Fishing Innate Immune System Properties through the Transcriptomic Single-Cell Data of Teleostei
Abstract
:Simple Summary
Abstract
1. Introduction
- (I)
- To perform a meta-analysis using currently available datasets for Danio rerio.
- (II)
- To obtain new scRNAseq data for nonmodel teleosts.
- (III)
- To conduct a comprehensive comparison between zebrafish and mammal IIS, including the evolutionary analysis of IIS-related genes, comparative genomics and transcriptomics, and systems biology approaches.
2. A Brief Overview of the Teleostei Innate Immune System
3. Methodology
- “single cell” (immune OR macrophage OR neutrophil OR eosinophil
- OR dendritic cells OR lymphocyte OR NK-like cells OR mast cells
- OR HSPC OR monocyte OR glia) AND “bony fish” [porgn:__txid7898]
- AND “Expression profiling by high throughput sequencing” [Filter].
- “single cell” (kidney OR embryo OR larvae OR intestine
- OR brain OR spleen OR glia OR head kidney OR liver OR heart
- OR neuronal retina OR trunk OR gills OR whole body OR thymus
- OR telencephalon OR blood OR HSPC OR spinal cord OR tail OR thyroid gland)
- AND “bony fish” [porgn:__txid7898]
- AND “Expression profiling by high throughput sequencing” [Filter].
4. Results
4.1. The Available Single-Cell Data for Adult Zebrafish
4.2. The Available Single-Cell Data for Immature Zebrafish
4.3. The Available Single-Cell Data for Other Teleostei
5. Future Directions
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IIS | innate immune system |
| PRRs | Pattern recognition receptors |
| scRNAseq | Single-cell RNA sequencing |
| HSCs | Hematopoietic stem cells |
| HSPCs | Hematopoietic stem and progenitor cells |
References
- Riera Romo, M.; Pérez-Martínez, D.; Castillo Ferrer, C. Innate immunity in vertebrates: An overview. Immunology 2016, 148, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Barreda, D.R.; Neely, H.R.; Flajnik, M.F. Evolution of myeloid cells. Myeloid Cells Health Dis. Synth. 2017, 4, 43–58. [Google Scholar] [CrossRef]
- Cacheiro, P.; Haendel, M.A.; Smedley, D. New models for human disease from the International Mouse Phenotyping Consortium. Mamm. Genome 2019, 30, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Katsandegwaza, B.; Horsnell, W.; Smith, K. Inflammatory bowel disease: A review of pre-clinical murine models of human disease. Int. J. Mol. Sci. 2022, 23, 9344. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Buffenstein, R. The unusual immune system of the naked mole-rat. Extraordinary Biol. Naked-Mole-Rat 2021, 1319, 315–327. [Google Scholar] [CrossRef]
- Chan, J.T.; Kadri, S.; Köllner, B.; Rebl, A.; Korytář, T. RNA-Seq of Single Fish Cells–Seeking Out the Leukocytes Mediating Immunity in Teleost Fishes. Front. Immunol. 2022, 13, 798712. [Google Scholar] [CrossRef] [PubMed]
- Rauta, P.R.; Nayak, B.; Das, S. Immune system and immune responses in fish and their role in comparative immunity study: A model for higher organisms. Immunol. Lett. 2012, 148, 23–33. [Google Scholar] [CrossRef]
- Stream, A.; Madigan, C.A. Zebrafish: An underutilized tool for discovery in host–microbe interactions. Trends Immunol. 2022, 43, 426–437. [Google Scholar] [CrossRef]
- Gomes, M.C.; Mostowy, S. The case for modeling human infection in zebrafish. Trends Microbiol. 2020, 28, 10–18. [Google Scholar] [CrossRef]
- Yu, Y.Y.; Ding, L.G.; Huang, Z.Y.; Xu, H.Y.; Xu, Z. Commensal bacteria-immunity crosstalk shapes mucosal homeostasis in teleost fish. Rev. Aquac. 2021, 13, 2322–2343. [Google Scholar] [CrossRef]
- Iwanami, N. Zebrafish as a model for understanding the evolution of the vertebrate immune system and human primary immunodeficiency. Exp. Hematol. 2014, 42, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Magnadóttir, B. Innate immunity of fish (overview). Fish Shellfish Immunol. 2006, 20, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, A.; Dhawale, S. Development of a method for extracting macrophages from zebrafish, Danio rerio and their use in assessing stress. Acta Ichthyol. Piscat. 2008, 38, 73–77. [Google Scholar] [CrossRef]
- Bavia, L.; Santiesteban-Lores, L.E.; Carneiro, M.C.; Prodocimo, M.M. Advances in the complement system of a teleost fish, Oreochromis niloticus. Fish Shellfish Immunol. 2022, 123, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Suresh, R.; Mosser, D.M. Pattern recognition receptors in innate immunity, host defense, and immunopathology. Adv. Physiol. Educ. 2013, 37, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Y.; Cao, X.; Jin, X.; Jin, T. Pattern recognition receptors in zebrafish provide functional and evolutionary insight into innate immune signaling pathways. Cell. Mol. Immunol. 2017, 14, 80–89. [Google Scholar] [CrossRef]
- Sahoo, B.R. Structure of fish Toll-like receptors (TLR) and NOD-like receptors (NLR). Int. J. Biol. Macromol. 2020, 161, 1602–1617. [Google Scholar] [CrossRef]
- Petit, J.; Bailey, E.C.; Wheeler, R.T.; De Oliveira, C.A.; Forlenza, M.; Wiegertjes, G.F. Studies into β-glucan recognition in fish suggests a key role for the C-type lectin pathway. Front. Immunol. 2019, 10, 280. [Google Scholar] [CrossRef]
- Liu, Z.; Ji, J.; Jiang, X.; Shao, T.; Fan, D.; Jiang, X.; Lin, A.; Xiang, L.; Shao, J. Characterization of cGAS homologs in innate and adaptive mucosal immunities in zebrafish gives evolutionary insights into cGAS-STING pathway. FASEB J. 2020, 34, 7786–7809. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Dijkstra, J.M. Major histocompatibility complex (MHC) genes and disease resistance in fish. Cells 2019, 8, 378. [Google Scholar] [CrossRef]
- Murdoch, C.C.; Rawls, J.F. Commensal microbiota regulate vertebrate innate immunity-insights from the zebrafish. Front. Immunol. 2019, 10, 2100. [Google Scholar] [CrossRef] [PubMed]
- Zapata, A.G. Lympho-Hematopoietic Microenvironments and Fish Immune System. Biology 2022, 11, 747. [Google Scholar] [CrossRef] [PubMed]
- Stosik, M.; Tokarz-Deptuła, B.; Deptuła, W. Haematopoiesis in zebrafish (Danio rerio). Front. Immunol. 2022, 13, 902941. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.; Chua, H.; Gong, Z.; Lam, T.; Sin, Y. Development and maturation of the immune system in zebrafish, Danio rerio: A gene expression profiling, in situ hybridization and immunological study. Dev. Comp. Immunol. 2004, 28, 9–28. [Google Scholar] [CrossRef]
- Kordon, A.O.; Karsi, A.; Pinchuk, L. Innate immune responses in fish: Antigen presenting cells and professional phagocytes. Turk. J. Fish. Aquat. Sci. 2018, 18, 1123–1139. [Google Scholar] [CrossRef]
- Rosowski, E.E. Determining macrophage versus neutrophil contributions to innate immunity using larval zebrafish. Dis. Model. Mech. 2020, 13, dmm041889. [Google Scholar] [CrossRef]
- Mokhtar, D.M.; Zaccone, G.; Alesci, A.; Kuciel, M.; Hussein, M.T.; Sayed, R.K. Main components of fish immunity: An overview of the fish immune system. Fishes 2023, 8, 93. [Google Scholar] [CrossRef]
- Var, S.R.; Byrd-Jacobs, C.A. Role of macrophages and microglia in zebrafish regeneration. Int. J. Mol. Sci. 2020, 21, 4768. [Google Scholar] [CrossRef]
- Sinha, R. Macrophage: A Key Player of Teleost Immune System. In Macrophages-Celebrating 140 Years of Discovery; IntechOpen: London, UK, 2022. [Google Scholar] [CrossRef]
- Wentzel, A.S.; Janssen, J.J.; De Boer, V.C.; Van Veen, W.G.; Forlenza, M.; Wiegertjes, G.F. Fish macrophages show distinct metabolic signatures upon polarization. Front. Immunol. 2020, 11, 152. [Google Scholar] [CrossRef]
- Wentzel, A.S.; Petit, J.; van Veen, W.G.; Fink, I.R.; Scheer, M.H.; Piazzon, M.C.; Forlenza, M.; Spaink, H.P.; Wiegertjes, G.F. Transcriptome sequencing supports a conservation of macrophage polarization in fish. Sci. Rep. 2020, 10, 13470. [Google Scholar] [CrossRef]
- Shwartz, A.; Goessling, W.; Yin, C. Macrophages in zebrafish models of liver diseases. Front. Immunol. 2019, 10, 2840. [Google Scholar] [CrossRef] [PubMed]
- Havixbeck, J.J.; Barreda, D.R. Neutrophil development, migration, and function in teleost fish. Biology 2015, 4, 715–734. [Google Scholar] [CrossRef] [PubMed]
- Reite, O.B.; Evensen, Ø. Inflammatory cells of teleostean fish: A review focusing on mast cells/eosinophilic granule cells and rodlet cells. Fish Shellfish Immunol. 2006, 20, 192–208. [Google Scholar] [CrossRef]
- Sfacteria, A.; Brines, M.; Blank, U. The mast cell plays a central role in the immune system of teleost fish. Mol. Immunol. 2015, 63, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Alesci, A.; Lauriano, E.R.; Aragona, M.; Capillo, G.; Pergolizzi, S. Marking vertebrates langerhans cells, from fish to mammals. Acta Histochem. 2020, 122, 151622. [Google Scholar] [CrossRef] [PubMed]
- Lugo-Villarino, G.; Balla, K.M.; Stachura, D.L.; Bañuelos, K.; Werneck, M.B.; Traver, D. Identification of dendritic antigen-presenting cells in the zebrafish. Proc. Natl. Acad. Sci. USA 2010, 107, 15850–15855. [Google Scholar] [CrossRef]
- Salinas, I.; Zhang, Y.A.; Sunyer, J.O. Mucosal immunoglobulins and B cells of teleost fish. Dev. Comp. Immunol. 2011, 35, 1346–1365. [Google Scholar] [CrossRef]
- Bilal, S.; Etayo, A.; Hordvik, I. Immunoglobulins in teleosts. Immunogenetics 2021, 73, 65–77. [Google Scholar] [CrossRef]
- Fischer, U.; Utke, K.; Somamoto, T.; Köllner, B.; Ototake, M.; Nakanishi, T. Cytotoxic activities of fish leucocytes. Fish Shellfish Immunol. 2006, 20, 209–226. [Google Scholar] [CrossRef]
- Yang, H.; Jia, H.; Zhao, Q.; Luo, K.Q. Visualization of natural killer cell-mediated killing of cancer cells at single-cell resolution in live zebrafish. Biosens. Bioelectron. 2022, 216, 114616. [Google Scholar] [CrossRef]
- Mali, P.; Sanyal, K.B.; Mukherjee, D.; Guchhait, A.; Dash, G. Nonspecific cytotoxic cells (NCC) in fish: A review. J. Interacad 2017, 21, 372–378. [Google Scholar]
- Teng, J.; Cui, M.Y.; Zhao, Y.; Chen, H.J.; Du, W.J.; Xue, L.Y.; Ji, X.S. Expression changes of non-specific cytotoxic cell receptor (NCCRP1) and proliferation and migration of NCCs post-Nocardia seriolae infection in Northern Snakehead. Dev. Comp. Immunol. 2023, 139, 104576. [Google Scholar] [CrossRef] [PubMed]
- Odaka, T.; Suetake, H.; Maeda, T.; Miyadai, T. Teleost basophils have IgM-dependent and dual Ig-independent degranulation systems. J. Immunol. 2018, 200, 2767–2776. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Troup, D.B.; Wilhite, S.E.; Ledoux, P.; Rudnev, D.; Evangelista, C.; Kim, I.F.; Soboleva, A.; Tomashevsky, M.; Marshall, K.A.; et al. NCBI GEO: Archive for high-throughput functional genomic data. Nucleic Acids Res. 2009, 37, D885–D890. [Google Scholar] [CrossRef] [PubMed]
- Papatheodorou, I.; Moreno, P.; Manning, J.; Fuentes, A.M.P.; George, N.; Fexova, S.; Fonseca, N.A.; Füllgrabe, A.; Green, M.; Huang, N.; et al. Expression Atlas update: From tissues to single cells. Nucleic Acids Res. 2020, 48, D77–D83. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Iyer, S.; Lobbardi, R.; Moore, J.C.; Chen, H.; Lareau, C.; Hebert, C.; Shaw, M.L.; Neftel, C.; Suva, M.L.; et al. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J. Exp. Med. 2017, 214, 2875–2887. [Google Scholar] [CrossRef]
- Baron, C.S.; Barve, A.; Muraro, M.J.; van der Linden, R.; Dharmadhikari, G.; Lyubimova, A.; de Koning, E.J.; van Oudenaarden, A. Cell type purification by single-cell transcriptome-trained sorting. Cell 2019, 179, 527–542. [Google Scholar] [CrossRef]
- Jiang, M.; Xiao, Y.; Ma, L.; Wang, J.; Chen, H.; Gao, C.; Liao, Y.; Guo, Q.; Peng, J.; Han, X.; et al. Characterization of the zebrafish cell landscape at single-cell resolution. Front. Cell Dev. Biol. 2021, 9, 743421. [Google Scholar] [CrossRef]
- Avagyan, S.; Henninger, J.; Mannherz, W.; Mistry, M.; Yoon, J.; Yang, S.; Weber, M.; Moore, J.; Zon, L. Resistance to inflammation underlies enhanced fitness in clonal hematopoiesis. Science 2021, 374, 768–772. [Google Scholar] [CrossRef]
- Avagyan, S.; Weber, M.C.; Ma, S.; Prasad, M.; Mannherz, W.P.; Yang, S.; Buenrostro, J.D.; Zon, L.I. Single-cell ATAC-seq reveals GATA2-dependent priming defect in myeloid and a maturation bottleneck in lymphoid lineages. Blood Adv. 2021, 5, 2673–2686. [Google Scholar] [CrossRef]
- Amanda, S.; Tan, T.K.; Ong, J.Z.L.; Theardy, M.S.; Wong, R.W.J.; Huang, X.Z.; Ali, M.Z.; Li, Y.; Gong, Z.; Inagaki, H.; et al. IRF4 drives clonal evolution and lineage choice in a zebrafish model of T-cell lymphoma. Nat. Commun. 2022, 13, 2420. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Yang, Q.; Zhang, S.; Millar, D.G.; Alpert, E.J.; Do, D.; Veloso, A.; Brunson, D.C.; Drapkin, B.J.; Stanzione, M.; et al. Single-cell imaging of T cell immunotherapy responses in vivo. J. Exp. Med. 2021, 218, e20210314. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yu, T.; Tan, X.; Jin, D.; Yang, W.; Zhang, J.; Dai, L.; He, Z.; Li, D.; Zhang, Y.; et al. Renal interstitial cells promote nephron regeneration by secreting prostaglandin E2. Elife 2023, 12, e81438. [Google Scholar] [CrossRef] [PubMed]
- Rubin, S.A.; Baron, C.S.; Pessoa Rodrigues, C.; Duran, M.; Corbin, A.F.; Yang, S.P.; Trapnell, C.; Zon, L.I. Single-cell analyses reveal early thymic progenitors and pre-B cells in zebrafish. J. Exp. Med. 2022, 219, e20220038. [Google Scholar] [CrossRef] [PubMed]
- Binder, V.; Li, W.; Faisal, M.; Oyman, K.; Calkins, D.L.; Shaffer, J.; Teets, E.M.; Sher, S.; Magnotte, A.; Belardo, A.; et al. Microenvironmental control of hematopoietic stem cell fate via CXCL8 and protein kinase C. Cell Rep. 2023, 42. [Google Scholar] [CrossRef] [PubMed]
- Hess, I.; Sagar; O’Meara, I.; Grün, D.; Schorpp, M.; Boehm, T. Stage-specific and cell type-specific requirements of ikzf1 during haematopoietic differentiation in zebrafish. Sci. Rep. 2022, 12, 21401. [Google Scholar] [CrossRef] [PubMed]
- Athanasiadis, E.I.; Botthof, J.G.; Andres, H.; Ferreira, L.; Lio, P.; Cvejic, A. Single-cell RNA-sequencing uncovers transcriptional states and fate decisions in haematopoiesis. Nat. Commun. 2017, 8, 2045. [Google Scholar] [CrossRef]
- Hernández, P.P.; Strzelecka, P.M.; Athanasiadis, E.I.; Hall, D.; Robalo, A.F.; Collins, C.M.; Boudinot, P.; Levraud, J.P.; Cvejic, A. Single-cell transcriptional analysis reveals ILC-like cells in zebrafish. Sci. Immunol. 2018, 3, eaau5265. [Google Scholar] [CrossRef]
- Kondera, E. Haematopoiesis and haematopoietic organs in fish. Anim. Sci. Genet. 2019, 15, 9–16. [Google Scholar] [CrossRef]
- Jiao, A.; Zhang, C.; Wang, X.; Sun, L.; Liu, H.; Su, Y.; Lei, L.; Li, W.; Ding, R.; Ding, C.; et al. Single-cell sequencing reveals the evolution of immune molecules across multiple vertebrate species. J. Adv. Res. 2023, in press. [Google Scholar] [CrossRef]
- Carmona, S.J.; Teichmann, S.A.; Ferreira, L.; Macaulay, I.C.; Stubbington, M.J.; Cvejic, A.; Gfeller, D. Single-cell transcriptome analysis of fish immune cells provides insight into the evolution of vertebrate immune cell types. Genome Res. 2017, 27, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Liu, S.; Chen, L.; Liu, Y.; Gu, C.; Ren, H.q.; Wu, B. Single-cell RNA sequencing reveals size-dependent effects of polystyrene microplastics on immune and secretory cell populations from zebrafish intestines. Environ. Sci. Technol. 2020, 54, 3417–3427. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, L.; Gu, W.; Liu, S.; Wu, B. Heterogeneity effects of nanoplastics and lead on zebrafish intestinal cells identified by single-cell sequencing. Chemosphere 2022, 289, 133133. [Google Scholar] [CrossRef] [PubMed]
- Hayot, G.; Massonot, M.; Keime, C.; Faure, E.; Golzio, C. Loss of autism-candidate CHD8 perturbs neural crest development and intestinal homeostatic balance. Life Sci. Alliance 2023, 6, e202201456. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhao, C.; Yang, Z.; Qu, R.; Li, Y.; Fan, Y.; Tang, J.; Xie, T.; Wen, Z. Cross-organ single-cell transcriptome profiling reveals macrophage and dendritic cell heterogeneity in zebrafish. Cell Rep. 2023, 42, 112793. [Google Scholar] [CrossRef]
- Sanz-Morejon, A.; Garcia-Redondo, A.B.; Reuter, H.; Marques, I.J.; Bates, T.; Galardi-Castilla, M.; Große, A.; Manig, S.; Langa, X.; Ernst, A.; et al. Wilms tumor 1b expression defines a pro-regenerative macrophage subtype and is required for organ regeneration in the zebrafish. Cell Rep. 2019, 28, 1296–1306. [Google Scholar] [CrossRef]
- Koth, J.; Wang, X.; Killen, A.C.; Stockdale, W.T.; Potts, H.G.; Jefferson, A.; Bonkhofer, F.; Riley, P.R.; Patient, R.K.; Göttgens, B.; et al. Runx1 promotes scar deposition and inhibits myocardial proliferation and survival during zebrafish heart regeneration. Development 2020, 147, dev186569. [Google Scholar] [CrossRef]
- Ma, H.; Liu, Z.; Yang, Y.; Feng, D.; Dong, Y.; Garbutt, T.A.; Hu, Z.; Wang, L.; Luan, C.; Cooper, C.D.; et al. Functional coordination of non-myocytes plays a key role in adult zebrafish heart regeneration. EMBO Rep. 2021, 22, e52901. [Google Scholar] [CrossRef]
- Xia, Y.; Duca, S.; Perder, B.; Dündar, F.; Zumbo, P.; Qiu, M.; Yao, J.; Cao, Y.; Harrison, M.R.; Zangi, L.; et al. Activation of a transient progenitor state in the epicardium is required for zebrafish heart regeneration. Nat. Commun. 2022, 13, 7704. [Google Scholar] [CrossRef]
- Botos, M.A.; Arora, P.; Chouvardas, P.; Mercader, N. Transcriptomic data meta-analysis reveals common and injury model specific gene expression changes in the regenerating zebrafish heart. Sci. Rep. 2023, 13, 5418. [Google Scholar] [CrossRef]
- Vliegenthart, A.B.; Tucker, C.S.; Del Pozo, J.; Dear, J.W. Zebrafish as model organisms for studying drug-induced liver injury. Br. J. Clin. Pharmacol. 2014, 78, 1217–1227. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Su, B.; Yu, J.; Wang, J.; Hu, H.; Ren, H.Q.; Wu, B. Combined effects of arsenic and 2,2-dichloroacetamide on different cell populations of zebrafish liver. Sci. Total Environ. 2022, 821, 152961. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.K.; DeRossi, C.; Alter, I.L.; Nayar, S.; Giri, M.; Zhang, C.; Cho, J.H.; Chu, J. Single-cell transcriptomics reveals conserved cell identities and fibrogenic phenotypes in zebrafish and human liver. Hepatol. Commun. 2022, 6, 1711–1724. [Google Scholar] [CrossRef] [PubMed]
- Guilliams, M.; Bonnardel, J.; Haest, B.; Vanderborght, B.; Wagner, C.; Remmerie, A.; Bujko, A.; Martens, L.; Thoné, T.; Browaeys, R.; et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 2022, 185, 379–396. [Google Scholar] [CrossRef] [PubMed]
- Oderberg, I.M.; Goessling, W. Biliary epithelial cells are facultative liver stem cells during liver regeneration in adult zebrafish. JCI Insight 2023, 8, e163929. [Google Scholar] [CrossRef]
- Pan, W.; Godoy, R.S.; Cook, D.P.; Scott, A.L.; Nurse, C.A.; Jonz, M.G. Single-cell transcriptomic analysis of neuroepithelial cells and other cell types of the gills of zebrafish (Danio rerio) exposed to hypoxia. Sci. Rep. 2022, 12, 10144. [Google Scholar] [CrossRef]
- Gillotay, P.; Shankar, M.; Haerlingen, B.; Eski, S.E.; Pozo-Morales, M.; Garteizgogeascoa, I.; Reinhardt, S.; Kränkel, A.; Bläsche, J.; Petzold, A.; et al. Single-cell transcriptome analysis reveals thyrocyte diversity in the zebrafish thyroid gland. EMBO Rep. 2020, 21, e50612. [Google Scholar] [CrossRef]
- Anderson, T.; Mo, J.; Gagarin, E.; Sherwood, D.; Blumenkrantz, M.; Mao, E.; Leon, G.; Levitz, H.; Chen, H.J.; Tseng, K.C.; et al. Ligament injury in adult zebrafish triggers ECM remodeling and cell dedifferentiation for scar-free regeneration. NPJ Regen. Med. 2023, 8, 51. [Google Scholar] [CrossRef]
- Alemany, A.; Florescu, M.; Baron, C.S.; Peterson-Maduro, J.; Van Oudenaarden, A. Whole-organism clone tracing using single-cell sequencing. Nature 2018, 556, 108–112. [Google Scholar] [CrossRef]
- Cronan, M.R.; Beerman, R.W.; Rosenberg, A.F.; Saelens, J.W.; Johnson, M.G.; Oehlers, S.H.; Sisk, D.M.; Smith, K.L.J.; Medvitz, N.A.; Miller, S.E.; et al. Macrophage epithelial reprogramming underlies mycobacterial granuloma formation and promotes infection. Immunity 2016, 45, 861–876. [Google Scholar] [CrossRef]
- Cronan, M.R.; Hughes, E.J.; Brewer, W.J.; Viswanathan, G.; Hunt, E.G.; Singh, B.; Mehra, S.; Oehlers, S.H.; Gregory, S.G.; Kaushal, D.; et al. A non-canonical type 2 immune response coordinates tuberculous granuloma formation and epithelialization. Cell 2021, 184, 1757–1774. [Google Scholar] [CrossRef] [PubMed]
- Robertson, T.F.; Hou, Y.; Schrope, J.; Shen, S.; Rindy, J.; Sauer, J.D.; Dinh, H.Q.; Huttenlocher, A. A tessellated lymphoid network provides whole-body T cell surveillance in zebrafish. Proc. Natl. Acad. Sci. USA 2023, 120, e2301137120. [Google Scholar] [CrossRef] [PubMed]
- Geirsdottir, L.; David, E.; Keren-Shaul, H.; Weiner, A.; Bohlen, S.C.; Neuber, J.; Balic, A.; Giladi, A.; Sheban, F.; Dutertre, C.A.; et al. Cross-species single-cell analysis reveals divergence of the primate microglia program. Cell 2019, 179, 1609–1622. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.; Rost, F.; Machate, A.; Reinhardt, S.; Lesche, M.; Weber, A.; Kuscha, V.; Dahl, A.; Rulands, S.; Brand, M. Single cell sequencing of radial glia progeny reveals the diversity of newborn neurons in the adult zebrafish brain. Development 2020, 147, dev185595. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhao, F.; Chen, B.; Cui, N.; Li, Z.; Qin, J.; Luo, L.; Zhao, C.; Li, L. Alterations in immune cell heterogeneities in the brain of aged zebrafish using single-cell resolution. Sci. China Life Sci. 2023, 66, 1358–1378. [Google Scholar] [CrossRef] [PubMed]
- Lyu, P.; Iribarne, M.; Serjanov, D.; Zhai, Y.; Hoang, T.; Campbell, L.; Boyd, P.; Palazzo, I.; Nagashima, M.; Silva, N.J.; et al. Common and divergent gene regulatory networks control injury-induced and developmental neurogenesis in zebrafish retina. bioRxiv 2023. [Google Scholar] [CrossRef]
- D’Gama, P.P.; Qiu, T.; Cosacak, M.I.; Rayamajhi, D.; Konac, A.; Hansen, J.N.; Ringers, C.; Acuña-Hinrichsen, F.; Hui, S.P.; Olstad, E.W.; et al. Diversity and function of motile ciliated cell types within ependymal lineages of the zebrafish brain. Cell Rep. 2021, 37, 109775. [Google Scholar] [CrossRef]
- Zambusi, A.; Novoselc, K.T.; Hutten, S.; Kalpazidou, S.; Koupourtidou, C.; Schieweck, R.; Aschenbroich, S.; Silva, L.; Yazgili, A.S.; van Bebber, F.; et al. TDP-43 condensates and lipid droplets regulate the reactivity of microglia and regeneration after traumatic brain injury. Nat. Neurosci. 2022, 25, 1608–1625. [Google Scholar] [CrossRef]
- Pandey, S.; Moyer, A.J.; Thyme, S.B. A single-cell transcriptome atlas of the maturing zebrafish telencephalon. Genome Res. 2023, 33, 658–671. [Google Scholar] [CrossRef]
- Morizet, D.; Foucher, I.; Alunni, A.; Bally-Cuif, L. Integrative single-cell transcriptomics clarifies adult neurogenesis and macroglia evolution. bioRxiv 2023. [Google Scholar] [CrossRef]
- Mitchell, D.M.; Sun, C.; Hunter, S.S.; New, D.D.; Stenkamp, D.L. Regeneration associated transcriptional signature of retinal microglia and macrophages. Sci. Rep. 2019, 9, 4768. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.J.; Dorman, L.C.; Vainchtein, I.D.; Horneck, N.C.; Molofsky, A.V. In situ and transcriptomic identification of microglia in synapse-rich regions of the developing zebrafish brain. Nat. Commun. 2021, 12, 5916. [Google Scholar] [CrossRef] [PubMed]
- Bise, T.; Pfefferli, C.; Bonvin, M.; Taylor, L.; Lischer, H.E.; Bruggmann, R.; Jaźwińska, A. The regeneration-responsive element careg monitors activation of Müller glia after MNU-induced damage of photoreceptors in the zebrafish retina. Front. Mol. Neurosci. 2023, 16, 1160707. [Google Scholar] [CrossRef] [PubMed]
- Celotto, L.; Rost, F.; Machate, A.; Bläsche, J.; Dahl, A.; Weber, A.; Hans, S.; Brand, M. Single cell RNA sequencing unravels the transcriptional network underlying zebrafish retina regeneration. Elife 2023, 12, RP86507. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.; Wang, J.; Boyd, P.; Wang, F.; Santiago, C.; Jiang, L.; Yoo, S.; Lahne, M.; Todd, L.J.; Jia, M.; et al. Gene regulatory networks controlling vertebrate retinal regeneration. Science 2020, 370, eabb8598. [Google Scholar] [CrossRef] [PubMed]
- Tsata, V.; Kroehne, V.; Wehner, D.; Rost, F.; Lange, C.; Hoppe, C.; Kurth, T.; Reinhardt, S.; Petzold, A.; Dahl, A.; et al. Reactive oligodendrocyte progenitor cells (re-) myelinate the regenerating zebrafish spinal cord. Development 2020, 147, dev193946. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.K.; Saraswathy, V.M.; Zhou, L.; McAdow, A.R.; Burris, B.; Butka, E.; Morris, S.A.; Dietmann, S.; Mokalled, M.H. Localized EMT reprograms glial progenitors to promote spinal cord repair. Dev. Cell 2021, 56, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Cigliola, V.; Shoffner, A.; Lee, N.; Ou, J.; Gonzalez, T.J.; Hoque, J.; Becker, C.J.; Han, Y.; Shen, G.; Faw, T.D.; et al. Spinal cord repair is modulated by the neurogenic factor Hb-egf under direction of a regeneration-associated enhancer. Nat. Commun. 2023, 14, 4857. [Google Scholar] [CrossRef]
- Xing, L.; Chai, R.; Wang, J.; Lin, J.; Li, H.; Wang, Y.; Lai, B.; Sun, J.; Chen, G. Expression of myelin transcription factor 1 and lamin B receptor mediate neural progenitor fate transition in the zebrafish spinal cord pMN domain. J. Biol. Chem. 2022, 298. [Google Scholar] [CrossRef]
- Fabian, P.; Tseng, K.C.; Thiruppathy, M.; Arata, C.; Chen, H.J.; Smeeton, J.; Nelson, N.; Crump, J.G. Lifelong single-cell profiling of cranial neural crest diversification in zebrafish. Nat. Commun. 2022, 13, 13. [Google Scholar] [CrossRef]
- Rougeot, J.; Torraca, V.; Zakrzewska, A.; Kanwal, Z.; Jansen, H.J.; Sommer, F.; Spaink, H.P.; Meijer, A.H. RNAseq profiling of leukocyte populations in zebrafish larvae reveals a cxcl11 chemokine gene as a marker of macrophage polarization during mycobacterial infection. Front. Immunol. 2019, 10, 832. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Liu, D.; Cui, G.; Ding, Y.; Ai, D.; Gao, S.; Zhang, Y.; Suo, S.; Wang, X.; Lv, P.; et al. A 3D atlas of hematopoietic stem and progenitor cell expansion by multi-dimensional RNA-seq analysis. Cell Rep. 2019, 27, 1567–1578. [Google Scholar] [CrossRef] [PubMed]
- Rawson, A.; Saxena, V.; Gao, H.; Hooks, J.; Xuei, X.; McGuire, P.; Hato, T.; Hains, D.S.; Anderson, R.M.; Schwaderer, A.L. A Pilot Single Cell Analysis of the Zebrafish Embryo Cellular Responses to Uropathogenic Escherichia coli Infection. Pathog. Immun. 2022, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Ratnayake, D.; Nguyen, P.D.; Rossello, F.J.; Wimmer, V.C.; Tan, J.L.; Galvis, L.A.; Julier, Z.; Wood, A.J.; Boudier, T.; Isiaku, A.I.; et al. Macrophages provide a transient muscle stem cell niche via NAMPT secretion. Nature 2021, 591, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Soto, R.A.; Najia, M.A.T.; Hachimi, M.; Frame, J.M.; Yette, G.A.; da Rocha, E.L.; Stankunas, K.; Daley, G.Q.; North, T.E. Sequential regulation of hemogenic fate and hematopoietic stem and progenitor cell formation from arterial endothelium by Ezh1/2. Stem Cell Rep. 2021, 16, 1718–1734. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Cheng, W.; Jia, M.; Chen, L.; Gu, C.; Ren, H.q.; Wu, B. Toxicity of perfluorooctanoic acid on zebrafish early embryonic development determined by single-cell RNA sequencing. J. Hazard. Mater. 2022, 427, 127888. [Google Scholar] [CrossRef]
- Ulloa, B.A.; Habbsa, S.S.; Potts, K.S.; Lewis, A.; McKinstry, M.; Payne, S.G.; Flores, J.C.; Nizhnik, A.; Norberto, M.F.; Mosimann, C.; et al. Definitive hematopoietic stem cells minimally contribute to embryonic hematopoiesis. Cell Rep. 2021, 36, 109703. [Google Scholar] [CrossRef]
- Wattrus, S.J.; Smith, M.L.; Rodrigues, C.P.; Hagedorn, E.J.; Kim, J.W.; Budnik, B.; Zon, L.I. Quality assurance of hematopoietic stem cells by macrophages determines stem cell clonality. Science 2022, 377, 1413–1419. [Google Scholar] [CrossRef]
- Denans, N.; Tran, N.T.; Swall, M.E.; Diaz, D.C.; Blanck, J.; Piotrowski, T. An anti-inflammatory activation sequence governs macrophage transcriptional dynamics during tissue injury in zebrafish. Nat. Commun. 2022, 13, 5356. [Google Scholar] [CrossRef]
- García-López, J.P.; Grimaldi, A.; Chen, Z.; Meneses, C.; Bravo-Tello, K.; Bresciani, E.; Banderas, A.; Burgess, S.M.; Hernández, P.P.; Feijoo, C.G. Ontogenetically distinct neutrophils differ in function and transcriptional profile in zebrafish. Nat. Commun. 2023, 14, 4942. [Google Scholar] [CrossRef]
- Keightley, M.C.; Carradice, D.P.; Layton, J.E.; Pase, L.; Bertrand, J.Y.; Wittig, J.G.; Dakic, A.; Badrock, A.P.; Cole, N.J.; Traver, D.; et al. The Pu. 1 target gene Zbtb11 regulates neutrophil development through its integrase-like HHCC zinc finger. Nat. Commun. 2017, 8, 14911. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, P.; Wang, J.; Ma, L.; E, W.; Suo, S.; Jiang, M.; Li, J.; Chen, H.; Sun, H.; et al. Construction of a cross-species cell landscape at single-cell level. Nucleic Acids Res. 2023, 51, 501–516. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, Y.; Wei, Y.; Zhao, Q.; Lou, X. blf and the drl cluster synergistically regulate cell fate commitment during zebrafish primitive hematopoiesis. Development 2022, 149, dev200919. [Google Scholar] [CrossRef] [PubMed]
- Farnsworth, D.R.; Saunders, L.M.; Miller, A.C. A single-cell transcriptome atlas for zebrafish development. Dev. Biol. 2020, 459, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Blaser, B.W.; Moore, J.L.; Hagedorn, E.J.; Li, B.; Riquelme, R.; Lichtig, A.; Yang, S.; Zhou, Y.; Tamplin, O.J.; Binder, V.; et al. CXCR1 remodels the vascular niche to promote hematopoietic stem and progenitor cell engraftment. J. Exp. Med. 2017, 214, 1011–1027. [Google Scholar] [CrossRef]
- Bresciani, E.; Carrington, B.; Yu, K.; Kim, E.M.; Zhen, T.; Guzman, V.S.; Broadbridge, E.; Bishop, K.; Kirby, M.; Harper, U.; et al. Redundant mechanisms driven independently by RUNX1 and GATA2 for hematopoietic development. Blood Adv. 2021, 5, 4949–4962. [Google Scholar] [CrossRef]
- Blokzijl-Franke, S.; Ponsioen, B.; Schulte-Merker, S.; Herbomel, P.; Kissa, K.; Choorapoikayil, S.; den Hertog, J. Phosphatidylinositol-3 kinase signaling controls survival and stemness of hematopoietic stem and progenitor cells. Oncogene 2021, 40, 2741–2755. [Google Scholar] [CrossRef]
- Solman, M.; Blokzijl-Franke, S.; Piques, F.; Yan, C.; Yang, Q.; Strullu, M.; Kamel, S.M.; Ak, P.; Bakkers, J.; Langenau, D.M.; et al. Inflammatory response in hematopoietic stem and progenitor cells triggered by activating SHP2 mutations evokes blood defects. Elife 2022, 11, e73040. [Google Scholar] [CrossRef]
- Xia, J.; Liu, M.; Zhu, C.; Liu, S.; Ai, L.; Ma, D.; Zhu, P.; Wang, L.; Liu, F. Activation of lineage competence in hemogenic endothelium precedes the formation of hematopoietic stem cell heterogeneity. Cell Res. 2023, 33, 448–463. [Google Scholar] [CrossRef]
- Schiavo, R.K.; Tamplin, O.J. Vascular endothelial growth factor c regulates hematopoietic stem cell fate in the dorsal aorta. Development 2022, 149, dev199498. [Google Scholar] [CrossRef]
- Jerison, E.R.; Quake, S.R. Heterogeneous T cell motility behaviors emerge from a coupling between speed and turning in vivo. Elife 2020, 9, e53933. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Kang, Z.; Xue, Y.; Ding, Y.; Gao, S.; Zhang, Y.; Lv, P.; Wang, X.; Ma, D.; Wang, L.; et al. A single-cell resolution developmental atlas of hematopoietic stem and progenitor cell expansion in zebrafish. Proc. Natl. Acad. Sci. USA 2021, 118, e2015748118. [Google Scholar] [CrossRef] [PubMed]
- Laplace-Builhé, B.; Barthelaix, A.; Assou, S.; Bohaud, C.; Pratlong, M.; Severac, D.; Tejedor, G.; Luz-Crawford, P.; Nguyen-Chi, M.; Mathieu, M.; et al. NRG1/ErbB signalling controls the dialogue between macrophages and neural crest-derived cells during zebrafish fin regeneration. Nat. Commun. 2021, 12, 6336. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.B.; Wang, J.; Hong, Y.; Li, H.; Fan, D.D.; Lin, A.F.; Xiang, L.X.; Shao, J.Z. Single-cell transcriptome profiling reveals diverse immune cell populations and their responses to viral infection in the spleen of zebrafish. FASEB J. 2023, 37, e22951. [Google Scholar] [CrossRef] [PubMed]
- Nayar, S.; Morrison, J.K.; Giri, M.; Gettler, K.; Chuang, L.s.; Walker, L.A.; Ko, H.M.; Kenigsberg, E.; Kugathasan, S.; Merad, M.; et al. A myeloid–stromal niche and gp130 rescue in NOD2-driven Crohn’s disease. Nature 2021, 593, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Kasper, D.M.; Hintzen, J.; Wu, Y.; Ghersi, J.J.; Mandl, H.K.; Salinas, K.E.; Armero, W.; He, Z.; Sheng, Y.; Xie, Y.; et al. The N-glycome regulates the endothelial-to-hematopoietic transition. Science 2020, 370, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Raj, B.; Wagner, D.E.; McKenna, A.; Pandey, S.; Klein, A.M.; Shendure, J.; Gagnon, J.A.; Schier, A.F. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 2018, 36, 442–450. [Google Scholar] [CrossRef]
- Marisca, R.; Hoche, T.; Agirre, E.; Hoodless, L.J.; Barkey, W.; Auer, F.; Castelo-Branco, G.; Czopka, T. Functionally distinct subgroups of oligodendrocyte precursor cells integrate neural activity and execute myelin formation. Nat. Neurosci. 2020, 23, 363–374. [Google Scholar] [CrossRef]
- Krylov, A.J.; Yu, S.; Newton, A.; He, J.; Jusuf, P.R. Heterogeneity in quiescent Müller glia in the uninjured zebrafish retina drive differential responses following photoreceptor ablation. Front. Mol. Neurosci. 2023, 16, 1087136. [Google Scholar] [CrossRef]
- D’Elia, K.P.; Hameedy, H.; Goldblatt, D.; Frazel, P.; Kriese, M.; Zhu, Y.; Hamling, K.R.; Kawakami, K.; Liddelow, S.A.; Schoppik, D.; et al. Determinants of motor neuron functional subtypes important for locomotor speed. Cell Rep. 2023, 42, 113049. [Google Scholar] [CrossRef]
- Tuttle, A.M.; Miller, L.N.; Royer, L.J.; Wen, H.; Kelly, J.J.; Calistri, N.L.; Heiser, L.M.; Nechiporuk, A.V. Single-cell analysis of Rohon-Beard neurons implicates Fgf signaling in axon maintenance and cell survival. bioRxiv 2023. [Google Scholar] [CrossRef]
- Bergen, V.; Lange, M.; Peidli, S.; Wolf, F.A.; Theis, F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 2020, 38, 1408–1414. [Google Scholar] [CrossRef] [PubMed]
- Peuß, R.; Box, A.C.; Chen, S.; Wang, Y.; Tsuchiya, D.; Persons, J.L.; Kenzior, A.; Maldonado, E.; Krishnan, J.; Scharsack, J.P.; et al. Adaptation to low parasite abundance affects immune investment and immunopathological responses of cavefish. Nat. Ecol. Evol. 2020, 4, 1416–1430. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Huang, J.; Wang, W.; Wang, Y.; Chen, H.; Wang, Q.; Zhang, Y.; Liu, Q.; Yang, D. Multi-tissue scRNA-seq reveals immune cell landscape of turbot (Scophthalmus maximus). Fundam. Res. 2022, 2, 550–561. [Google Scholar] [CrossRef]
- Mu, D.; Yang, J.; Jiang, Y.; Wang, Z.; Chen, W.; Huang, J.; Zhang, Y.; Liu, Q.; Yang, D. Single-cell transcriptomic analysis reveals neutrophil as orchestrator during β-glucan–induced trained immunity in a teleost fish. J. Immunol. 2022, 209, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhu, C.; Gao, X.; Zheng, Y.; Zhu, X.; Jiang, H.; Wei, W.; Jiang, Q.; Zhang, X. Single-cell transcriptome analysis reveals a cellular immune response in freshwater dark sleeper (Odontobutis potamophila) after infection with Aeromonas veronii. Front. Physiol. 2023, 14, 1201914. [Google Scholar] [CrossRef]
- Liu, F.; Yuwono, C.; Tay, A.C.Y.; Wehrhahn, M.C.; Riordan, S.M.; Zhang, L. Analysis of global Aeromonas veronii genomes provides novel information on source of infection and virulence in human gastrointestinal diseases. BMC Genom. 2022, 23, 166. [Google Scholar] [CrossRef]
- Perdiguero, P.; Morel, E.; Díaz-Rosales, P.; Tafalla, C. Individual B cells transcribe multiple rearranged immunoglobulin light chains in teleost fish. Iscience 2021, 24, 102615. [Google Scholar] [CrossRef]
- Taylor, R.S.; Ruiz Daniels, R.; Dobie, R.; Naseer, S.; Clark, T.C.; Henderson, N.C.; Boudinot, P.; Martin, S.A.; Macqueen, D.J. Single cell transcriptomics of Atlantic salmon (Salmo salar L.) liver reveals cellular heterogeneity and immunological responses to challenge by Aeromonas salmonicida. Front. Immunol. 2022, 13, 984799. [Google Scholar] [CrossRef]
- Parker, J.; Guslund, N.C.; Jentoft, S.; Roth, O. Characterization of pipefish immune cell populations through single-cell transcriptomics. Front. Immunol. 2022, 13, 131. [Google Scholar] [CrossRef]
- Bobrovskikh, A.; Doroshkov, A.; Mazzoleni, S.; Cartenì, F.; Giannino, F.; Zubairova, U. A sight on single-cell transcriptomics in plants through the prism of cell-based computational modeling approaches: Benefits and challenges for data analysis. Front. Genet. 2021, 12, 652974. [Google Scholar] [CrossRef] [PubMed]
- Zappia, L.; Phipson, B.; Oshlack, A. Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. PLoS Comput. Biol. 2018, 14, e1006245. [Google Scholar] [CrossRef]
- Quatredeniers, M.; Serafin, A.S.; Benmerah, A.; Rausell, A.; Saunier, S.; Viau, A. Meta-analysis of single-cell and single-nucleus transcriptomics reveals kidney cell type consensus signatures. Sci. Data 2023, 10, 361. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Stuart, T.; Kowalski, M.H.; Choudhary, S.; Hoffman, P.; Hartman, A.; Srivastava, A.; Molla, G.; Madad, S.; Fernandez-Granda, C.; et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 2023. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.r.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Looney, A.P.; Liu, L.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R.P.; Wolters, P.J.; Abate, A.R.; et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019, 20, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Speer, T.; Dimmeler, S.; Schunk, S.J.; Fliser, D.; Ridker, P.M. Targeting innate immunity-driven inflammation in CKD and cardiovascular disease. Nat. Rev. Nephrol. 2022, 18, 762–778. [Google Scholar] [CrossRef]
- Bahrar, H.; Bekkering, S.; Stienstra, R.; Netea, M.; Riksen, N. Innate immune memory in cardiometabolic disease. Cardiovasc. Res. online ahead of print. 2023. [Google Scholar] [CrossRef]
- Nati, M.; Chung, K.J.; Chavakis, T. The role of innate immune cells in nonalcoholic fatty liver disease. J. Innate Immun. 2022, 14, 31–41. [Google Scholar] [CrossRef]
- Amersfoort, J.; Eelen, G.; Carmeliet, P. Immunomodulation by endothelial cells—partnering up with the immune system? Nat. Rev. Immunol. 2022, 22, 576–588. [Google Scholar] [CrossRef]
- Niu, J.; Huang, Y.; Liu, X.; Zhang, Z.; Tang, J.; Wang, B.; Lu, Y.; Cai, J.; Jian, J. Single-cell RNA-seq reveals different subsets of non-specific cytotoxic cells in teleost. Genomics 2020, 112, 5170–5179. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xu, Z.; Zeng, H.; Sun, N.; Wu, B.; Wang, C.; Bo, J.; Li, L.; Dong, Y.; He, S. FishDB: An integrated functional genomics database for fishes. BMC Genom. 2020, 21, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Iwama, R.E.; Moran, Y. Origins and diversification of animal innate immune responses against viral infections. Nat. Ecol. Evol. 2023, 7, 182–193. [Google Scholar] [CrossRef] [PubMed]
- De Mitcheson, Y.S.; Liu, M. Functional hermaphroditism in teleosts. Fish Fish. 2008, 9, 1–43. [Google Scholar] [CrossRef]
- Smith, N.C.; Rise, M.L.; Christian, S.L. A comparison of the innate and adaptive immune systems in cartilaginous fish, ray-finned fish, and lobe-finned fish. Front. Immunol. 2019, 10, 2292. [Google Scholar] [CrossRef] [PubMed]
- Hastings, P.A.; Walker, H.J.; Galland, G.R. Fishes: A Guide to Their Diversity; University of California Press: Oakland, CA, USA, 2015. [Google Scholar]
- Wootton, R.J. Ecology of Teleost Fishes; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; Volume 1. [Google Scholar]
- Karvonen, A.; Lundsgaard-Hansen, B.; Jokela, J.; Seehausen, O. Differentiation in parasitism among ecotypes of whitefish segregating along depth gradients. Oikos 2013, 122, 122–128. [Google Scholar] [CrossRef]
- Kough, A.S.; Paris, C.B.; Behringer, D.C.; Butler IV, M.J. Modelling the spread and connectivity of waterborne marine pathogens: The case of PaV1 in the Caribbean. ICES J. Mar. Sci. 2015, 72, i139–i146. [Google Scholar] [CrossRef]
- Behringer, D.C.; Karvonen, A.; Bojko, J. Parasite avoidance behaviours in aquatic environments. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170202. [Google Scholar] [CrossRef]
- Raby, L.; Völkel, P.; Le Bourhis, X.; Angrand, P.O. Genetic engineering of zebrafish in cancer research. Cancers 2020, 12, 2168. [Google Scholar] [CrossRef]
- Baines, C.; Meitern, R.; Kreitsberg, R.; Sepp, T. Comparative study of the evolution of cancer gene duplications across fish. Evol. Appl. 2022, 15, 1834–1845. [Google Scholar] [CrossRef]
- Reznick, D.; Ghalambor, C.; Nunney, L. The evolution of senescence in fish. Mech. Ageing Dev. 2002, 123, 773–789. [Google Scholar] [CrossRef] [PubMed]
- Sauer, D.J.; Heidinger, B.J.; Kittilson, J.D.; Lackmann, A.R.; Clark, M.E. No evidence of physiological declines with age in an extremely long-lived fish. Sci. Rep. 2021, 11, 9065. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.D.; Wu, D.; Meydani, S.N. Age-associated alterations in immune function and inflammation. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2022, 118, 110576. [Google Scholar] [CrossRef] [PubMed]
- Mogilenko, D.A.; Shchukina, I.; Artyomov, M.N. Immune ageing at single-cell resolution. Nat. Rev. Immunol. 2022, 22, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Shebzukhov, Y.; Holtze, S.; Hirseland, H.; Schäfer, H.; Radbruch, A.; Hildebrandt, T.; Grützkau, A. Identification of cross-reactive antibodies for the detection of lymphocytes, myeloid cells and haematopoietic precursors in the naked mole rat. Eur. J. Immunol. 2019, 49, 2103–2110. [Google Scholar] [CrossRef]
- Gorshkova, E.A.; Gubernatorova, E.O.; Dvorianinova, E.M.; Yurakova, T.R.; Marey, M.V.; Averina, O.A.; Holtze, S.; Hildebrandt, T.B.; Dmitriev, A.A.; Drutskaya, M.S.; et al. Macrophages from naked mole-rat possess distinct immunometabolic signatures upon polarization. Front. Immunol. 2023, 14, 1172467. [Google Scholar] [CrossRef]
- Schuhmacher, L.N.; Husson, Z.; St. John Smith, E. The naked mole-rat as an animal model in biomedical research: Current perspectives. Anim. Physiol. 2015, 2015, 137–148. [Google Scholar] [CrossRef]
- Artwohl, J.; Ball-Kell, S.; Valyi-Nagy, T.; Wilson, S.P.; Lu, Y.; Park, T.J. Extreme susceptibility of African naked mole rats (Heterocephalus glaber) to experimental infection with herpes simplex virus type 1. Comp. Med. 2009, 59, 83–90. [Google Scholar]
- Hoseinifar, S.H.; Ringø, E.; Shenavar Masouleh, A.; Esteban, M.Á. Probiotic, prebiotic and synbiotic supplements in sturgeon aquaculture: A review. Rev. Aquac. 2016, 8, 89–102. [Google Scholar] [CrossRef]
- Guo, H.; Dixon, B. Understanding acute stress-mediated immunity in teleost fish. Fish Shellfish Immunol. Rep. 2021, 2, 100010. [Google Scholar] [CrossRef]
- Buchmann, K. Control of parasitic diseases in aquaculture. Parasitology 2022, 149, 1985–1997. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.H.; Schiapparelli, L.M.; Ma, Y.; Yokota, S.; Atkins, M.; Xia, X.; Cameron, E.G.; Huang, T.; Saturday, S.; Sun, C.B.; et al. Quantitative transportomics identifies Kif5a as a major regulator of neurodegeneration. Elife 2022, 11, e68148. [Google Scholar] [CrossRef] [PubMed]
- Bohaud, C.; De La Cruz, J.; Terraza, C.; Barthelaix, A.; Laplace-Builhé, B.; Jorgensen, C.; Arribat, Y.; Djouad, F. Lactate metabolism coordinates macrophage response and regeneration in zebrafish. Theranostics 2022, 12, 3995–4009. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, P.; Morel, E.; Tafalla, C. Diversity of Rainbow Trout Blood B Cells Revealed by Single Cell RNA Sequencing. Biology 2021, 10, 511. [Google Scholar] [CrossRef]
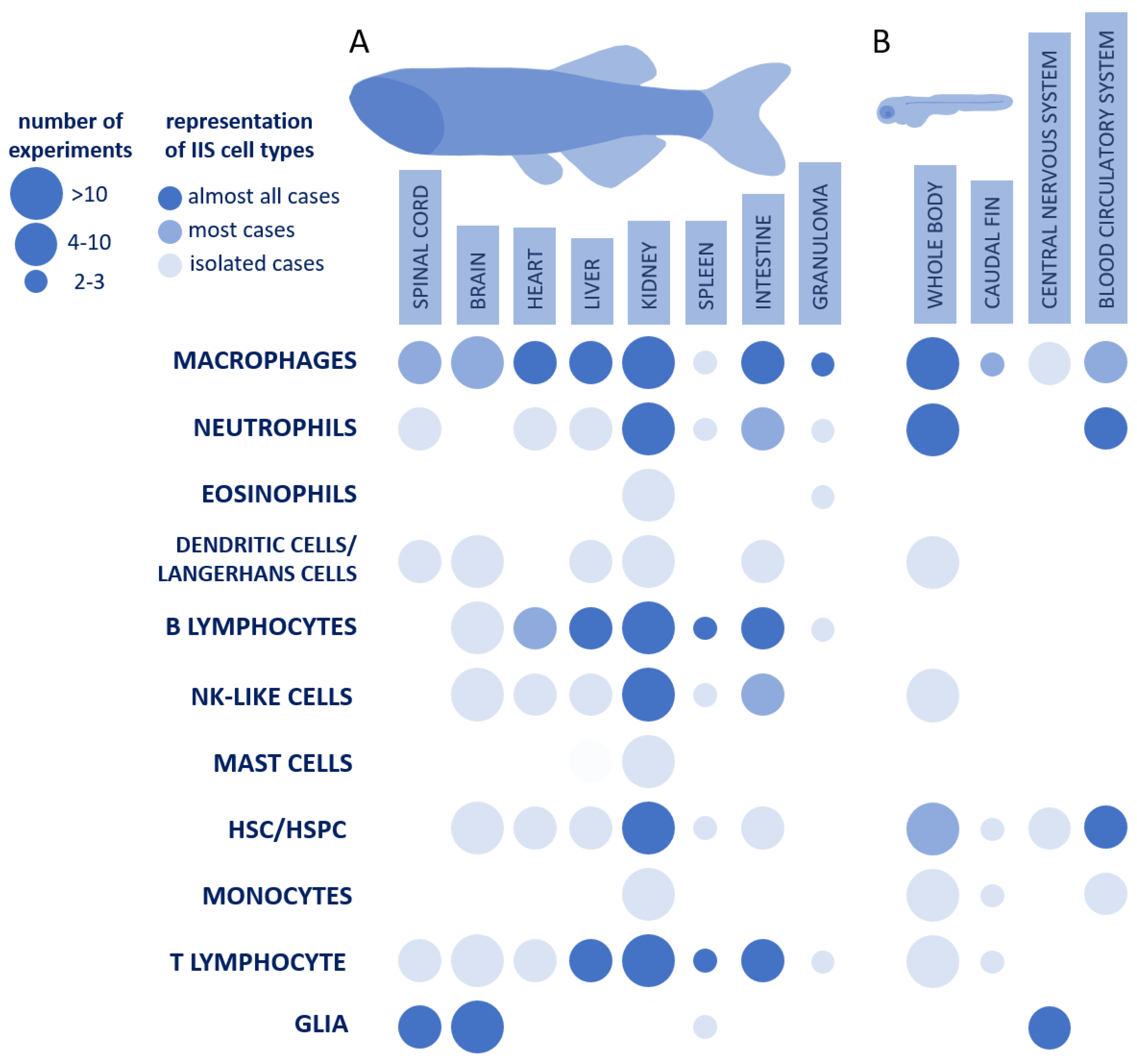
| Tissues | Identifiers of Experiments and Their Links |
|---|---|
| Whole body (14) | GSE68920, GSE120503, GSE160038, GSE162979, GSE173972, GSE176853, GSE182213, GSE191029, GSE196553, GSE198571, GSE202193, GSE209884, GSE239880, GSE239949 |
| Central nervous system (8) | E-CURD-123, GSE105010, GSE132166, GSE164772, GSE212314, GSE218107, GSE240026, GSE241296 |
| Blood circulatory system (7) | GSE92542, GSE158099, GSE166900, GSE167787, GSE186423, GSE186427, GSE186565 |
| Caudal fin (3) | GSE137770, GSE146404, GSE158851 |
| Granuloma (1) | GSE81913 |
| Endothelium of the neural crest (1) | GSE135246 |
| Intestine (1) | GSE150498 |
| Spleen (1) | GSE211396 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bobrovskikh, A.V.; Zubairova, U.S.; Doroshkov, A.V. Fishing Innate Immune System Properties through the Transcriptomic Single-Cell Data of Teleostei. Biology 2023, 12, 1516. https://doi.org/10.3390/biology12121516
Bobrovskikh AV, Zubairova US, Doroshkov AV. Fishing Innate Immune System Properties through the Transcriptomic Single-Cell Data of Teleostei. Biology. 2023; 12(12):1516. https://doi.org/10.3390/biology12121516
Chicago/Turabian StyleBobrovskikh, Aleksandr V., Ulyana S. Zubairova, and Alexey V. Doroshkov. 2023. "Fishing Innate Immune System Properties through the Transcriptomic Single-Cell Data of Teleostei" Biology 12, no. 12: 1516. https://doi.org/10.3390/biology12121516
APA StyleBobrovskikh, A. V., Zubairova, U. S., & Doroshkov, A. V. (2023). Fishing Innate Immune System Properties through the Transcriptomic Single-Cell Data of Teleostei. Biology, 12(12), 1516. https://doi.org/10.3390/biology12121516









