Identification of Potential Hub Genes Related to Aflatoxin B1, Liver Fibrosis and Hepatocellular Carcinoma via Integrated Bioinformatics Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source and Identification of DEGs
2.2. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
2.3. Protein-Protein Interaction (PPI) and Hub Gene Discovery
2.4. Gene Set Enrichment Analysis (GSEA)
2.5. Statistical Analysis
3. Results
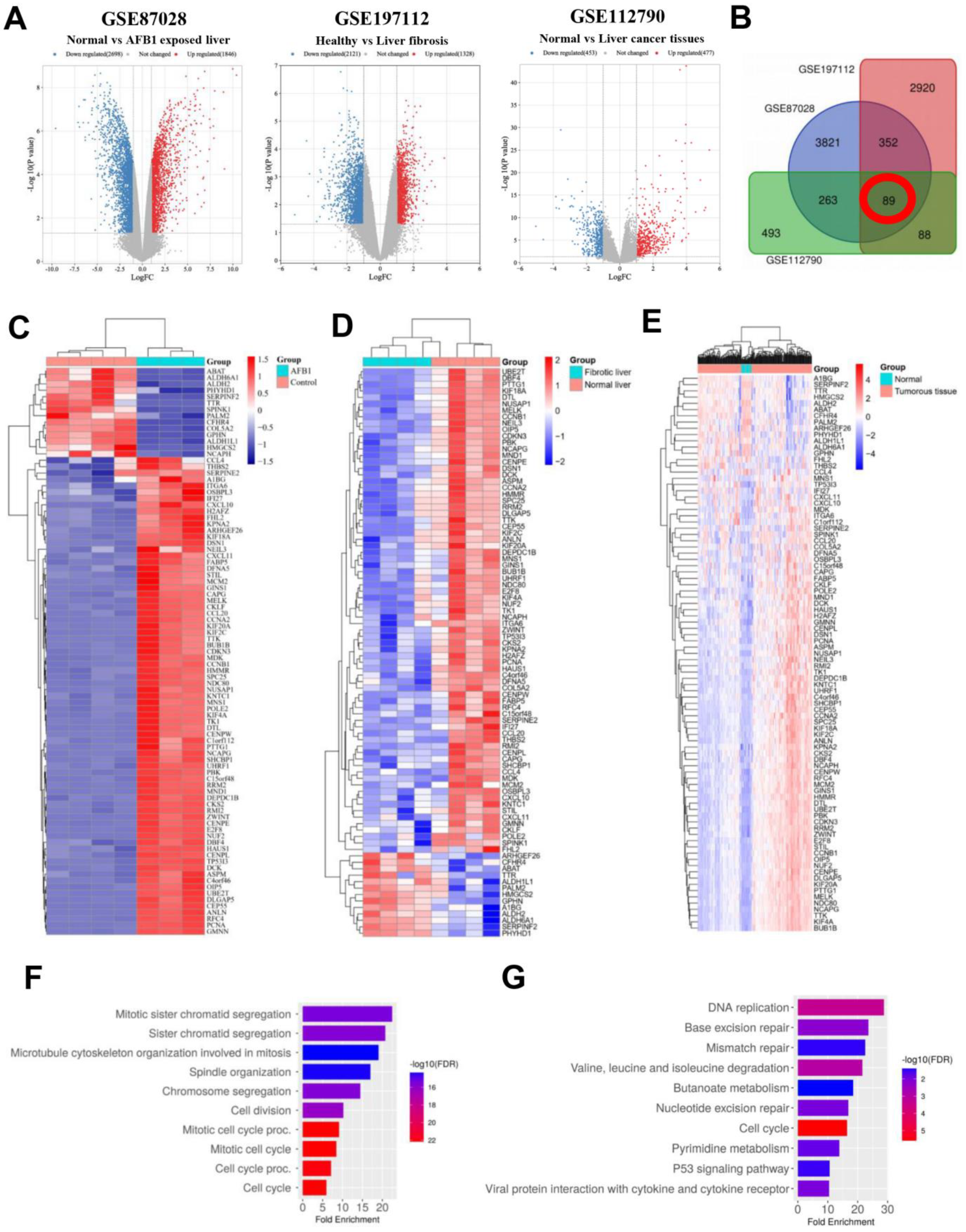
3.1. Identification and Analysis of DEGs
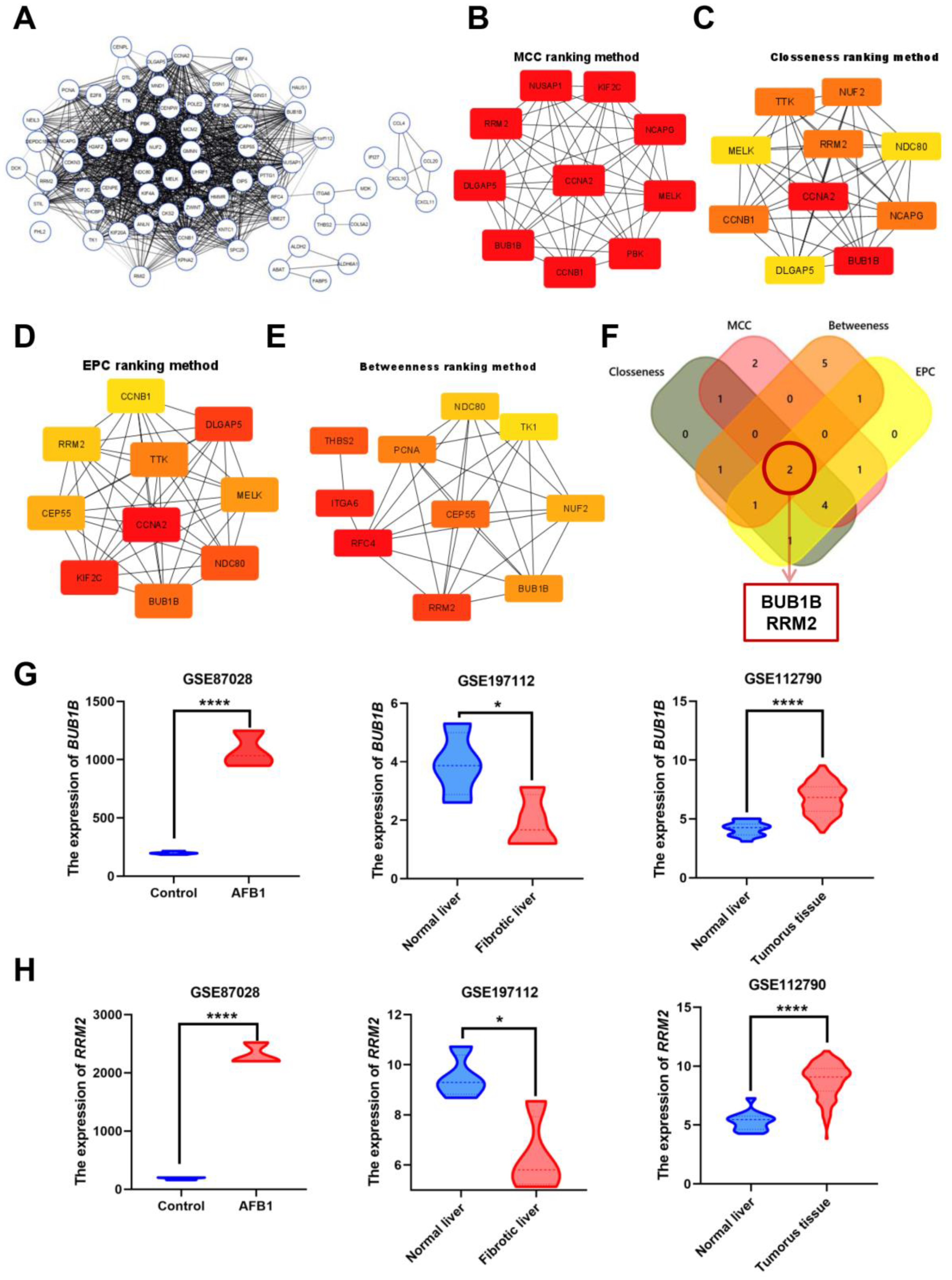
3.2. Identification and Analysis of Hub Genes
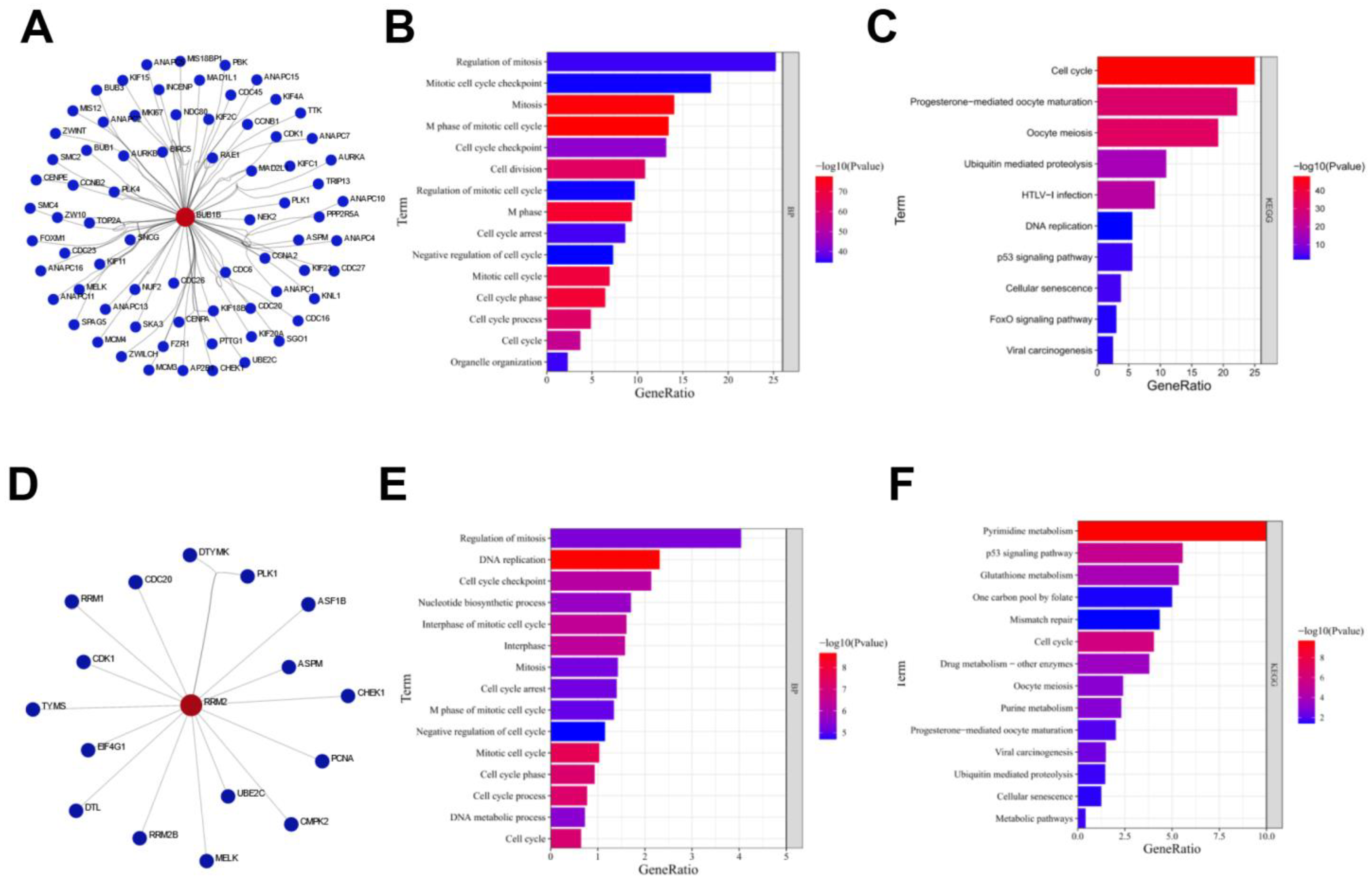
3.3. Functional Enrichment Analysis of Proteins Interacting with BUB1B or RRM2
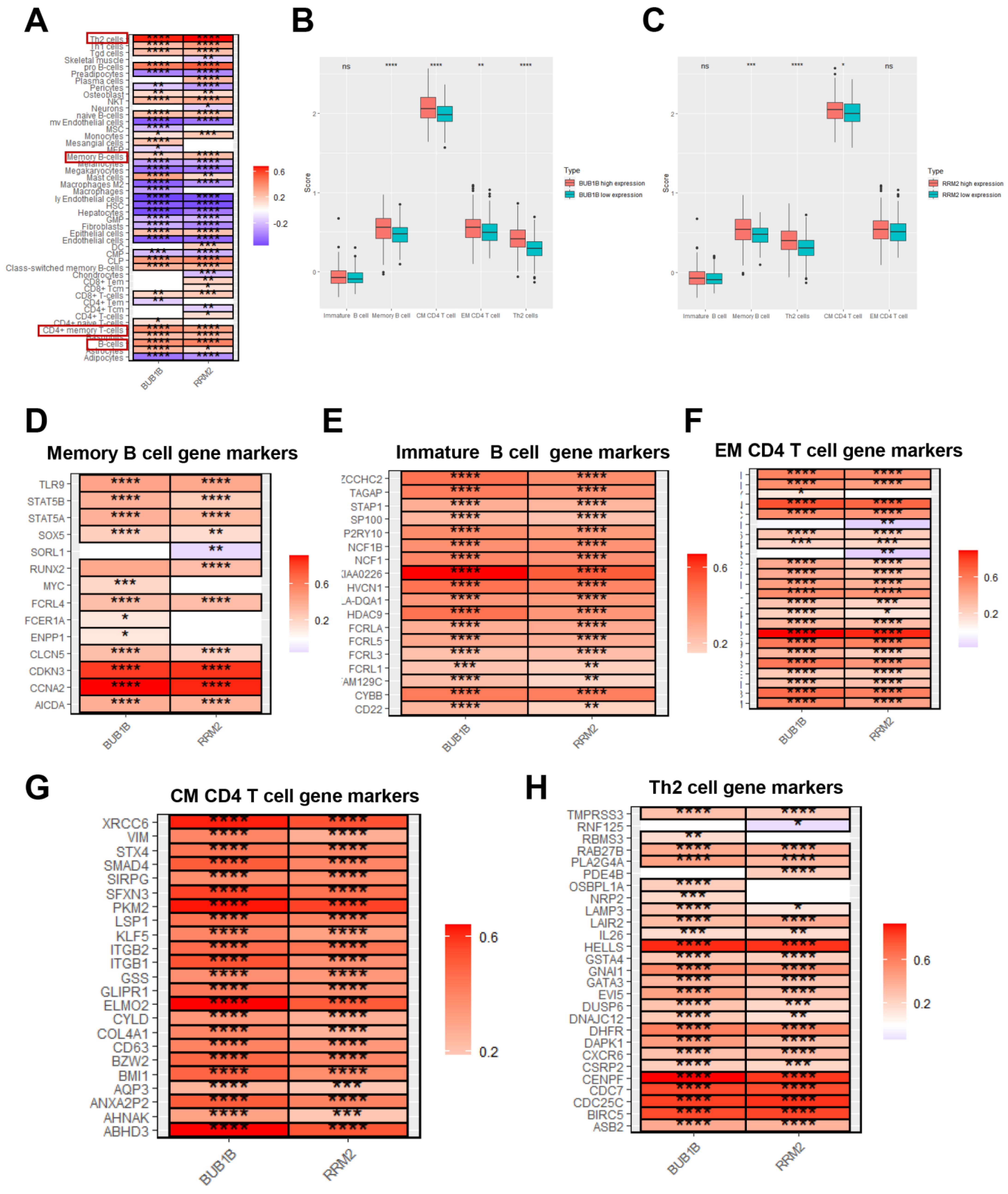
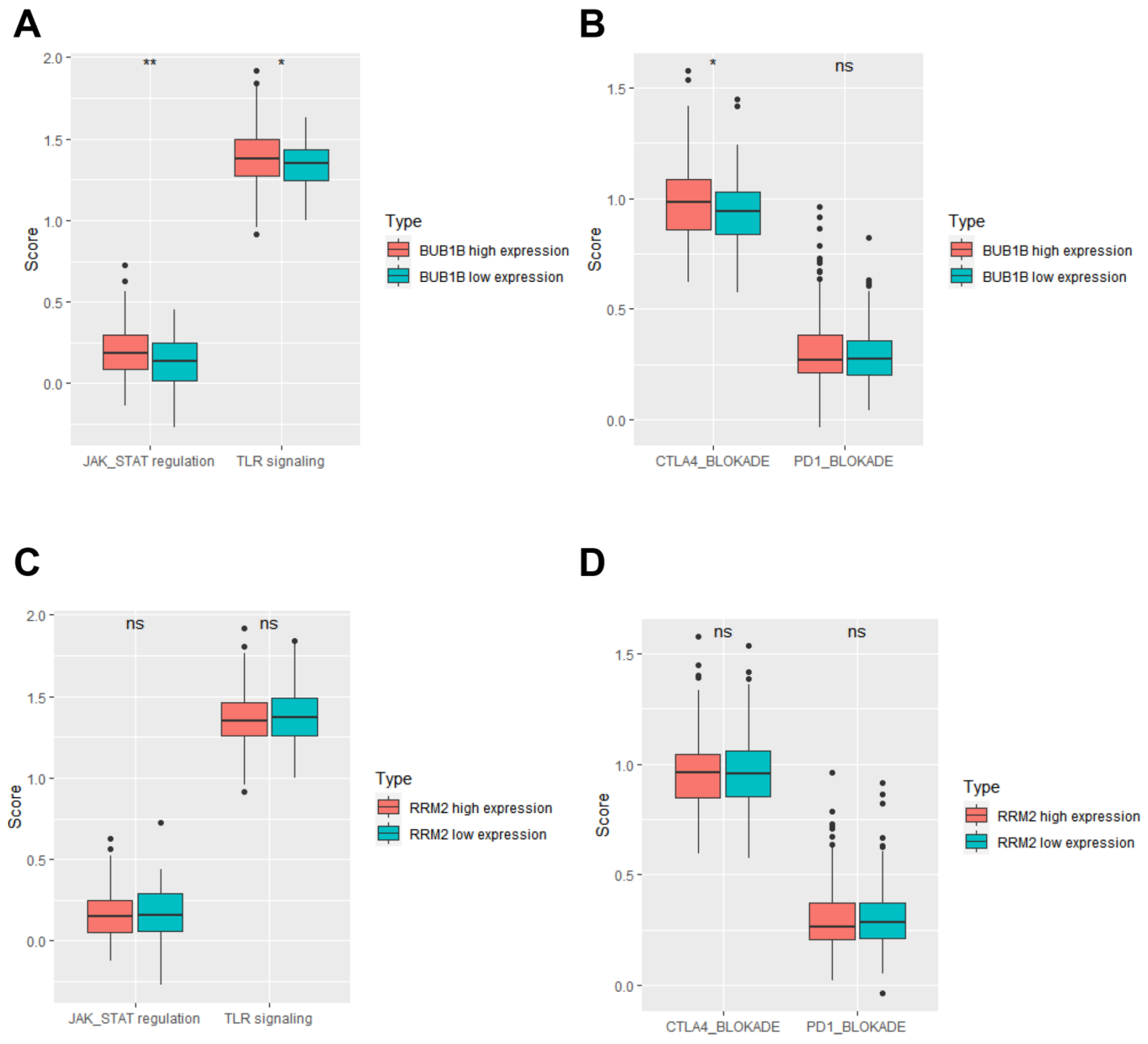
3.4. BUB1B and RRM2 Are Related to Immune Cell Infiltration
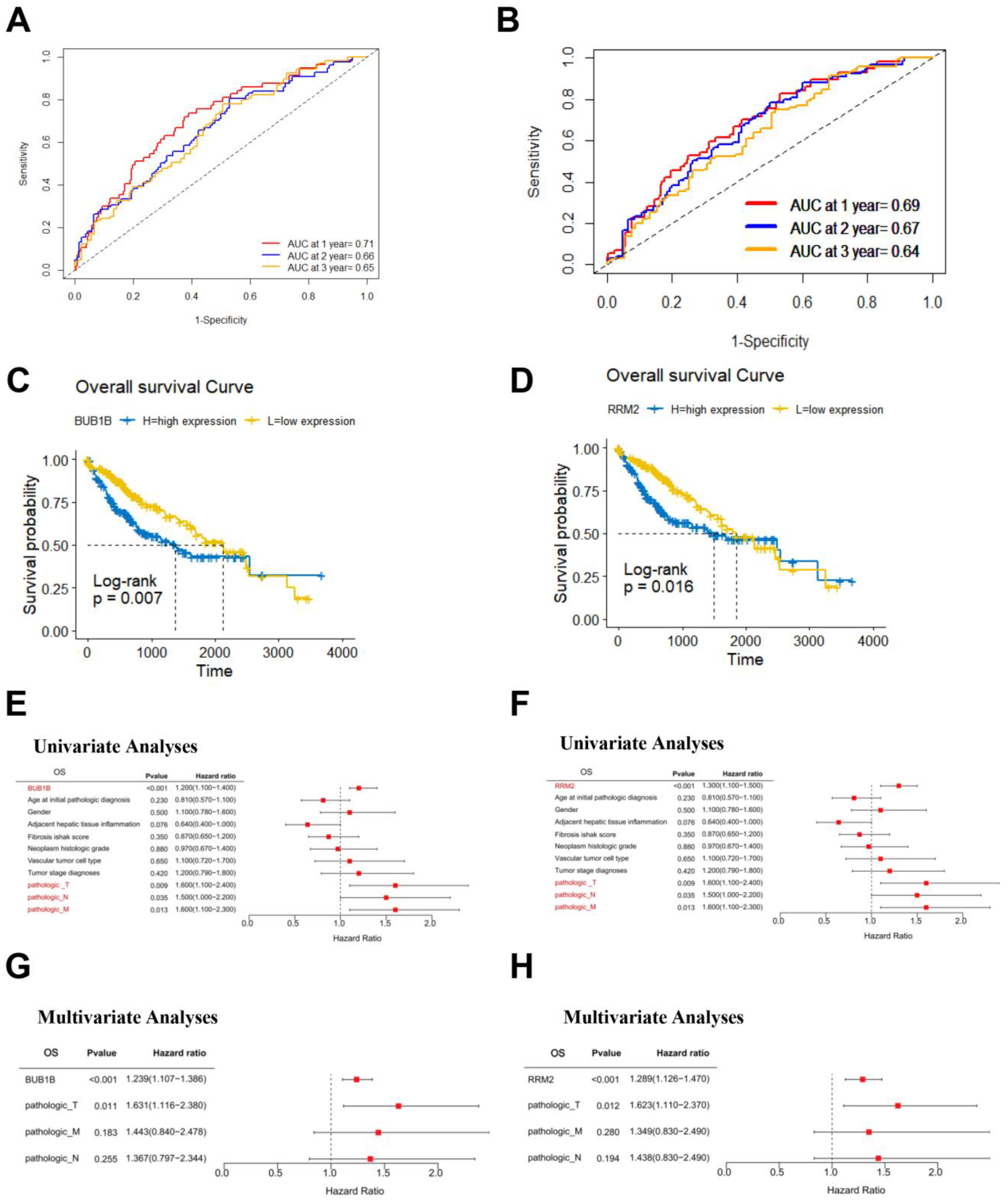
3.5. Survival Analysis of BUB1B and RRM2 in TCGA-LIHC Cohort
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFB1 | aflatoxin b1 |
| DEG | differentially expressed genes |
| HCC | hepatocellular carcinoma |
| PPI | protein-protein interaction |
| GEO | Gene Expression Omnibus |
| TCGA | The Cancer Genome Atlas |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO-BP | Gene ontology biological process |
| BUB1B | Mitotic Checkpoint Serine/Threonine Kinase B |
| RRM2 | Ribonucleotide Reductase Regulatory Subunit M2 |
References
- Tsochatzis, E.A.; Bosch, J.; Burroughs, A.K. Liver cirrhosis. Lancet 2014, 383, 1749–1761. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.C.; Carvajal, M.; Garner, R.C. Does aflatoxin exposure in the United Kingdom constitute a cancer risk? Environ. Health Perspect. 1993, 99, 99–105.4. [Google Scholar] [CrossRef]
- Wang, J.S.; Groopman, J.D. DNA damage by mycotoxins. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1999, 424, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Eom, S.Y.; Yim, D.H.; Zhang, Y.; Yun, J.K.; Moon, S.I.; Yun, h.; Kim, h. Dietary aflatoxin B1 intake, genetic polymorphisms of CYP1A2, CYP2E1, EPHX1, GSTM1, and GSTT1, and gastric cancer risk in Korean. Cancer Causes Control. 2013, 24, 1963–1972. [Google Scholar] [CrossRef]
- Cui, A.; Hua, h.; Shao, T.; Song, P.; Kong, Q.; Luo, T.; Jiang, Y. Aflatoxin B1 induces Src phosphorylation and stimulates lung cancer cell migration. Tumor Biol. 2015, 36, 6507–6513. [Google Scholar] [CrossRef]
- Yip, K.Y.; Wan, M.L.Y.; Wong, A.S.T.; Korach, K.S.; El-Nezami, h. Combined low-dose zearalenone and aflatoxin B1 on cell growth and cell-cycle progression in breast cancer MCF-7 cells. Toxicol. Lett. 2017, 281, 139–151. [Google Scholar] [CrossRef]
- Costa, J.; Lima, N.; Santos, C. An overview on possible links between aflatoxin B1 exposure and gallbladder cancer. Mycotoxin Res. 2021, 37, 205–214. [Google Scholar] [CrossRef]
- Kucukcakan, B.; Hayrulai-Musliu, Z. Challenging role of dietary aflatoxin B1 exposure and hepatitis B infection on risk of hepatocellular carcinoma. Open Access Maced. J. Med. Sci. 2015, 3, 363–369. [Google Scholar] [CrossRef]
- Montalto, G.; Cervello, M.; Giannitrapani, L.; Dantona, F.; Terranova, A.; Castagnetta, L.A. Epidemiology, risk factors, and natural history of hepatocellular carcinoma. Ann. NY Acad. Sci. 2002, 963, 13–20. [Google Scholar] [CrossRef]
- Madden, C.R.; Finegold, M.J.; Slagle, B.L. Altered DNA mutation spectrum in aflatoxin b1-treated transgenic mice that express the hepatitis B virus x protein. J. Virol. 2002, 76, 11770–11774. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef]
- Xiao, Y.; Sun, Y.; Liu, G.; Zhao, J.; Gao, Y.; Yeh, S.; Gong, L.; Chang, C. Androgen receptor (AR)/miR-520f-3p/SOX9 signaling is involved in altering hepatocellular carcinoma (HCC) cell sensitivity to the Sorafenib therapy under hypoxia via increasing cancer stem cells phenotype. Cancer Lett. 2019, 444, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Coussens, L.M.; Zitvogel, L.; Palucka, A.K. Neutralizing Tumor-Promoting Chronic Inflammation: A Magic Bullet? Science 2013, 339, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Kalafati, L.; Kourtzelis, I.; Schulte-Schrepping, J.; Li, X.; Hatzioannou, A.; Grinenko, T.; Hagag, E.; Sinha, A.; Has, C.; Dietz, S.; et al. Innate Immune Training of Granulopoiesis Promotes Anti-tumor Activity. Cell 2020, 183, 771–785.e12. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Stewart, P.A.; Welsh, E.A.; Slebos, R.J.C.; Fang, B.; Izumi, V.; Chambers, M.; Zhang, G.; Cen, L.; Pettersson, F.; Zhang, Y.; et al. Proteogenomic landscape of squamous cell lung cancer. Nat. Commun. 2019, 10, 1–17. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer Immunoediting: Integrating Immunity’s Roles in Cancer Suppression and Promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef]
- Machida, Y.J.; Dutta, A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007, 21, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Komura, K.; Inamoto, T.; Tsujino, T.; Matsui, Y.; Konuma, T.; Nishimura, K.; Uchimoto, T.; Tsutsumi, T.; Matsunaga, T.; Maenosono, R.; et al. Increased BUB1B/BUBR1 expression contributes to aberrant DNA repair activity leading to resistance to DNA-damaging agents. Oncogene 2021, 40, 6210–6222. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zheng, J.; Chen, S.; Huang, B.; Li, G.; Feng, Z.; Wang, J.; Xu, S. RRM2 promotes the progression of human glioblastoma. J. Cell. Physiol. 2018, 233, 6759–6767. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.N.; Wilson, E.A.; Malla, R.; Elford, h.L.; Faridi, J.S. Targeting Ribonucleotide Reductase M2 and NF-κB Activation with Didox to Circumvent Tamoxifen Resistance in Breast Cancer. Mol. Cancer Ther. 2015, 14, 2411–2421. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Amin, A.R.; Wang, D.; Koenig, L.; Nannapaneni, S.; Chen, Z.; Wang, Z.; Sica, G.; Deng, X.; Chen, Z.; et al. RRM2 Regulates Bcl-2 in Head and Neck and Lung Cancers: A Potential Target for Cancer Therapy. Clin. Cancer Res. 2013, 19, 3416–3428. [Google Scholar] [CrossRef]
- Wang, N.; Li, Y.; Zhou, J. Downregulation of ribonucleotide reductase subunits M2 induces apoptosis and G1 arrest of cervical cancer cells. Oncol. Lett. 2018, 15, 3719–3725. [Google Scholar] [CrossRef]
- Kops, G.J.P.L.; Foltz, D.R.; Cleveland, D.W. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. USA 2004, 101, 8699–8704. [Google Scholar] [CrossRef]
- Dvorak, h.F. Tumors: Wounds that do not heal-redux. Cancer Immunol. Res. 2015, 3, 1–11. [Google Scholar] [CrossRef]
- Rybinski, B.; Franco-Barraza, J.; Cukierman, E. The wound healing, chronic fibrosis, and cancer progression triad. Physiol. Genom. 2014, 46, 223–244. [Google Scholar] [CrossRef]
- Qiu, J.; Zhang, S.; Wang, P.; Wang, h.; Sha, B.; Peng, h.; Ju, Z.; Rao, J.; Lu, L. BUB1B promotes hepatocellular carcinoma progression via activation of the mTORC1 signaling pathway. Cancer Med. 2020, 9, 8159–8172. [Google Scholar] [CrossRef]
- Giannelli, G.; Koudelkova, P.; Dituri, F.; Mikulits, W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J. Hepatol. 2016, 65, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Feng, J.; Wu, X.; Chu, W.; Zhang, Y.; Li, P. Bioinformatics Analysis of Candidate Genes and Pathways Related to Hepatocellular Carcinoma in China: A Study Based on Public Databases. Pathol. Oncol. Res. 2021, 27, 588532. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Huang, S.; Berger, E.; Liu, L.; Gross, N.; Heinzmann, F.; Ringelhan, M.; Connor, T.O.; Stadler, M.; Meister, M.; et al. Kupffer Cell-Derived Tnf Triggers Cholangiocellular Tumorigenesis through JNK due to Chronic Mitochondrial Dysfunction and ROS. Cancer Cell 2017, 31, 771–789.e6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shi, M.; Chen, T.; Zhang, B. Characterization of the Immune Cell Infiltration Landscape in Head and Neck Squamous Cell Carcinoma to Aid Immunotherapy. Mol. Ther. Nucleic Acids 2020, 22, 298–309. [Google Scholar] [CrossRef]
- Vesely, M.D.; Kershaw, M.H.; Schreiber, R.D.; Smyth, M.J. Natural Innate and Adaptive Immunity to Cancer. Annu. Rev. Immunol. 2011, 29, 235–271. [Google Scholar] [CrossRef] [PubMed]
- Kythreotou, A.; Siddique, A.; Mauri, F.A.; Bower, M.; Pinato, D.J. PD-L1. J. Clin. Pathol. 2018, 71, 189–194. [Google Scholar] [CrossRef]
- Wang, S.-S.; Liu, W.; Ly, D.; Xu, h.; Qu, L.; Zhang, L. Tumor-infiltrating B cells: Their role and application in anti-tumor immunity in lung cancer. Cell. Mol. Immunol. 2018, 16, 6–18. [Google Scholar] [CrossRef]
- Petitprez, F.; Meylan, M.; De Reyniès, A.; Sautes-Fridman, C.; Fridman, W.H. The Tumor Microenvironment in the Response to Immune Checkpoint Blockade Therapies. Front. Immunol. 2020, 11, 784. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamdy, H.; Yang, Y.; Cheng, C.; Liu, Q. Identification of Potential Hub Genes Related to Aflatoxin B1, Liver Fibrosis and Hepatocellular Carcinoma via Integrated Bioinformatics Analysis. Biology 2023, 12, 205. https://doi.org/10.3390/biology12020205
Hamdy H, Yang Y, Cheng C, Liu Q. Identification of Potential Hub Genes Related to Aflatoxin B1, Liver Fibrosis and Hepatocellular Carcinoma via Integrated Bioinformatics Analysis. Biology. 2023; 12(2):205. https://doi.org/10.3390/biology12020205
Chicago/Turabian StyleHamdy, Hayam, Yi Yang, Cheng Cheng, and Qizhan Liu. 2023. "Identification of Potential Hub Genes Related to Aflatoxin B1, Liver Fibrosis and Hepatocellular Carcinoma via Integrated Bioinformatics Analysis" Biology 12, no. 2: 205. https://doi.org/10.3390/biology12020205
APA StyleHamdy, H., Yang, Y., Cheng, C., & Liu, Q. (2023). Identification of Potential Hub Genes Related to Aflatoxin B1, Liver Fibrosis and Hepatocellular Carcinoma via Integrated Bioinformatics Analysis. Biology, 12(2), 205. https://doi.org/10.3390/biology12020205





