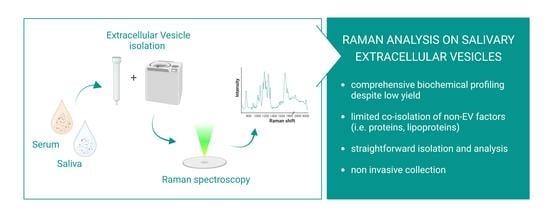
Biochemical Characterization of Human Salivary Extracellular Vesicles as a Valuable Source of Biomarkers
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Subject Recruitment and Sample Collection
2.2. Isolation of Extracellular Vesicles
2.3. EV Size Distribution
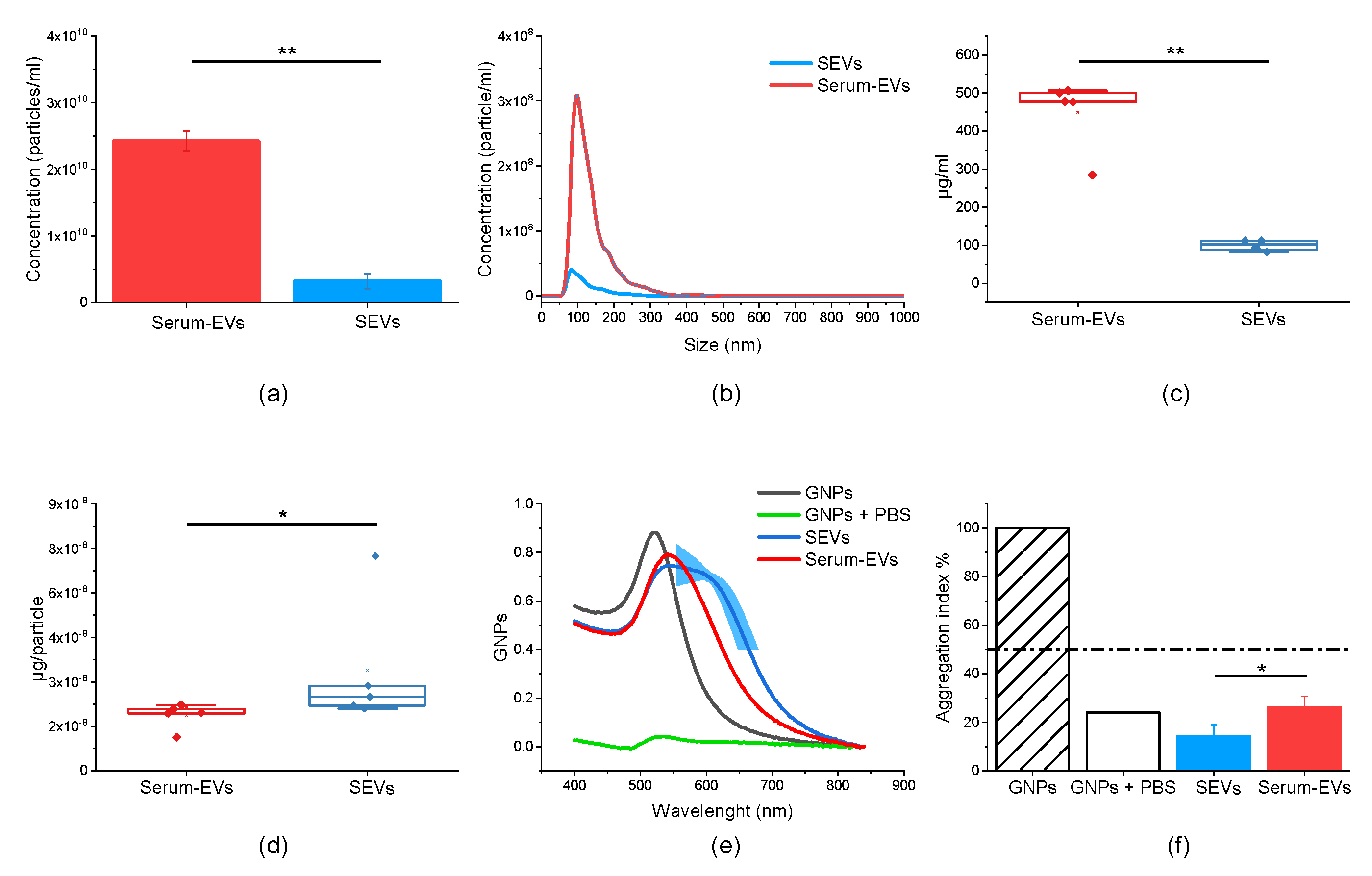
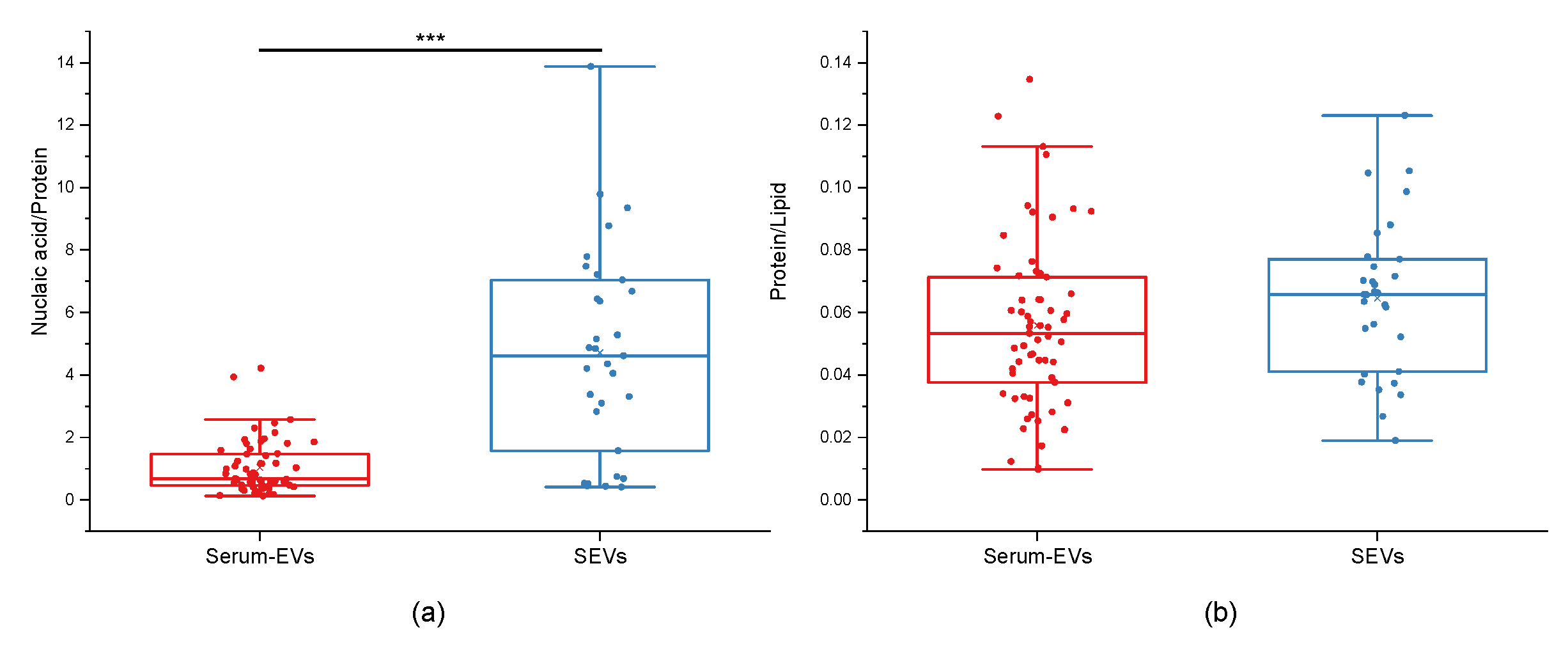
2.4. EVs Purity Assessment
2.5. Raman Analysis
2.6. Statistical Analysis
3. Results
3.1. EV Physico-Chemical Characterization
3.2. Raman Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.F. Extracellular Vesicles and Neurodegenerative Diseases. J. Neurosci. 2019, 39, 9269–9273. [Google Scholar] [CrossRef] [PubMed]
- Martins, T.S.; Vaz, M.; Henriques, A.G. A Review on Comparative Studies Addressing Exosome Isolation Methods from Body Fluids. Anal. Bioanal. Chem. 2022, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Clayton, A.; Boilard, E.; Buzas, E.I.; Cheng, L.; Falcón-Perez, J.M.; Gardiner, C.; Gustafson, D.; Gualerzi, A.; Hendrix, A.; Hoffman, A.; et al. Considerations towards a Roadmap for Collection, Handling and Storage of Blood Extracellular Vesicles. J. Extracell. Vesicles 2019, 8, 1647027. [Google Scholar] [CrossRef]
- Jamaly, S.; Ramberg, C.; Olsen, R.; Latysheva, N.; Webster, P.; Sovershaev, T.; Brækkan, S.K.; Hansen, J.-B. Impact of Preanalytical Conditions on Plasma Concentration and Size Distribution of Extracellular Vesicles Using Nanoparticle Tracking Analysis. Sci. Rep. 2018, 8, 17216. [Google Scholar] [CrossRef]
- Carlomagno, C.; Banfi, P.I.; Gualerzi, A.; Picciolini, S.; Volpato, E.; Meloni, M.; Lax, A.; Colombo, E.; Ticozzi, N.; Verde, F.; et al. Human Salivary Raman Fingerprint as Biomarker for the Diagnosis of Amyotrophic Lateral Sclerosis. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Carlomagno, C.; Bertazioli, D.; Gualerzi, A.; Picciolini, S.; Banfi, P.I.; Lax, A.; Messina, E.; Navarro, J.; Bianchi, L.; Caronni, A.; et al. COVID-19 Salivary Raman Fingerprint: Innovative Approach for the Detection of Current and Past SARS-CoV-2 Infections. Sci. Rep. 2021, 11, 4943. [Google Scholar] [CrossRef]
- Carlomagno, C.; Gualerzi, A.; Picciolini, S.; Rodà, F.; Banfi, P.I.; Lax, A.; Bedoni, M. Characterization of the COPD Salivary Fingerprint through Surface Enhanced Raman Spectroscopy: A Pilot Study. Diagnostics 2021, 11, 508. [Google Scholar] [CrossRef]
- Carlomagno, C.; Bertazioli, D.; Gualerzi, A.; Picciolini, S.; Andrico, M.; Rodà, F.; Meloni, M.; Banfi, P.I.; Verde, F.; Ticozzi, N.; et al. Identification of the Raman Salivary Fingerprint of Parkinson’s Disease Through the Spectroscopic—Computational Combinatory Approach. Front. Neurosci. 2021, 15, 704963. [Google Scholar] [CrossRef]
- Kaczor-Urbanowicz, K.E.; Carreras-Presas, C.M.; Aro, K.; Tu, M.; Garcia-Godoy, F.; Wong, D.T.W. Saliva Diagnostics—Current Views and Directions. Exp. Biol. Med. 2017, 242, 459–472. [Google Scholar] [CrossRef]
- Dawes, C. Circadian Rhythms in Human Salivary Flow Rate and Composition. J. Physiol. 1972, 220, 529–545. [Google Scholar] [CrossRef]
- Hu, S.; Arellano, M.; Boontheung, P.; Wang, J.; Zhou, H.; Jiang, J.; Elashoff, D.; Wei, R.; Loo, J.A.; Wong, D.T. Salivary Proteomics for Oral Cancer Biomarker Discovery. Clin. Cancer Res. 2008, 14, 6246–6252. [Google Scholar] [CrossRef]
- Hu, S.; Wang, J.; Meijer, J.; Ieong, S.; Xie, Y.; Yu, T.; Zhou, H.; Henry, S.; Vissink, A.; Pijpe, J.; et al. Salivary Proteomic and Genomic Biomarkers for Primary Sjögren’s Syndrome. Arthritis Rheum 2007, 56, 3588–3600. [Google Scholar] [CrossRef]
- Zhang, L.; Xiao, H.; Karlan, S.; Zhou, H.; Gross, J.; Elashoff, D.; Akin, D.; Yan, X.; Chia, D.; Karlan, B.; et al. Discovery and Preclinical Validation of Salivary Transcriptomic and Proteomic Biomarkers for the Non-Invasive Detection of Breast Cancer. PLoS ONE 2010, 5, e15573. [Google Scholar] [CrossRef]
- Xiao, H.; Zhang, Y.; Kim, Y.; Kim, S.; Kim, J.J.; Kim, K.M.; Yoshizawa, J.; Fan, L.-Y.; Cao, C.-X.; Wong, D.T.W. Differential Proteomic Analysis of Human Saliva Using Tandem Mass Tags Quantification for Gastric Cancer Detection. Sci. Rep. 2016, 6, 22165. [Google Scholar] [CrossRef]
- Winck, F.V.; Prado Ribeiro, A.C.; Ramos Domingues, R.; Ling, L.Y.; Riaño-Pachón, D.M.; Rivera, C.; Brandão, T.B.; Gouvea, A.F.; Santos-Silva, A.R.; Coletta, R.D.; et al. Insights into Immune Responses in Oral Cancer through Proteomic Analysis of Saliva and Salivary Extracellular Vesicles. Sci. Rep. 2015, 5, 16305. [Google Scholar] [CrossRef]
- Xiao, H.; Wong, D.T.W. Proteomic Analysis of Microvesicles in Human Saliva by Gel Electrophoresis with Liquid Chromatography-Mass Spectrometry. Anal. Chim. Acta 2012, 723, 61–67. [Google Scholar] [CrossRef]
- Deutsch, O.; Fleissig, Y.; Zaks, B.; Krief, G.; Aframian, D.J.; Palmon, A. An Approach to Remove Alpha Amylase for Proteomic Analysis of Low Abundance Biomarkers in Human Saliva. Electrophoresis 2008, 29, 4150–4157. [Google Scholar] [CrossRef]
- Sun, Y.; Xia, Z.; Shang, Z.; Sun, K.; Niu, X.; Qian, L.; Fan, L.-Y.; Cao, C.-X.; Xiao, H. Facile Preparation of Salivary Extracellular Vesicles for Cancer Proteomics. Sci. Rep. 2016, 6, 24669. [Google Scholar] [CrossRef]
- Li, K.; Lin, Y.; Luo, Y.; Xiong, X.; Wang, L.; Durante, K.; Li, J.; Zhou, F.; Guo, Y.; Chen, S.; et al. A Signature of Saliva-Derived Exosomal Small RNAs as Predicting Biomarker for Esophageal Carcinoma: A Multicenter Prospective Study. Mol. Cancer 2022, 21, 21. [Google Scholar] [CrossRef]
- Cheng, Y.; Pereira, M.; Raukar, N.; Reagan, J.L.; Queseneberry, M.; Goldberg, L.; Borgovan, T.; LaFrance, W.C.; Dooner, M.; Deregibus, M.; et al. Potential Biomarkers to Detect Traumatic Brain Injury by the Profiling of Salivary Extracellular Vesicles. J. Cell. Physiol. 2019, 234, 14377–14388. [Google Scholar] [CrossRef] [PubMed]
- Devic, I.; Hwang, H.; Edgar, J.S.; Izutsu, K.; Presland, R.; Pan, C.; Goodlett, D.R.; Wang, Y.; Armaly, J.; Tumas, V.; et al. Salivary α-Synuclein and DJ-1: Potential Biomarkers for Parkinson’s Disease. Brain 2011, 134, e178. [Google Scholar] [CrossRef] [PubMed]
- Rani, K.; Mukherjee, R.; Singh, E.; Kumar, S.; Sharma, V.; Vishwakarma, P.; Bharti, P.S.; Nikolajeff, F.; Dinda, A.K.; Goyal, V.; et al. Neuronal Exosomes in Saliva of Parkinson’s Disease Patients: A Pilot Study. Park. Relat. Disord. 2019, 67, 21–23. [Google Scholar] [CrossRef] [PubMed]
- Gualerzi, A.; Picciolini, S.; Carlomagno, C.; Rodà, F.; Bedoni, M. Biophotonics for Diagnostic Detection of Extracellular Vesicles. Adv. Drug Deliv. Rev. 2021, 174, 229–249. [Google Scholar] [CrossRef] [PubMed]
- Carlomagno, C.; Cabinio, M.; Picciolini, S.; Gualerzi, A.; Baglio, F.; Bedoni, M. SERS-based Biosensor for Alzheimer Disease Evaluation through the Fast Analysis of Human Serum. J. Biophotonics. 2020. [Google Scholar] [CrossRef]
- Krafft, C.; Popp, J. The Many Facets of Raman Spectroscopy for Biomedical Analysis. Anal. Bioanal. Chem. 2015, 407, 699–717. [Google Scholar] [CrossRef]
- Hardy, M.; Kelleher, L.; Gomes, P.d.C.; Buchan, E.; Chu, H.O.M.; Oppenheimer, P.G. Methods in Raman Spectroscopy for Saliva Studies—A Review. Appl. Spectrosc. Rev. 2022, 57, 177–233. [Google Scholar] [CrossRef]
- Zendrini, A.; Paolini, L.; Busatto, S.; Radeghieri, A.; Romano, M.; Wauben, M.H.M.; van Herwijnen, M.J.C.; Nejsum, P.; Borup, A.; Ridolfi, A.; et al. Augmented COlorimetric NANoplasmonic (CONAN) Method for Grading Purity and Determine Concentration of EV Microliter Volume Solutions. Front. Bioeng. Biotechnol. 2020, 7, 452. [Google Scholar] [CrossRef]
- Maiolo, D.; Paolini, L.; Di Noto, G.; Zendrini, A.; Berti, D.; Bergese, P.; Ricotta, D. Colorimetric Nanoplasmonic Assay To Determine Purity and Titrate Extracellular Vesicles. Anal. Chem. 2015, 87, 4168–4176. [Google Scholar] [CrossRef]
- Turkevich, J.; Stevenson, P.C.; Hillier, J. A Study of the Nucleation and Growth Processes in the Synthesis of Colloidal Gold. Discuss. Faraday Soc. 1951, 11, 55. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.K.; Aveyard, J.; Fernig, D.G. Determination of Size and Concentration of Gold Nanoparticles from UV−Vis Spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef]
- Gualerzi, A.; Niada, S.; Giannasi, C.; Picciolini, S.; Morasso, C.; Vanna, R.; Rossella, V.; Masserini, M.; Bedoni, M.; Ciceri, F.; et al. Raman Spectroscopy Uncovers Biochemical Tissue-Related Features of Extracellular Vesicles from Mesenchymal Stromal Cells. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Gualerzi, A.; Kooijmans, S.A.A.; Niada, S.; Picciolini, S.; Brini, A.T.; Camussi, G.; Bedoni, M. Raman Spectroscopy as a Quick Tool to Assess Purity of Extracellular Vesicle Preparations and Predict Their Functionality. J. Extracell. Vesicles 2019, 8, 1568780. [Google Scholar] [CrossRef]
- Gualerzi, A.; Picciolini, S.; Carlomagno, C.; Terenzi, F.; Ramat, S.; Sorbi, S.; Bedoni, M. Raman Profiling of Circulating Extracellular Vesicles for the Stratification of Parkinson’s Patients. Nanomed. Nanotechnol. Biol. Med. 2019, 22, 102097. [Google Scholar] [CrossRef]
- Mihály, J.; Deák, R.; Szigyártó, I.C.; Bóta, A.; Beke-Somfai, T.; Varga, Z. Characterization of Extracellular Vesicles by IR Spectroscopy: Fast and Simple Classification Based on Amide and C[Sbnd]H Stretching Vibrations. Biochim. Et Biophys. Acta Biomembr. 2017, 1859, 459–466. [Google Scholar] [CrossRef]
- Tatischeff, I.; Larquet, E.; Falcón-Pérez, J.M.; Turpin, P.-Y.; Kruglik, S.G. Fast Characterisation of Cell-Derived Extracellular Vesicles by Nanoparticles Tracking Analysis, Cryo-Electron Microscopy, and Raman Tweezers Microspectroscopy. J. Extracell. Vesicles 2012, 1, 19179. [Google Scholar] [CrossRef]
- Krafft, C.; Wilhelm, K.; Eremin, A.; Nestel, S.; von Bubnoff, N.; Schultze-Seemann, W.; Popp, J.; Nazarenko, I. A Specific Spectral Signature of Serum and Plasma-Derived Extracellular Vesicles for Cancer Screening. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 835–841. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Ricciardi, A.; Piuri, G.; Porta, M.D.; Mazzucchelli, S.; Bonizzi, A.; Truffi, M.; Sevieri, M.; Allevi, R.; Corsi, F.; Cazzola, R.; et al. Raman Spectroscopy Characterization of the Major Classes of Plasma Lipoproteins. Vib. Spectrosc. 2020, 109, 103073. [Google Scholar] [CrossRef]
- Rastogi, S.; Rani, K.; Kumar, S. Progression of Cognitive Impairment to Alzheimer’s Disease: Through the Lens of Salivary Extracellular Vesicles. J. Exp. Neurosci. 2021, 16, 263310552110586. [Google Scholar] [CrossRef]
- Yuana, Y.; Böing, A.N.; Grootemaat, A.E.; van der Pol, E.; Hau, C.M.; Cizmar, P.; Buhr, E.; Sturk, A.; Nieuwland, R. Handling and Storage of Human Body Fluids for Analysis of Extracellular Vesicles. J. Extracell. Vesicles 2015, 4, 29260. [Google Scholar] [CrossRef] [PubMed]
- Radeghieri, A.; Alacqua, S.; Zendrini, A.; Previcini, V.; Todaro, F.; Martini, G.; Ricotta, D.; Bergese, P. Active Antithrombin Glycoforms Are Selectively Physiosorbed on Plasma Extracellular Vesicles. J. Extracell. Biol. 2022, 1, e57. [Google Scholar] [CrossRef]
- Rai, A.; Fang, H.; Claridge, B.; Simpson, R.J.; Greening, D.W. Proteomic Dissection of Large Extracellular Vesicle Surfaceome Unravels Interactive Surface Platform. J. Extracell. Vesicles 2021, 10, e12164. [Google Scholar] [CrossRef] [PubMed]
- Kesimer, M.; Scull, M.; Brighton, B.; DeMaria, G.; Burns, K.; O’Neal, W.; Pickles, R.J.; Sheehan, J.K. Characterization of Exosome-like Vesicles Released from Human Tracheobronchial Ciliated Epithelium: A Possible Role in Innate Defense. FASEB J. 2009, 23, 1858–1868. [Google Scholar] [CrossRef]

| Serum | Saliva | ||
|---|---|---|---|
| cm−1 | Assignment | cm−1 | Assignment |
| 760 | Tryptophan | 618 | C-C twisting (protein) |
| 937 | C-C stretch | 760 | Tryptophan |
| 1002 | Phenylalanine | 787 | Phosphatidylserine |
| 1064 | Lipids | 898 | Monosaccharides |
| 1131 | Fatty acid | 971 | C-C stretch |
| 1207 | Hydroxyproline, Tyrosine | 1060 | Lipids |
| 1440 | CH2 lipids | 1209 | Tryptophan and phenylalanine mode |
| 1600–1800 | Amide I | 1290–1400 | CH-bending |
| 2700–3500 | Stretching vibrations of CH, NH, OH | 2700–3500 | Stretching vibrations of CH, NH, OH |
| 2800–3050 | Contributions from acyl chains | 2800–3050 | Contributions from acyl chains |
| 2929–2940 | CH2 asymmetric stretch | 2929–2940 | CH2 asymmetric stretch |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mangolini, V.; Gualerzi, A.; Picciolini, S.; Rodà, F.; Del Prete, A.; Forleo, L.; Rossetto, R.A.; Bedoni, M. Biochemical Characterization of Human Salivary Extracellular Vesicles as a Valuable Source of Biomarkers. Biology 2023, 12, 227. https://doi.org/10.3390/biology12020227
Mangolini V, Gualerzi A, Picciolini S, Rodà F, Del Prete A, Forleo L, Rossetto RA, Bedoni M. Biochemical Characterization of Human Salivary Extracellular Vesicles as a Valuable Source of Biomarkers. Biology. 2023; 12(2):227. https://doi.org/10.3390/biology12020227
Chicago/Turabian StyleMangolini, Valentina, Alice Gualerzi, Silvia Picciolini, Francesca Rodà, Angela Del Prete, Luana Forleo, Rudy Alexander Rossetto, and Marzia Bedoni. 2023. "Biochemical Characterization of Human Salivary Extracellular Vesicles as a Valuable Source of Biomarkers" Biology 12, no. 2: 227. https://doi.org/10.3390/biology12020227
APA StyleMangolini, V., Gualerzi, A., Picciolini, S., Rodà, F., Del Prete, A., Forleo, L., Rossetto, R. A., & Bedoni, M. (2023). Biochemical Characterization of Human Salivary Extracellular Vesicles as a Valuable Source of Biomarkers. Biology, 12(2), 227. https://doi.org/10.3390/biology12020227