The Enrichment of miRNA-Targeted mRNAs in Translationally Less Active over More Active Polysomes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Evolutionarily Conserved miRNA Binding Site Count
2.3. Polysome Profiling
2.4. The Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) Gene Sets
2.5. Statistical Analysis
2.6. LOESS Regression and the Adjustment of the mRNA Abundance Log-Ratio with Open Reading Frame (ORF) Length
3. Results
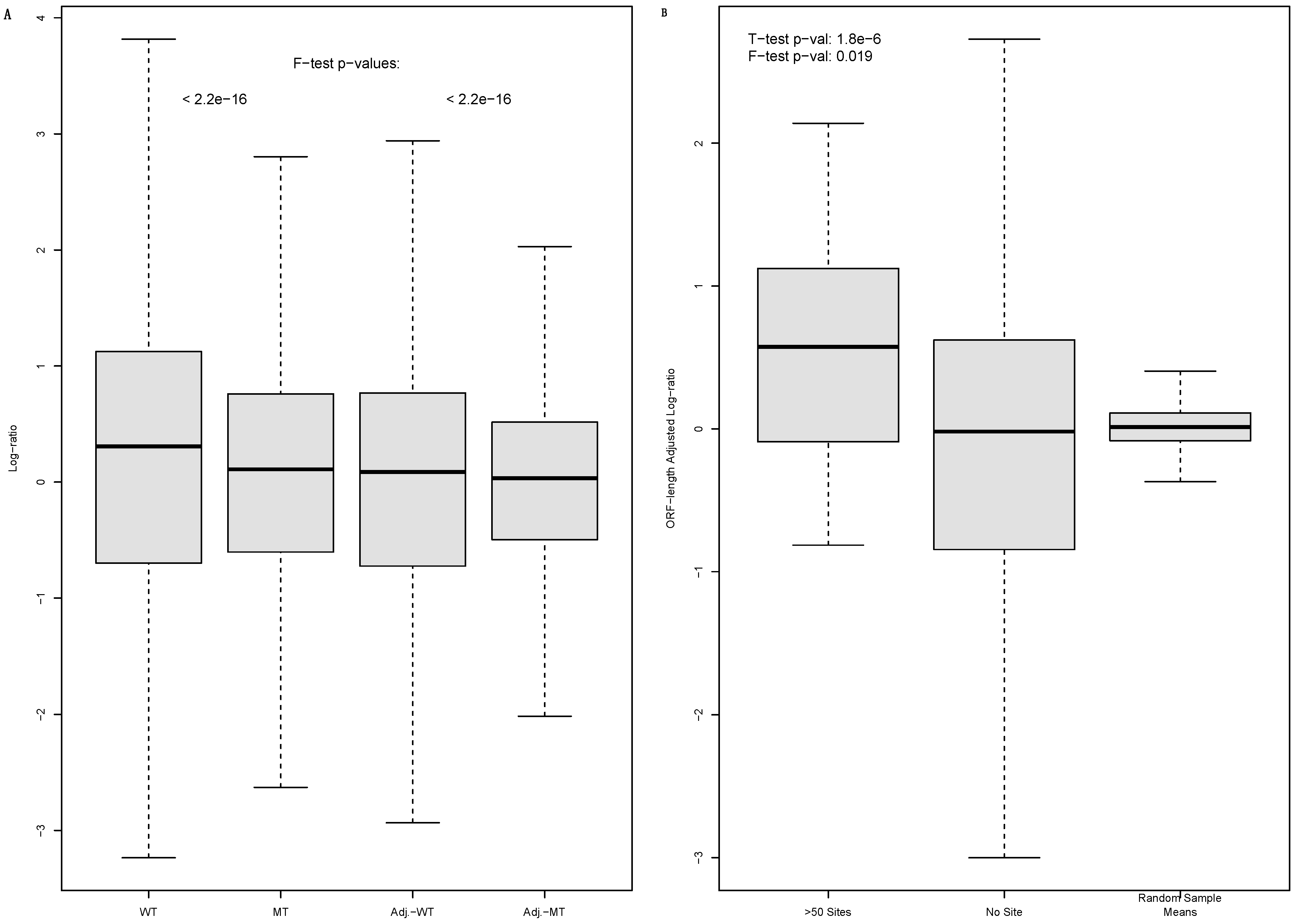
3.1. The Enrichment of MiRNA-Targeted mRNAs in Translationally Less-Active Light Polysomes Suggested by the Comparative WT–Mutant Cell Analysis
3.2. The Inconsistency with Raw Light to Heavy Polysome mRNA Abundance Log-Ratios of WT Cells
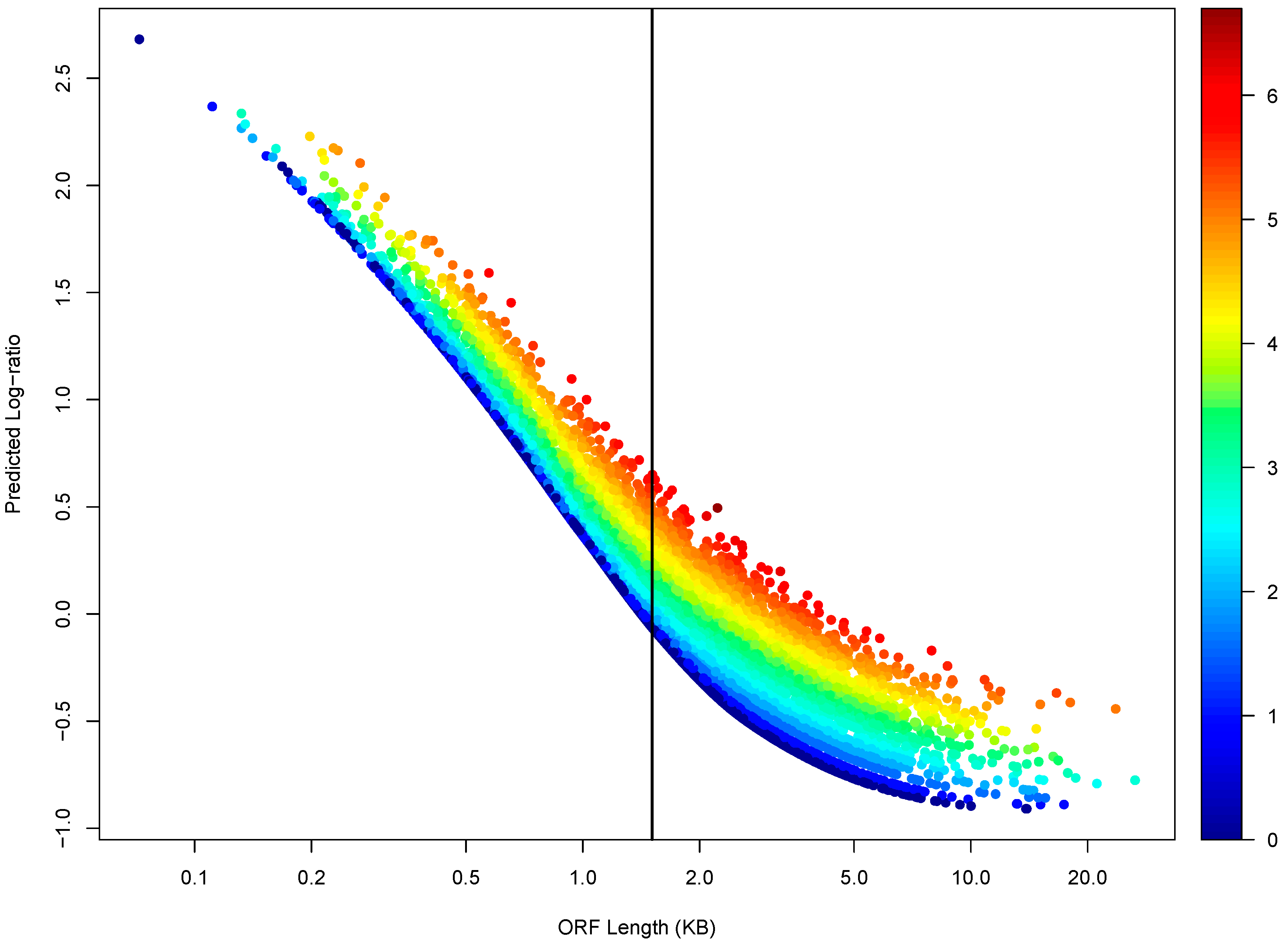
3.3. ORF Length as a Determinant of Light-to-Heavy Polysome mRNA Abundance Log-Ratio and an Effect of DICER1 Disruption on the Log-Ratio
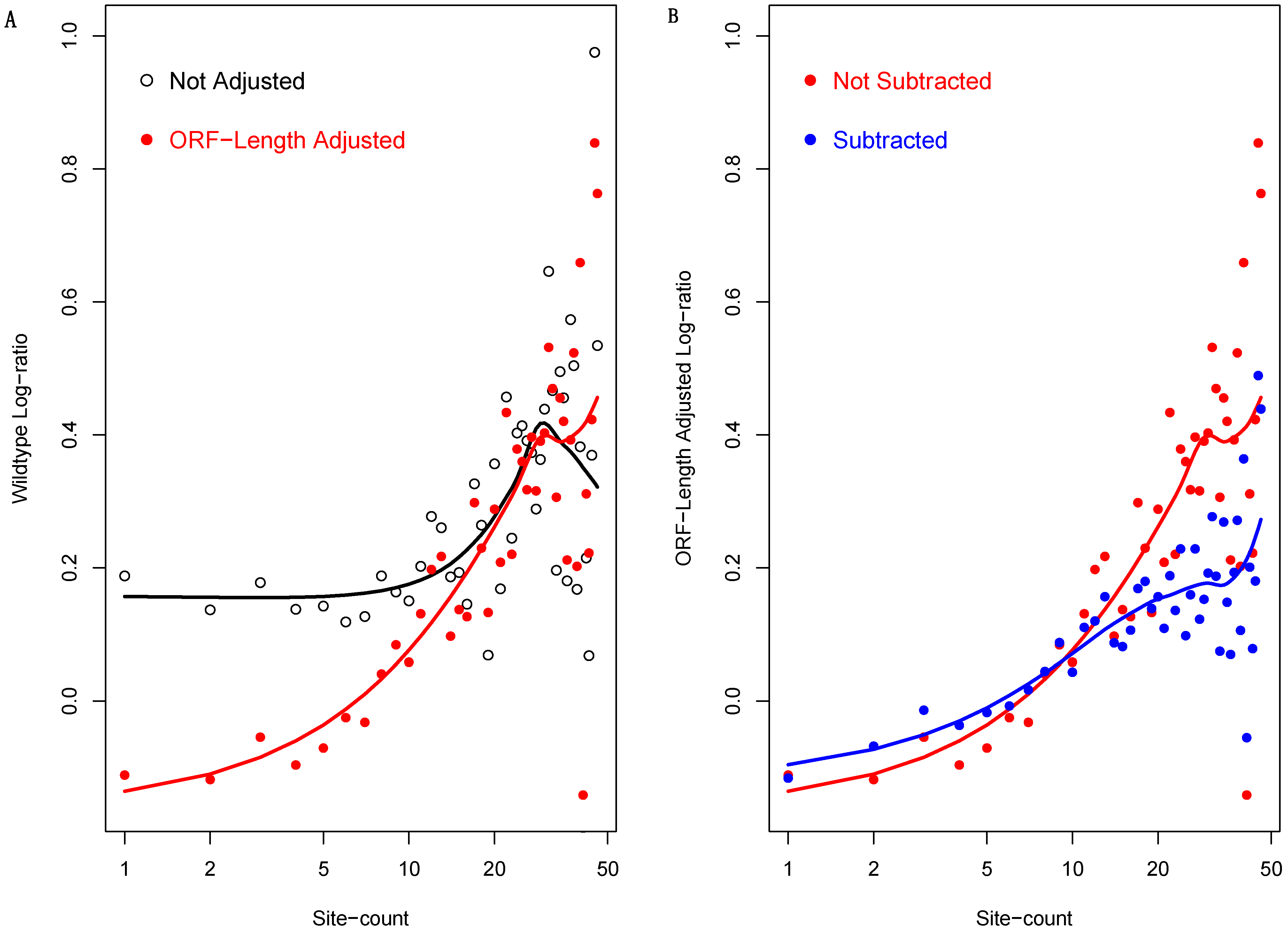
3.4. The ORF-Length-Adjusted Light to Heavy Polysome mRNA Abundance Log-Ratio
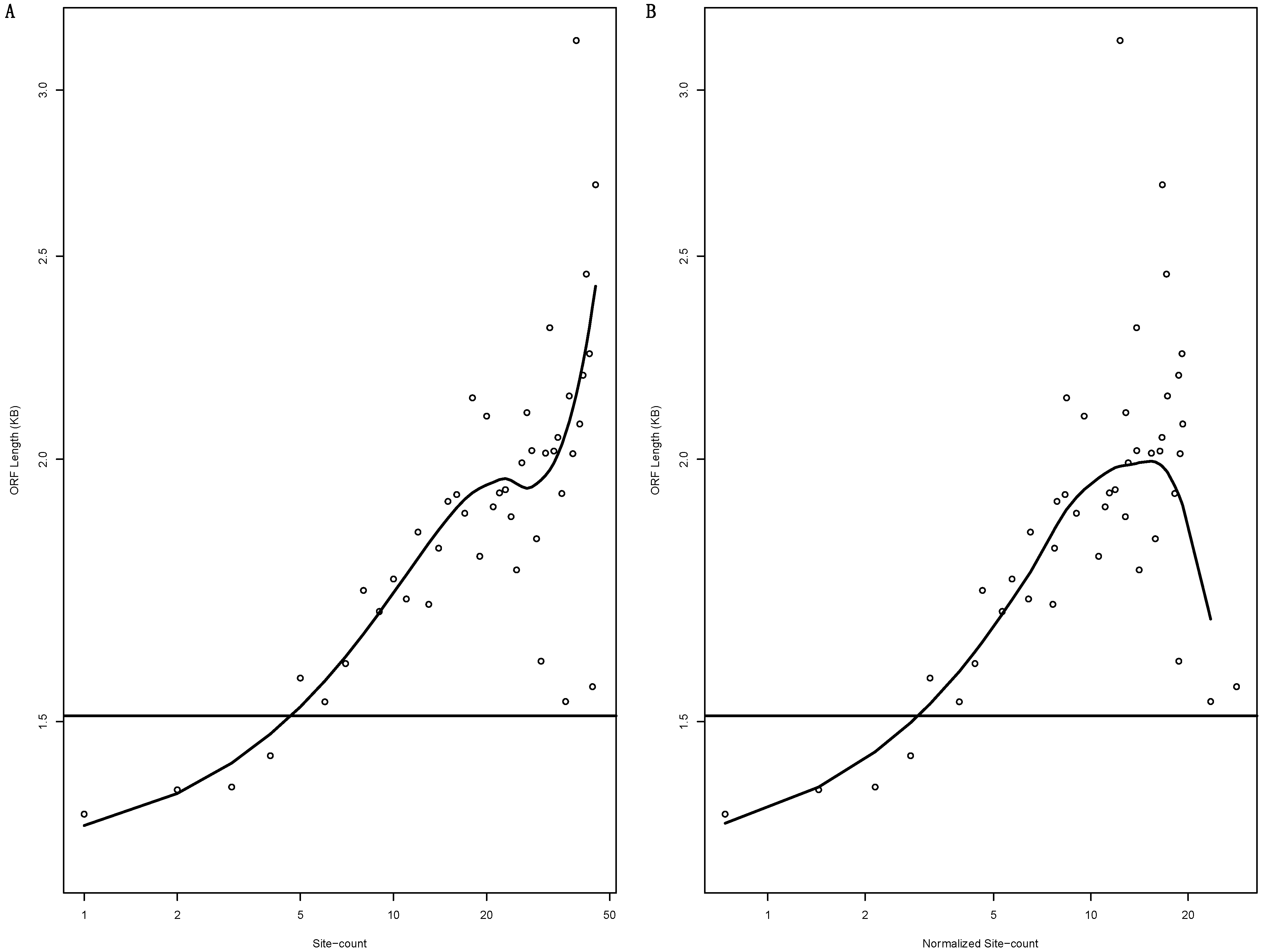
3.5. MiRNA-Targeted mRNAs Tend to Have Longer ORFs
3.6. Incorporating ORF Length into the Analysis
3.7. The Enrichment of Protein Kinase and Transcription Factor in Light-Polysome-Enriched mRNAs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 2019, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Blandino, G.; Fazi, F.; Donzelli, S.; Kedmi, M.; Sas-Chen, A.; Muti, P.; Strano, S.; Yarden, Y. Tumor suppressor microRNAs: A novel non-coding alliance against cancer. FEBS Lett. 2014, 588, 2639–2652. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Ziebarth, J.D.; Cui, Y. SomamiR: A database for somatic mutations impacting microRNA function in cancer. Nucleic Acids Res. 2013, 41, D977–D982. [Google Scholar] [CrossRef]
- Bhaumik, P.; Gopalakrishnan, C.; Kamaraj, B.; Purohit, R. Single nucleotide polymorphisms in microRNA binding sites: Implications in colorectal cancer. Sci. World J. 2014, 2014, 547154. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Haase, A.D.; Jaskiewicz, L.; Zhang, H.; Laine, S.; Sack, R.; Gatignol, A.; Filipowicz, W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005, 6, 961–967. [Google Scholar] [CrossRef] [PubMed]
- MacRae, I.J.; Ma, E.; Zhou, M.; Robinson, C.V.; Doudna, J.A. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. USA 2008, 105, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.S.; Lai, E.C. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell 2011, 43, 892–903. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef]
- Xie, M.; Li, M.; Vilborg, A.; Lee, N.; Shu, M.D.; Yartseva, V.; Sestan, N.; Steitz, J.A. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell 2013, 155, 1568–1580. [Google Scholar] [CrossRef]
- Yang, J.S.; Maurin, T.; Robine, N.; Rasmussen, K.D.; Jeffrey, K.L.; Chandwani, R.; Papapetrou, E.P.; Sadelain, M.; O’Carroll, D.; Lai, E.C. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 15163–15168. [Google Scholar] [CrossRef]
- Kim, Y.K.; Kim, B.; Kim, V.N. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E1881–E1889. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Jannot, G.; Bajan, S.; Giguere, N.J.; Bouasker, S.; Banville, I.H.; Piquet, S.; Hutvagner, G.; Simard, M.J. The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO Rep. 2011, 12, 581–586. [Google Scholar] [CrossRef]
- Kim, J.; Krichevsky, A.; Grad, Y.; Hayes, G.D.; Kosik, K.S.; Church, G.M.; Ruvkun, G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. USA 2004, 101, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Maroney, P.A.; Yu, Y.; Fisher, J.; Nilsen, T.W. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006, 13, 1102–1107. [Google Scholar] [CrossRef] [PubMed]
- Molotski, N.; Soen, Y. Differential association of microRNAs with polysomes reflects distinct strengths of interactions with their mRNA targets. RNA 2012, 18, 1612–1623. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.T.; Hatzigeorgiou, A.G.; Mourelatos, Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA 2004, 10, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Nottrott, S.; Simard, M.J.; Richter, J.D. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006, 13, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- Brattain, M.G.; Fine, W.D.; Khaled, F.M.; Thompson, J.; Brattain, D.E. Heterogeneity of malignant cells from a human colonic carcinoma. Cancer Res. 1981, 41, 1751–1756. [Google Scholar] [PubMed]
- Boyd, D.D.; Levine, A.E.; Brattain, D.E.; McKnight, M.K.; Brattain, M.G. Comparison of growth requirements of two human intratumoral colon carcinoma cell lines in monolayer and soft agarose. Cancer Res. 1988, 48, 2469–2474. [Google Scholar] [PubMed]
- Cummins, J.M.; He, Y.; Leary, R.J.; Pagliarini, R.; Diaz, L.A., Jr.; Sjoblom, T.; Barad, O.; Bentwich, Z.; Szafranska, A.E.; Labourier, E.; et al. The colorectal microRNAome. Proc. Natl. Acad. Sci. USA 2006, 103, 3687–3692. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Wang, D.; Karamyshev, A.L. Next Generation Sequencing (NGS) Application in Multiparameter Gene Expression Analysis. Methods Mol. Biol. 2020, 2102, 17–34, Correction in Methods Mol. Biol. 2020, 2102, C1. [Google Scholar] [PubMed]
- Tian, S.; Wang, J.; Zhang, F.; Wang, D. Comparative Analysis of microRNA Binding Site Distribution and microRNA-Mediated Gene Expression Repression of Oncogenes and Tumor Suppressor Genes. Genes 2022, 13, 481. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, T.; Gill, A.; Hilliard, T.; Chen, F.; Karamyshev, A.L.; Zhang, F. Uncovering the cellular capacity for intensive and specific feedback self-control of the argonautes and MicroRNA targeting activity. Nucleic Acids Res. 2020, 48, 4681–4697. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Padawer, T.; Leighty, R.E.; Wang, D. Duplicate gene enrichment and expression pattern diversification in multicellularity. Nucleic Acids Res. 2012, 40, 7597–7605. [Google Scholar] [CrossRef]
- Guo, Z.; Jiang, W.; Lages, N.; Borcherds, W.; Wang, D. Relationship between gene duplicability and diversifiability in the topology of biochemical networks. BMC Genom. 2014, 15, 577. [Google Scholar] [CrossRef]
- Barabasi, A.L.; Albert, R. Emergence of scaling in random networks. Science 1999, 286, 509–512. [Google Scholar] [CrossRef]
- Karev, G.P.; Wolf, Y.I.; Rzhetsky, A.Y.; Berezovskaya, F.S.; Koonin, E.V. Birth and death of protein domains: A simple model of evolution explains power law behavior. BMC Evol. Biol. 2002, 2, 18. [Google Scholar] [CrossRef] [PubMed]
- Searls, D.B. The language of genes. Nature 2002, 420, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Wuchty, S. Scale-free behavior in protein domain networks. Mol. Biol. Evol. 2001, 18, 1694–1702. [Google Scholar] [CrossRef] [PubMed]
- Das, A.B. Small-world networks of prognostic genes associated with lung adenocarcinoma development. Genomics 2020, 112, 4078–4088. [Google Scholar] [CrossRef] [PubMed]
- Khanin, R.; Wit, E. How scale-free are biological networks. J. Comput. Biol. 2006, 13, 810–818. [Google Scholar] [CrossRef] [PubMed]
- Lima-Mendez, G.; van Helden, J. The powerful law of the power law and other myths in network biology. Mol. Biosyst. 2009, 5, 1482–1493. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, D. The Pattern of microRNA Binding Site Distribution. Genes 2017, 8, 296. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Consortium, T.G.O.; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Berg, J.M.; Stryer, L.; Tymoczko, J.L.; Gatto, G.J. Biochemistry; Macmillan Learning: New York, NY, USA, 2015. [Google Scholar]
- Jewett, M.C.; Miller, M.L.; Chen, Y.; Swartz, J.R. Continued protein synthesis at low [ATP] and [GTP] enables cell adaptation during energy limitation. J. Bacteriol. 2009, 191, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, F.; Schwarzl, T.; Valcarcel, J.; Hentze, M.W. RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 2021, 22, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Corley, M.; Burns, M.C.; Yeo, G.W. How RNA-Binding Proteins Interact with RNA: Molecules and Mechanisms. Mol. Cell 2020, 78, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M., Jr.; Jungkamp, A.C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Tat, T.T.; Maroney, P.A.; Chamnongpol, S.; Coller, J.; Nilsen, T.W. Cotranslational microRNA mediated messenger RNA destabilization. eLife 2016, 5, e12880. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, S.W.; Guo, H.; McGeary, S.E.; Rodriguez-Mias, R.A.; Shin, C.; Baek, D.; Hsu, S.H.; Ghoshal, K.; Villen, J.; Bartel, D.P. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 2014, 56, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef]
- Brent, R.; Bruck, J. 2020 computing: Can computers help to explain biology? Nature 2006, 440, 416–417. [Google Scholar] [CrossRef]
- Condon, A.; Kirchner, H.; Lariviere, D.; Marshall, W.; Noireaux, V.; Tlusty, T.; Fourmentin, E. Will biologists become computer scientists? A truly interdisciplinary effort by computer scientists and biologists to understand how cells process information may yield new insights for both fields. EMBO Rep. 2018, 19, e46628. [Google Scholar] [CrossRef] [PubMed]
- Partridge, D. A logical alternative for biological computing. Nature 2006, 441, 25. [Google Scholar] [CrossRef] [PubMed]
- Wang, D. Discrepancy between mRNA and protein abundance: Insight from information retrieval process in computers. Comput. Biol. Chem. 2008, 32, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Gribskov, M. Examining the architecture of cellular computing through a comparative study with a computer. J. R. Soc. Interface 2005, 2, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.G. "Molecular gene": Interpretation in the right context. Biol. Philos. 2005, 20, 453–464. [Google Scholar] [CrossRef]
- Thunders, M.; Delahunt, B. Gene of the month: DICER1: Ruler and controller. J. Clin. Pathol. 2021, 74, 69–72. [Google Scholar] [CrossRef]
- Gonzalez, I.A.; Stewart, D.R.; Schultz, K.A.P.; Field, A.P.; Hill, D.A.; Dehner, L.P. DICER1 tumor predisposition syndrome: An evolving story initiated with the pleuropulmonary blastoma. Mod. Pathol. 2022, 35, 4–22. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Priest, J.R.; Duchaine, T.F. DICER1: Mutations, microRNAs and mechanisms. Nat. Rev. Cancer 2014, 14, 662–672. [Google Scholar] [CrossRef]


| Rank | KEGG | GO MF |
|---|---|---|
| 1 | MAPK_signaling_pathway | DNA_binding_transcription_factor_activity |
| 2 | chronic_myeloid_leukemia | cis_regulatory_region_sequence_specific_DNA_binding |
| 3 | neurotrophin_signaling_pathway | sequence_specific_DNA_binding |
| 4 | renal_cell_carcinoma | protein_kinase_activity |
| 5 | ERBb_signaling_pathway | protein_serine_threonine_kinase_activity |
| 6 | glioma | transcription_regulator_activity |
| 7 | pancreatic_cancer | protein_serine_kinase_activity |
| 8 | phosphatidylinositol_signaling_system | G-protein_coupled_receptor_activity |
| 9 | glycosaminoglycan_biosynthesis_keratan_sulfate | molecular_transducer_activity |
| 10 | melanogenesis | voltage_gated_potassium_channel_activity |
| 11 | chemokine_signaling_pathway | nucleoside_triphosphatase_regulator_activity |
| 12 | non_small_cell_lung_cancer | kinase_activity |
| 13 | melanoma | DNA_binding_transcription_activator_activity |
| 14 | neuroactive_ligand_receptor_interaction | delayed_rectifier_potassium_channel_activity |
| 15 | hedgehog_signaling_pathway | receptor_tyrosine_kinase_binding |
| 16 | acute_myeloid_leukemia | UDP_glycosyltransferase_activity |
| 17 | GnRH_signaling_pathway | GTPase_activator_activity |
| 18 | Fc_epsilon_RI_signaling_pathway | phosphatidylinositol_3_kinase_regulator_activity |
| 19 | Fc_gamma_R_mediated_phagocytosis | phosphatase_regulator_activity |
| 20 | colorectal_cancer | DNA_binding_transcription_repressor_activity |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Tian, S.; Tikhonova, E.B.; Karamyshev, A.L.; Wang, J.J.; Zhang, F.; Wang, D. The Enrichment of miRNA-Targeted mRNAs in Translationally Less Active over More Active Polysomes. Biology 2023, 12, 1536. https://doi.org/10.3390/biology12121536
Wang T, Tian S, Tikhonova EB, Karamyshev AL, Wang JJ, Zhang F, Wang D. The Enrichment of miRNA-Targeted mRNAs in Translationally Less Active over More Active Polysomes. Biology. 2023; 12(12):1536. https://doi.org/10.3390/biology12121536
Chicago/Turabian StyleWang, Tingzeng, Shuangmei Tian, Elena B. Tikhonova, Andrey L. Karamyshev, Jing J. Wang, Fangyuan Zhang, and Degeng Wang. 2023. "The Enrichment of miRNA-Targeted mRNAs in Translationally Less Active over More Active Polysomes" Biology 12, no. 12: 1536. https://doi.org/10.3390/biology12121536
APA StyleWang, T., Tian, S., Tikhonova, E. B., Karamyshev, A. L., Wang, J. J., Zhang, F., & Wang, D. (2023). The Enrichment of miRNA-Targeted mRNAs in Translationally Less Active over More Active Polysomes. Biology, 12(12), 1536. https://doi.org/10.3390/biology12121536






