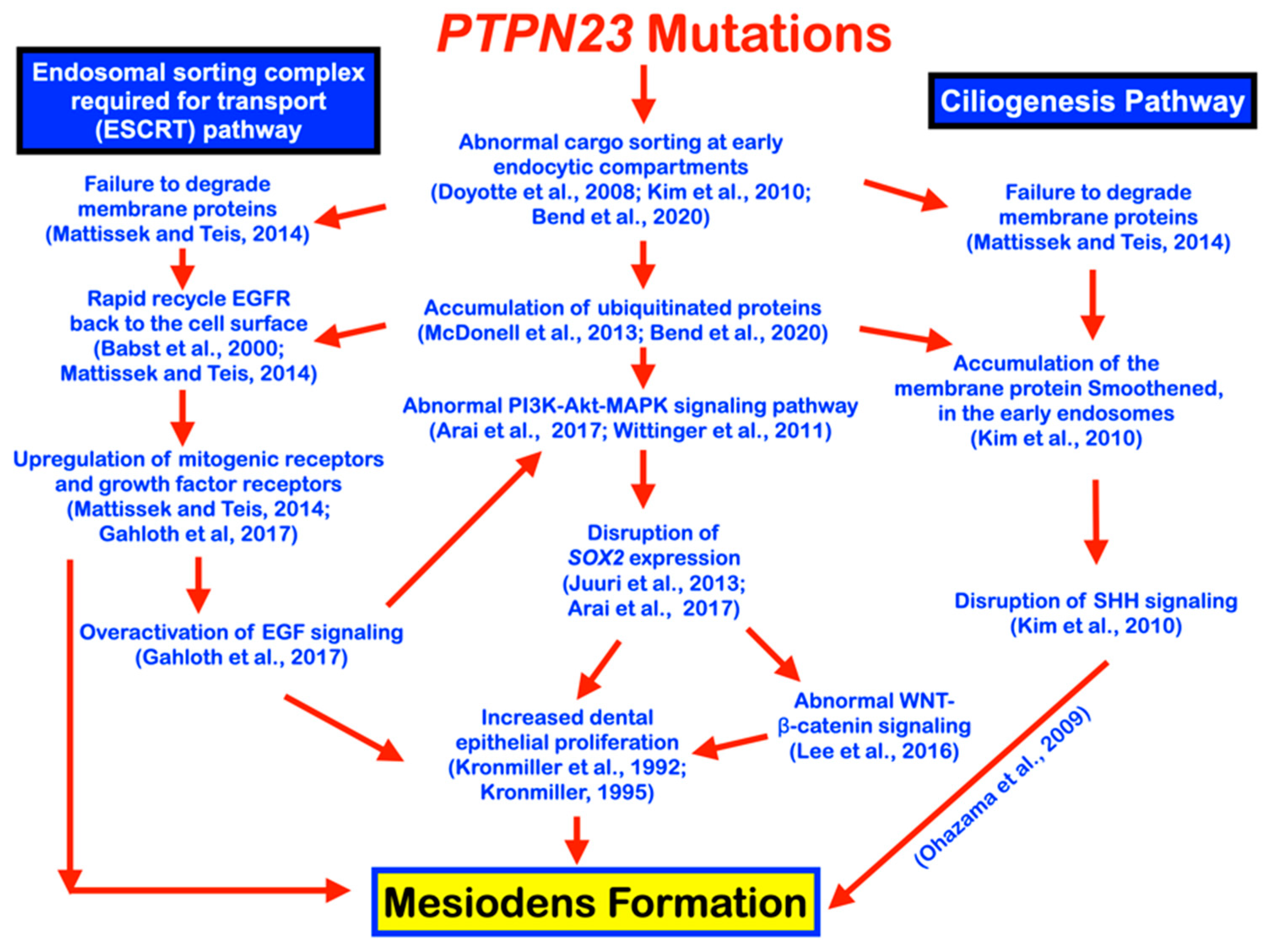
4. Discussion
Limitations of the Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bolk, L. Die Uberzahligen oberen incisivi des Menschen. Dtsch. Mscf. 2hk 1917, 35, 185. [Google Scholar]
- Thilander, B.; Myrberg, N. The prevalence of malocclusion in Swedish schoolchildren. Eur. J. Oral Sci. 1973, 81, 12–21. [Google Scholar] [CrossRef]
- Gündüz, K.; Çelenk, P.; Zengin, Z.; Sümer, P. Mesiodens: A radiographic study in children. J. Oral Sci. 2008, 50, 287–291. [Google Scholar] [CrossRef]
- Kositbowornchai, S.; Keinprasit, C.; Poomat, N. Prevalence and distribution of dental anomalies in pretreatment orthodontic Thai patients. Khon Kaen Dent. J. 2010, 13, 92–100. [Google Scholar]
- Mukhopadhyay, S. Mesiodens: A clinical and radiographic study in children. J. Indian Soc. Pedod. Prev. Dent. 2011, 29, 34–38. [Google Scholar] [CrossRef]
- Kazanci, F.; Celikoglu, M.; Miloglu, O.; Yildirim, H.; Ceylan, I. The frequency and characteristics of mesiodens in a Turkish patient population. Eur. J. Dent. 2011, 5, 361–365. [Google Scholar] [CrossRef]
- Lara, T.S.; Lancia, M.; Silva Filho, O.G.d.; Garib, D.G.; Ozawa, T.O. Prevalence of mesiodens in orthodontic patients with deciduous and mixed dentition and its association with other dental anomalies. Dent. Press J. Orthod. 2013, 18, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Goksel, S.; Agirgol, E.; Karabas, H.C.; Ozcan, I. Evaluation of Prevalence and Positions of Mesiodens Using Cone-Beam Computed Tomography. J. Oral Maxillofac. Res. 2018, 9, e1. [Google Scholar] [CrossRef] [PubMed]
- Aren, G.; Erdem, A.P.; Onur, Ö.D.; Ak, G. The prevelance of mesiodens in a group of non-syndromic Turkish children: A radiographic study. Eur. Oral Res. 2018, 52, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, K.; Venkataraghavan, K.; Kunjappan, S.; Ramesh, M. Mesiodens: A clinical and radiographic study of 82 teeth in 55 children below 14 years. J. Pharm. Bioallied Sci. 2013, 5, S60. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.D.; Guimarães Cabral, L.A.; Martins Gomes, A.P.; Moraes, E. Supernumerary mesiodentes with familial character: A clinical report. Quintessence Int. 1995, 26, 343–345. [Google Scholar] [PubMed]
- Sykaras, S.N. Mesiodens in primary and permanent dentitions: Report of a case. Oral Surg. Oral Med. Oral Pathol. 1975, 39, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Asaumi, J.-I.; Shibata, Y.; Yanagi, Y.; Hisatomi, M.; Matsuzaki, H.; Konouchi, H.; Kishi, K. Radiographic examination of mesiodens and their associated complications. Dentomaxillofac. Radiol. 2004, 33, 125–127. [Google Scholar] [CrossRef] [PubMed]
- von Arx, T. Anterior maxillary supernumerary teeth: A clinical and radiographic study. Aust. Dent. J. 1992, 37, 189–195. [Google Scholar] [CrossRef]
- Khandelwal, V.; Nayak, A.; Naveen, R.; Ninawe, N.; Nayak, P.; Prasad, S.S. Prevalence of mesiodens among six-to seventeen-year-old school going children of Indore. J. Indian Soc. Pedod. Prev. Dent. 2011, 29, 288–293. [Google Scholar] [CrossRef]
- Mitchell, L. Supernumerary teeth. Dent. Update 1989, 16, 65–66, 68–69. [Google Scholar]
- Anthonappa, R.; King, N.; Rabie, A. Aetiology of supernumerary teeth: A literature review. Eur. Arch. Paediatr. Dent. 2013, 14, 279–288. [Google Scholar] [CrossRef]
- Peterkova, R.; Hovorakova, M.; Peterka, M.; Lesot, H. Three-dimensional analysis of the early development of the dentition. Aust. Dent. J. 2014, 59, 55–80. [Google Scholar] [CrossRef]
- Sedano, H.O.; Gorlin, R.J. Familial occurrence of mesiodens. Oral Surg. Oral Med. Oral Pathol. 1969, 27, 360–361. [Google Scholar] [CrossRef]
- Cadenat, H.; Combelles, R.; Fabert, G.; Clouet, M. Mesiodens and heredity. Rev. Stomatol. Chir. Maxillofac. 1977, 78, 341–346. [Google Scholar]
- Desai, R.S.; Shah, N.P. Multiple supernumerary teeth in two brothers: A case report. J. Oral Pathol. Med. 1998, 27, 411–413. [Google Scholar] [CrossRef] [PubMed]
- Marya, C.M.; Kumar, B. Familial occurrence of mesiodentes with unusual findings. Quintessence Int. 1998, 29, 49–51. [Google Scholar] [PubMed]
- Gallas, M.M.; Garcia, A. Retention of permanent incisors by mesiodens: A family affair. Br. Dent. J. 2000, 188, 63–64. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Hosomichi, K.; Yamaguchi, T.; Yano, K.; Funatsu, T.; Adel, M.; Haga, S.; Maki, K.; Tajima, A. Whole-exome sequencing analysis of supernumerary teeth occurrence in Japanese individuals. Hum. Genome Var. 2017, 4, hgv201646. [Google Scholar] [CrossRef] [PubMed]
- Mason, C.; Rule, D. Midline supernumeraries: A family affair. Dent. Update 1995, 22, 34–35. [Google Scholar] [PubMed]
- Sharma, A. Familial occurence of mesiodens--a case report. J. Indian Soc. Pedod. Prev. Dent. 2003, 21, 84–85. [Google Scholar] [PubMed]
- Severin, E.; Purcărea, R.; Albu, C.; Albu, D.F.; Ionescu, E.; Stanciu, D. Familial mesiodens. Rom. J. Stomatol. 2009, 55, 293–296. [Google Scholar]
- Hunstadbråten, K. Anomalies in twins. Quintessenz 1965, 16, 71–72. [Google Scholar]
- Schön, F. Supernumerary incisors in uniovular twins, and their treatment by means of electrosurgery. Quintessence Int. Dent. Dig. 1974, 5, 13–18. [Google Scholar]
- Bucci, E.; Martina, R. True hyperdontia in monochorial twins. Clinical case. Arch. Stomatol. 1975, 16, 305–313. [Google Scholar]
- Carton, A.; Rees, R. Mirror image dental anomalies in identical twins. Br. Dent. J. 1987, 162, 193–194. [Google Scholar] [CrossRef]
- Choi, W.; Chang, R.; Chuang, S. Bilateral mesiodentes of identical twins--a case report. Zhonghua Ya Yi Xue Hui Za Zhi 1990, 9, 116–121. [Google Scholar] [PubMed]
- Beere, D.; Hargreaves, J.; Sperber, G.; Cleaton-Jones, P. Mirror image supplemental primary incisor teeth in twins: Case report and review. Pediatr. Dent. 1990, 12, 390–392. [Google Scholar] [PubMed]
- Seddon, R.P.; Johnstone, S.C.; Smith, P.B. Mesiodentes in twins: A case report and a review of the literature. Int. J. Paediatr. Dent. 1997, 7, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Brand, A.; Akhavan, M.; Tong, H.; Kooka, Y.; Zernik, J.H. Orthodontic, genetic, and periodontal considerations in the treatment of impacted maxillary central incisors: A study of twins. Am. J. Orthod. Dentofac. Orthop. 2000, 117, 68–74. [Google Scholar] [CrossRef]
- Townsend, G.; Richards, L.; Hughes, T.; Pinkerton, S.; Schwerdt, W. Epigenetic influences may explain dental differences in monozygotic twin pairs. Aust. Dent. J. 2005, 50, 95–100. [Google Scholar] [CrossRef]
- Sharma, A. A rare case of concomitant hypo-hyperdontia in identical twins. J. Indian Soc. Pedod. Prev. Dent. 2008, 26, S79–S81. [Google Scholar]
- Babacan, H.; Öztürk, F.; Polat, H.B. Identical unerupted maxillary incisors in monozygotic twins. Am. J. Orthod. Dentofac. Orthop. 2010, 138, 498–509. [Google Scholar] [CrossRef]
- Gurgel, C.V.; Cota, A.L.S.A.L.S.; Kobayashi, T.Y.; Silva, S.M.B.; Machado, M.A.A.M.; Rios, D.; Garib, D.G.; Oliveira, T.M. Bilateral mesiodens in monozygotic twins: 3D diagnostic and management. Case Rep. Dent. 2013, 2013, 193614. [Google Scholar] [CrossRef]
- Reddy, G.S.M.; Mahajan, B.; Desai, R.S. Mesiodens In Twins: A Case Report. Indian J. Dent. Sci. 2013, 5, 088–089. [Google Scholar]
- Nance, W.; Warburg, M.; Bixler, D.; Helveston, E. Congenital X-linked cataract, dental anomalies and brachymetacarpalia. Birth Defects Orig. Artic. Ser. 1974, 10, 285–291. [Google Scholar]
- Bixler, D.; Higgins, M.; Hartsfield, J., Jr. The Nance-Horan syndrome: A rare X-linked ocular-dental trait with expression in heterozygous females. Clin. Genet. 1984, 26, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Walpole, I.R.; Hockey, A.; Nicoll, A. The Nance-Horan syndrome. J. Med. Genet. 1990, 27, 632. [Google Scholar] [CrossRef] [PubMed]
- Gjørup, H.; Haubek, D.; Jacobsen, P.; Ostergaard, J.R. Nance–Horan syndrome—The oral perspective on a rare disease. Am. J. Med. Genet. A 2017, 173, 88–98. [Google Scholar] [CrossRef]
- Chan, M.Y.; Shifteh, K.; Shanske, A.L. Mesiodens, a new microform of holoprosencephaly? Am. J. Med. Genet. A 2009, 149, 268–271. [Google Scholar] [CrossRef]
- Kantaputra, P.; Jatooratthawichot, P.; Chintakanon, K.; Intachai, W.; Pradermdutsadeeporn, P.; Adisornkanj, P.; Tongsima, S.; Ngamphiw, C.; Olsen, B.; Tucker, A.S. Mutations in LRP6 highlight the role of WNT signaling in oral exostoses and dental anomalies. Arch. Oral Biol. 2022, 142, 105514. [Google Scholar] [CrossRef] [PubMed]
- Kantaputra, P.N.; Guven, Y.; Tripuwabhrut, K.; Adisornkanj, P.; Hatsadaloi, A.; Kaewgahya, M.; Olsen, B.; Ngamphiw, C.; Jatooratthawichot, P.; Tongsima, S. Mutations in LRP5 and BMP4 are associated with mesiodens, tooth agenesis, root malformation, and oral exostoses. Clin. Genet. 2022, 102, 333–338. [Google Scholar] [CrossRef]
- Kantaputra, P.N.; Tripuwabhrut, K.; Jatooratthawichot, P.; Adisornkanj, P.; Hatsadaloi, A.; Nop Porntrakoolsaree, N.; Kaewgaya, M.; Olsen, B.; Tongsima, S.; Ngamphiw, C.; et al. Genetic variants in the WLS are associated with dental anomalies, torus palatinus, and torus mandibularis. Eur. J. Orthod. 2022. Epub ahead of printing. [Google Scholar] [CrossRef]
- Kantaputra, P.; Jatooratthawichot, P.; Kottege, N.; Anthonappa, R.P.; Kaewgahya, M.; Tongsima, S.; Ngamphiw, C.; Cairns, J.R.K.; Predes, D.; He, X. DKK1 is a strong candidate for mesiodens and taurodontism. Clin. Genet. 2023. Epub ahead of printing. [Google Scholar] [CrossRef]
- Kantaputra, P.; Jatooratthawichot, P.; Adisornkanj, P.; Kitsadayurach, P.; Kaewgahya, M.; Olsen, B.; Ohazama, A.; Ngamphiw, C.; Tongsima, S.; Cox, T.C.; et al. Rare variants in LRP4 are associated with mesiodens, root maldevelopment, and oral exostoses in humans. Biology 2023, 12, 220. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The ensembl variant effect predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, C.; Li, C.; Boerwinkle, E. dbNSFP v3. 0: A one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat. 2016, 37, 235–241. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Gouw, M.; Michael, S.; Sámano-Sánchez, H.; Pancsa, R.; Glavina, J.; Diakogianni, A.; Valverde, J.A.; Bukirova, D.; Čalyševa, J.; et al. ELM—The eukaryotic linear motif resource in 2020. Nucleic Acids Res. 2019, 48, D296–D306. [Google Scholar] [CrossRef]
- Gingras, M.C.; Kharitidi, D.; Chénard, V.; Uetani, N.; Bouchard, M.; Tremblay, M.L.; Pause, A. Expression analysis and essential role of the putative tyrosine phosphatase His-domain-containing protein tyrosine phosphatase (HD-PTP). Int. J. Dev. Biol. 2009, 53, 1069–1074. [Google Scholar] [CrossRef]
- Doyotte, A.; Mironov, A.; McKenzie, E.; Woodman, P. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 6308–6313. [Google Scholar] [CrossRef]
- Burdon, K.P.; McKay, J.D.; Sale, M.M.; Russell-Eggitt, I.M.; Mackey, D.A.; Wirth, M.G.; Elder, J.E.; Nicoll, A.; Clarke, M.P.; FitzGerald, L.M.; et al. Mutations in a novel gene, NHS, cause the pleiotropic effects of Nance-Horan syndrome, including severe congenital cataract, dental anomalies, and mental retardation. Am. J. Hum. Genet. 2003, 73, 1120–1130. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Hwang, J.; Kim, H.-S.; Kwon, H.J.; Kim, S.; Lee, J.H.; Lee, J.H. Genetic alterations in mesiodens as revealed by targeted NGS and gene co-occurrence network analysis. Oral Dis. 2017, 23, 966–972. [Google Scholar] [CrossRef]
- Bend, R.; Cohen, L.; Carter, M.T.; Lyons, M.J.; Niyazov, D.; Mikati, M.A.; Rojas, S.K.; Person, R.E.; Si, Y.; Wentzensen, I.M. Phenotype and mutation expansion of the PTPN23 associated disorder characterized by neurodevelopmental delay and structural brain abnormalities. Eur. J. Hum. Genet. 2020, 28, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Toyooka, S.-I.; Ouchida, M.; Jitsumori, Y.; Tsukuda, K.; Sakai, A.; Nakamura, A.; Shimizu, N.; Shimizu, K. HD-PTP: A novel protein tyrosine phosphatase gene on human chromosome 3p21.3. Biochem. Biophys. Res. Commun. 2000, 278, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Khalaf-Nazzal, R.; Fasham, J.; Ubeyratna, N.; Evans, D.J.; Leslie, J.S.; Warner, T.T.; Al-Hijawi, F.; Alshaer, S.; Baker, W.; Turnpenny, P.D. Final Exon Frameshift Biallelic PTPN23 Variants Are Associated with Microcephalic Complex Hereditary Spastic Paraplegia. Brain Sci. 2021, 11, 614. [Google Scholar] [CrossRef] [PubMed]
- Husedzinovic, A.; Neumann, B.; Reymann, J.; Draeger-Meurer, S.; Chari, A.; Erfle, H.; Fischer, U.; Gruss, O.J. The catalytically inactive tyrosine phosphatase HD-PTP/PTPN23 is a novel regulator of SMN complex localization. Mol. Biol. Cell 2015, 26, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Barr, A.J.; Ugochukwu, E.; Lee, W.H.; King, O.N.; Filippakopoulos, P.; Alfano, I.; Savitsky, P.; Burgess-Brown, N.A.; Müller, S.; Knapp, S. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell 2009, 136, 352–363. [Google Scholar] [CrossRef]
- Gingras, M.C.; Zhang, Y.L.; Kharitidi, D.; Barr, A.J.; Knapp, S.; Tremblay, M.L.; Pause, A. HD-PTP is a catalytically inactive tyrosine phosphatase due to a conserved divergence in its phosphatase domain. PLoS ONE 2009, 4, e5105. [Google Scholar] [CrossRef]
- Tabernero, L.; Woodman, P. Dissecting the role of His domain protein tyrosine phosphatase/PTPN23 and ESCRTs in sorting activated epidermal growth factor receptor to the multivesicular body. Biochem. Soc. Trans. 2018, 46, 1037–1046. [Google Scholar] [CrossRef]
- Gahloth, D.; Levy, C.; Heaven, G.; Stefani, F.; Wunderley, L.; Mould, P.; Cliff, M.J.; Bella, J.; Fielding, A.J.; Woodman, P. Structural basis for selective interaction between the ESCRT regulator HD-PTP and UBAP1. Structure 2016, 24, 2115–2126. [Google Scholar] [CrossRef]
- McDonell, L.M.; Mirzaa, G.M.; Alcantara, D.; Schwartzentruber, J.; Carter, M.T.; Lee, L.J.; Clericuzio, C.L.; Graham, J.M.; Morris-Rosendahl, D.J.; Polster, T. Mutations in STAMBP, encoding a deubiquitinating enzyme, cause microcephaly–capillary malformation syndrome. Nat. Genet. 2013, 45, 556–562. [Google Scholar] [CrossRef]
- Ali, N.; Zhang, L.; Taylor, S.; Mironov, A.; Urbé, S.; Woodman, P. Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr. Biol. 2013, 23, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Babst, M.; Odorizzi, G.; Estepa, E.J.; Emr, S.D. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 2000, 1, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Mattissek, C.; Teis, D. The role of the endosomal sorting complexes required for transport (ESCRT) in tumorigenesis. Mol. Membr. Biol. 2014, 31, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Kyuuma, M.; Sugamura, K. Endosomal sorting complex required for transport proteins in cancer pathogenesis, vesicular transport, and non-endosomal functions. Cancer Sci. 2008, 99, 1293–1303. [Google Scholar] [CrossRef] [PubMed]
- Raiborg, C.; Bache, K.G.; Gillooly, D.J.; Madshus, I.H.; Stang, E.; Stenmark, H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 2002, 4, 394–398. [Google Scholar] [CrossRef]
- Sigismund, S.; Woelk, T.; Puri, C.; Maspero, E.; Tacchetti, C.; Transidico, P.; Di Fiore, P.P.; Polo, S. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. USA 2005, 102, 2760–2765. [Google Scholar] [CrossRef] [PubMed]
- Gahloth, D.; Heaven, G.; Jowitt, T.A.; Mould, A.P.; Bella, J.; Baldock, C.; Woodman, P.; Tabernero, L. The open architecture of HD-PTP phosphatase provides new insights into the mechanism of regulation of ESCRT function. Sci. Rep. 2017, 7, 9151. [Google Scholar] [CrossRef]
- Tanase, C.-A. Histidine domain-protein tyrosine phosphatase interacts with Grb2 and GrpL. PLoS ONE 2010, 5, e14339. [Google Scholar] [CrossRef]
- Kronmiller, J.E. Spatial distribution of epidermal growth-factor transcripts and effects of exogenous epidermal growth factor on the pattern of the mouse dental lamina. Arch. Oral Biol. 1995, 40, 137–143. [Google Scholar] [CrossRef]
- Kronmiller, J.E.; Upholt, W.B.; Kollar, E.J. Alteration of murine odontogenic patterning and prolongation of expression of epidermal growth factor mRNA by retinol in vitro. Arch. Oral Biol. 1992, 37, 129–138. [Google Scholar] [CrossRef]
- Seow, W.K.; Brown, J.; Romaniuk, K. The Nance-Horan syndrome of dental anomalies, congenital cataracts, microphthalmia, and anteverted pinna: Case report. Pediatr. Dent. 1985, 7, 307–311. [Google Scholar] [PubMed]
- Brooks, S.P.; Coccia, M.; Tang, H.R.; Kanuga, N.; Machesky, L.M.; Bailly, M.; Cheetham, M.E.; Hardcastle, A.J. The Nance–Horan syndrome protein encodes a functional WAVE homology domain (WHD) and is important for co-ordinating actin remodelling and maintaining cell morphology. Hum. Mol. Genet. 2010, 19, 2421–2432. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Ang, S.L.; Shaw, M.; Mackey, D.A.; Gécz, J.; McAvoy, J.W.; Craig, J.E. Nance–Horan syndrome protein, NHS, associates with epithelial cell junctions. Hum. Mol. Genet. 2006, 15, 1972–1983. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Burdon, K.P.; Dave, A.; Jamieson, R.V.; Yaron, Y.; Billson, F.; Van Maldergem, L.; Lorenz, B.; Gécz, J.; Craig, J.E. Novel causative mutations in patients with Nance-Horan syndrome and altered localization of the mutant NHS-A protein isoform. Mol. Vis. 2008, 14, 1856. [Google Scholar]
- Akamatsu, M.; Vasan, R.; Serwas, D.; Ferrin, M.A.; Rangamani, P.; Drubin, D.G. Principles of self-organization and load adaptation by the actin cytoskeleton during clathrin-mediated endocytosis. Elife 2020, 9, e49840. [Google Scholar] [CrossRef]
- Arai, C.; Yoshizaki, K.; Miyazaki, K.; Saito, K.; Yamada, A.; Han, X.; Funada, K.; Fukumoto, E.; Haruyama, N.; Iwamoto, T. Nephronectin plays critical roles in Sox2 expression and proliferation in dental epithelial stem cells via EGF-like repeat domains. Sci. Rep. 2017, 7, 45181. [Google Scholar] [CrossRef]
- Juuri, E.; Jussila, M.; Seidel, K.; Holmes, S.; Wu, P.; Richman, J.; Heikinheimo, K.; Chuong, C.-M.; Arnold, K.; Hochedlinger, K. Sox2 marks epithelial competence to generate teeth in mammals and reptiles. Development 2013, 140, 1424–1432. [Google Scholar] [CrossRef]
- Wittinger, M.; Vanhara, P.; El-Gazzar, A.; Savarese-Brenner, B.; Pils, D.; Anees, M.; Grunt, T.W.; Sibilia, M.; Holcmann, M.; Horvat, R. hVps37A Status affects prognosis and cetuximab sensitivity in ovarian cancer. Clin. Cancer Res. 2011, 17, 7816–7827. [Google Scholar] [CrossRef]
- Swiatnicki, M.R.; Rennhack, J.P.; Ortiz, M.M.; Hollern, D.P.; Perry, A.V.; Kubiak, R.; Riveria Riveria, S.M.; O’Reilly, S.; Andrechek, E.R. Elevated phosphorylation of EGFR in NSCLC due to mutations in PTPRH. PLoS Genet. 2022, 18, e1010362. [Google Scholar] [CrossRef]
- Saito, K.; Takahashi, K.; Huang, B.; Asahara, M.; Kiso, H.; Togo, Y.; Tsukamoto, H.; Mishima, S.; Nagata, M.; Iida, M.; et al. Loss of Stemness, EMT, and Supernumerary Tooth Formation in Cebpb−/−Runx2+/− Murine Incisors. Sci. Rep. 2018, 8, 5169. [Google Scholar] [CrossRef]
- Chacon-Camacho, O.F.; Fuerte-Flores, B.I.; Ricardez-Marcial, E.F.; Zenteno, J.C. SOX2 anophthalmia syndrome and dental anomalies. Am. J. Med. Genet. A 2015, 167, 2830–2833. [Google Scholar] [CrossRef] [PubMed]
- Numakura, C.; Kitanaka, S.; Kato, M.; Ishikawa, S.; Hamamoto, Y.; Katsushima, Y.; Kimura, T.; Hayasaka, K. Supernumerary impacted teeth in a patient with SOX2 anophthalmia syndrome. Am. J. Med. Genet. A 2010, 152, 2355–2359. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Jung, S.Y.; Wu, Z.; Zhang, S.; Jung, H.S. Sox2 maintains epithelial cell proliferation in the successional dental lamina. Cell Prolif. 2020, 53, e12729. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-J.; Kim, E.-J.; Otsu, K.; Harada, H.; Jung, H.-S. Sox2 contributes to tooth development via Wnt signaling. Cell Tissue Res. 2016, 365, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.J.; Rasch, L.J.; Cooper, R.L.; Metscher, B.D.; Johanson, Z.; Fraser, G.J. Sox2+ progenitors in sharks link taste development with the evolution of regenerative teeth from denticles. Proc. Natl. Acad. Sci. USA 2016, 113, 14769–14774. [Google Scholar] [CrossRef] [PubMed]
- Xavier, G.M.; Patist, A.L.; Healy, C.; Pagrut, A.; Carreno, G.; Sharpe, P.T.; Martinez-Barbera, J.P.; Thavaraj, S.; Cobourne, M.T.; Andoniadou, C.L. Activated WNT signaling in postnatal SOX2-positive dental stem cells can drive odontoma formation. Sci. Rep. 2015, 5, 14479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yuan, G.; Liu, H.; Lin, H.; Wan, C.; Chen, Z. Expression pattern of Sox2 during mouse tooth development. Gene Expr. Patterns 2012, 12, 273–281. [Google Scholar] [CrossRef]
- Gahloth, D.; Levy, C.; Walker, L.; Wunderley, L.; Mould, A.P.; Taylor, S.; Woodman, P.; Tabernero, L. Structural basis for specific interaction of TGFβ signaling regulators SARA/Endofin with HD-PTP. Structure 2017, 25, 1011–1024.e1014. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.E.; Heynen-Genel, S.; Suyama, E.; Ono, K.; Lee, K.; Ideker, T.; Aza-Blanc, P.; Gleeson, J.G. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature 2010, 464, 1048–1051. [Google Scholar] [CrossRef]
- Maxfield, F.R.; McGraw, T.E. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 2004, 5, 121–132. [Google Scholar] [CrossRef]
- Ohazama, A.; Haycraft, C.J.; Seppala, M.; Blackburn, J.; Ghafoor, S.; Cobourne, M.; Martinelli, D.C.; Fan, C.-M.; Peterkova, R.; Lesot, H. Primary cilia regulate Shh activity in the control of molar tooth number. Development 2009, 136, 897–903. [Google Scholar] [CrossRef] [PubMed]






| Authors | Ethnic | Number of Patients | Prevalence (%) | Radiographic Examination | Male:Female |
|---|---|---|---|---|---|
| Thilander and Myrberg, 1973 [2] | Swedish | 6398 | 0.9 | yes | Not mentioned |
| von Arx, 1992 [14] | Swiss | Not mentioned | Not mentioned | yes | 2.6:1 |
| Asaumi et al., 2004 [13] | Japanese | Not mentioned | Not mentioned | yes | 2.8:1 |
| Gunduz et al., 2008 [3] | Turkish | 23000 | 0.3 | yes | 2.1:1 |
| Kositbowornchai et al., 2010 [4] | Thai | 570 | 1.05 | yes | Not mentioned |
| Mukhopadhyay, 2011 [5] | Indian | 7932 | 0.8 | yes | 1.78:1 |
| Kazanci et al., 2011 [6] | Turkish | 3351 | 0.3 | yes | 1.5:1 |
| Lara et al., 2013 [7] | Brazilian | 1995 | 1.5 | yes | 1.5:1 |
| Ramesh et al., 2013 [10] | Indian | Not mentioned | Not mentioned | yes | 2.05:1 |
| Goksel et al., 2018 [8] | Turkish | 1902 | 5.04 | yes | 1.9:1 |
| Aren et al., 2018 [9] | Turkish | 58142 | 0.1 | yes | Not mentioned |
| Authors | N (Patients) | N (Teeth) | Number | % | Unerupted % | Orientation | % |
|---|---|---|---|---|---|---|---|
| Asaumi et al., 2004 [13] | 200 | 256 | Single | 73 | Not mentioned | Vertical | 27 |
| Double | 26 | Inverted | 67 | ||||
| >3 | 1 | Transverse | 6 | ||||
| Gunduz et al., 2008 [3] | 69 | 85 | Single | 76.8 | 78.8 | Vertical | 55.2 |
| Double | 23.1 | Inverted | 37.6 | ||||
| Transverse | 7 | ||||||
| Mukhopadhyay, 2011 [5] | 64 | 78 | Single | 78.1 | 53.8 | Vertical | 62.8 |
| Double | 21.9 | Inverted | 30.8 | ||||
| Transverse | 6.4 | ||||||
| Kazanci et al., 2011 [6] | 10 | 12 | Single | 80 | 66.7 | Vertical | 58.3 |
| Double | 20 | Inverted | 33.3 | ||||
| >3 | 0 | Transverse | 8.4 | ||||
| Lara et al., 2013 [7] | 30 | 36 | Single | 80 | 75 | Vertical | 75 |
| Double | 20 | Inverted | 13.9 | ||||
| >3 | 0 | Transverse | 11.1 | ||||
| Goksel et al., 2018 [8] | 101 | 130 | Single | 76.23 | 78.46 | Vertical | 63.1 |
| Double | 18.81 | Inverted | 20 | ||||
| >3 | 4.95 | Transverse | 16.9 | ||||
| Aren et al., 2018 [9] | 59 | 83 | Single | 61 | 68.7 | Vertical | 77.1 |
| Double | 37.3 | Inverted | 22.9 | ||||
| >3 | 1 | Transverse | 0 |
| Types of Familial Occurrences | References | Descriptions |
|---|---|---|
| Siblings | Sedano and Gorlin, 1969 [19] | 2 siblings affected (normal parents) |
| Cadenat et al., 1977 [20] | Discordant dizygotic twin (parents’ phenotype: N/A) | |
| Almeida et al., 1995 [11] | 3 siblings affected (parents’ phenotype: N/A) | |
| Desai and Shah, 1998 [21] | 2 siblings affected (parents’ phenotype: N/A) | |
| Marya and Kumar, 1998 [22] | 2 siblings affected (parents’ phenotype: N/A) | |
| Gallas and Garcia, 2000 [23] | 2 siblings affected (parents’ phenotype: N/A) | |
| Takahashi et al., 2017 [24] | 2 siblings affected (parents’ phenotype: N/A) | |
| Monozygotic twins | Hunstadbråten, 1965 [28] | Mirror imaged discordant |
| Schön, 1974 [29] | Concordant | |
| Bucci and Martina, 1975 [30] | Discordant | |
| Carton and Rees, 1987 [31] | Mirror imaged discordant | |
| Choi et al., 1990 [32] | Near concordant | |
| Beere et al.,1990 [33] | Mirror imaged discordant | |
| Seddon et al., 1997 [34] | Near concordant | |
| Brand et al., 2000 [35] | Mirror imaged discordant | |
| Townsend et al., 2005 [36] | 8 pairs of discordant | |
| Sharma, 2008 [37] | Discordant | |
| Babacan et al., 2010 [38] | Concordant | |
| Gurgel et al., 2013 [39] | Concordant | |
| Reddy et al., 2013 [40] | Discordant | |
| More than one generation | Mason and Rule, 1995 [25] | Father, mother and 2 sons |
| Sharma, 2003 [26] | Father and daughter | |
| Severin et al., 2009 [27] | Family 1: Mother, 2 daughters, uncle, and cousin | |
| Family 2: Father, daughter, and son | ||
| Family 1: Mother and son | ||
| Takahashi et al., 2017 [24] | Family 2: Father and son | |
| Family 3: Mother and 3 sons | ||
| Skip generation | Severin et al., 2009 [27] | Grandmother and grandson |
| Families | Patients | Ethnic | Gender | Phenotypes | Orientation | Eruption | PTPN23 Mutations |
|---|---|---|---|---|---|---|---|
| Family 1 | II-14 | Hmong | Male | Double mesiodentes | Normal | Erupted | c.1807G>A; p.Glu603Lys; rs141113890 |
| II-15 | Hmong | Female | Single mesiodens | Normal | Erupted | ||
| II-11 | Hmong | Male | Double mesiodentes | Inverted | Unerupted | ||
| II-12 | Hmong | Female | Single mesiodens | Inverted | Unerupted | ||
| I-5 | Hmong | Male | Single mesiodens | Normal | Erupted | ||
| II-8 | Hmong | Male | Single mesiodens | Normal | Erupted | ||
| II-9 | Hmong | Male | Single mesiodens | Normal | Erupted | ||
| II-1 | Hmong | Female | Single mesiodens | Normal | Erupted | Not available | |
| I-9 | Hmong | Female | Unaffected | - | - | c.1807G>A; p.Glu603Lys; rs141113890 | |
| I-1 | Hmong | Male | Unaffected | - | - | ||
| I-8 | Hmong | Male | Unaffected | - | - | ||
| I-11 | Hmong | Female | Unaffected | - | - | ||
| II-4 | Hmong | Male | Unaffected | - | - | ||
| II-6 | Hmong | Female | Unaffected | - | - | ||
| I-10 | Hmong | Male | Unaffected | - | - | No mutation | |
| I-3 | Hmong | Female | Unaffected | - | - | ||
| I-7 | Hmong | Female | Unaffected | - | - | ||
| II-2 | Hmong | Male | Unaffected | - | - | ||
| II-3 | Hmong | Male | Unaffected | - | - | ||
| II-5 | Hmong | Male | Unaffected | - | - | ||
| II-7 | Hmong | Female | Unaffected | - | - | ||
| II-10 | Hmong | Male | Unaffected | - | - | ||
| II-13 | Hmong | Female | Unaffected | - | - | ||
| Male: Female with mesiodens = 1.67:1 (5:3) | Single: Double mesiodens cases = 6:2 (3:1) | Inverted mesiodens cases = 25% (2/8) | Unerupted mesiodens cases = 25% (2/8) | Penetrance = 53.84% (7/13) | |||
| Family 2 | Unrelated patient 1 | Thai | Female | Single mesiodens | Normal | Erupted | c.2248C>G p.Pro750Ala rs199549354 |
| Family 3 | Unrelated patient 2 | Thai | Male | Double mesiodentes | Normal | Erupted | c.3298C>T p.Arg1100Cys rs201946361 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adisornkanj, P.; Chanprasit, R.; Eliason, S.; Fons, J.M.; Intachai, W.; Tongsima, S.; Olsen, B.; Arold, S.T.; Ngamphiw, C.; Amendt, B.A.; et al. Genetic Variants in Protein Tyrosine Phosphatase Non-Receptor Type 23 Are Responsible for Mesiodens Formation. Biology 2023, 12, 393. https://doi.org/10.3390/biology12030393
Adisornkanj P, Chanprasit R, Eliason S, Fons JM, Intachai W, Tongsima S, Olsen B, Arold ST, Ngamphiw C, Amendt BA, et al. Genetic Variants in Protein Tyrosine Phosphatase Non-Receptor Type 23 Are Responsible for Mesiodens Formation. Biology. 2023; 12(3):393. https://doi.org/10.3390/biology12030393
Chicago/Turabian StyleAdisornkanj, Ploy, Rajit Chanprasit, Steven Eliason, Juan M. Fons, Worrachet Intachai, Sissades Tongsima, Bjorn Olsen, Stefan T. Arold, Chumpol Ngamphiw, Brad A. Amendt, and et al. 2023. "Genetic Variants in Protein Tyrosine Phosphatase Non-Receptor Type 23 Are Responsible for Mesiodens Formation" Biology 12, no. 3: 393. https://doi.org/10.3390/biology12030393
APA StyleAdisornkanj, P., Chanprasit, R., Eliason, S., Fons, J. M., Intachai, W., Tongsima, S., Olsen, B., Arold, S. T., Ngamphiw, C., Amendt, B. A., Tucker, A. S., & Kantaputra, P. (2023). Genetic Variants in Protein Tyrosine Phosphatase Non-Receptor Type 23 Are Responsible for Mesiodens Formation. Biology, 12(3), 393. https://doi.org/10.3390/biology12030393