Dynamics of Methane-Consuming Biomes from Wieliczka Formation: Environmental and Enrichment Studies
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
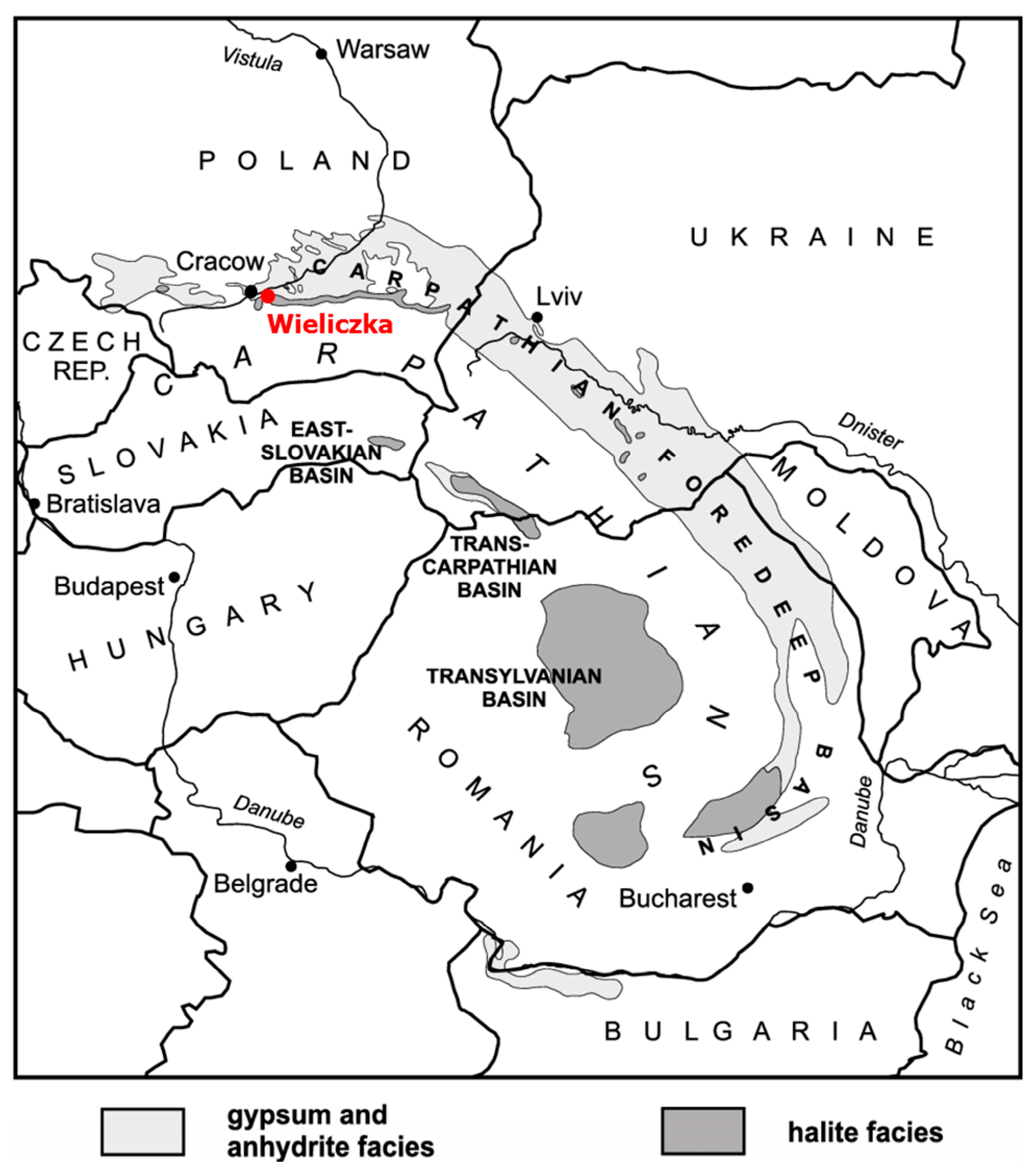
2.1. Study Site and Samples Collection
2.2. Determination of Methanotrophic Activity of Environmental Sample
2.3. Enrichment of Methane-Oxidizing Bacterial Community
2.4. Bacterial Diversity and Molecular Techniques
2.5. Bioinformatic and Statistic Analysis
2.6. Data Availability
3. Results and Discussion
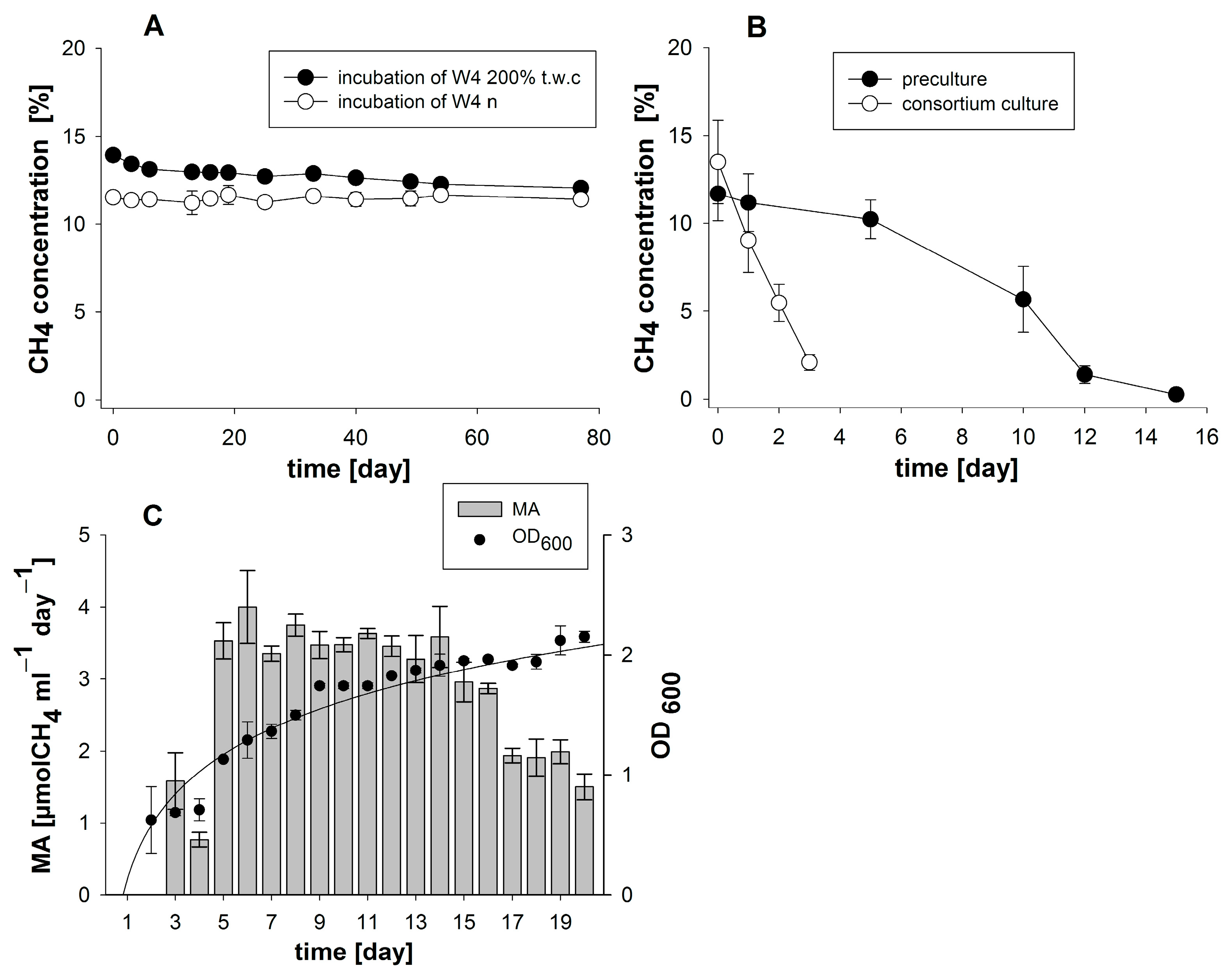
3.1. Enrichment and Characterization of a Methane-Oxidizing Bacterial Community
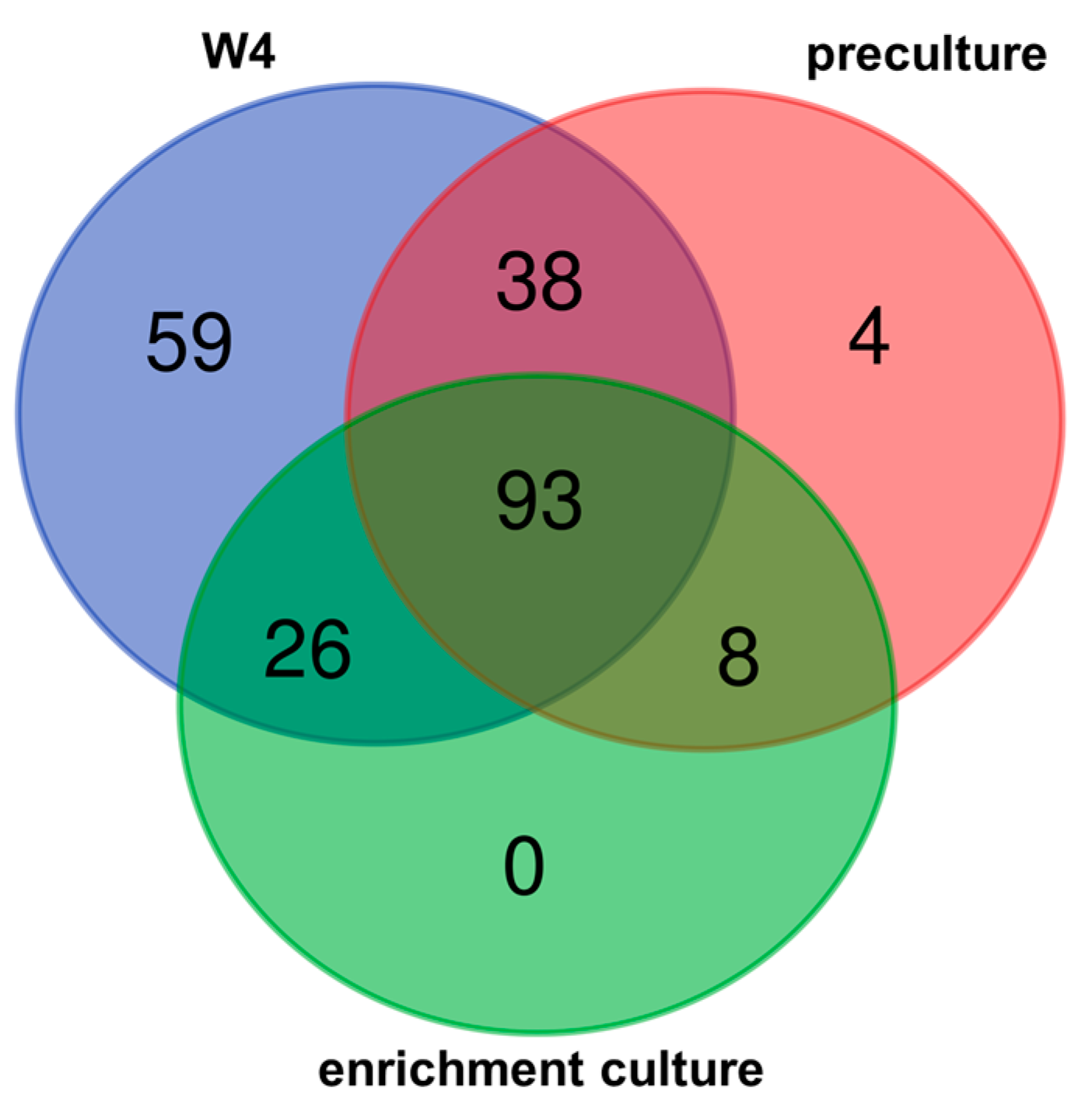
3.2. Bacterial Diversity
3.3. Changing Biodiversity of the Methanotrophic Community
3.4. Enrichment Culture Composition
3.5. The Importance of Microorganism Community in Biotechnology
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolińska, A.; Pytlak, A.; Stepniewska, Z.; Kuźniar, A.; Piasecki, C. Identification of methanotrophic bacteria community in the Jastrzebie-Moszczenica coal mine by fluorescence in situ hybridization and PCR techniques. Pol. J. Environ. Stud. 2013, 22, 275–282. [Google Scholar]
- Pytlak, A.; Stepniewska, Z.; Kuźniar, A.; Szafranek-Nakonieczna, A.; Wolińska, A.; Banach, A. Potential for Aerobic Methane Oxidation in Carboniferous Coal Measures. Geomicrobiol. J. 2014, 31, 737–747. [Google Scholar] [CrossRef]
- Szafranek-Nakonieczna, A.; Zheng, Y.; Słowakiewicz, M.; Pytlak, A.; Polakowski, C.; Kubaczyński, A.; Bieganowski, A.; Banach, A.; Wolińska, A.; Stępniewska, Z. Methanogenic potential of lignites in Poland. Int. J. Coal Geol. 2018, 196, 201–210. [Google Scholar] [CrossRef]
- Whittenbury, R.; Phillips, K.C.; Wilkinson, J.F. Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 1970, 61, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.F.U.; Xu, H.J.; Colin Murrell, J.; Crombie, A. Facultative methanotrophs—Diversity, genetics, molecular ecology and biotechnological potential: A mini-review. Microbiology 2020, 166, 894–908. [Google Scholar] [CrossRef]
- Stein, L.Y.; Yoon, S.; Semrau, J.D.; DiSpirito, A.A.; Crombie, A.; Murrell, J.C.; Vuilleumier, S.; Kalyuzhnaya, M.G.; Op Den Camp, H.J.M.; Bringel, F.; et al. Genome sequence of the obligate methanotroph Methylosinus trichosporium strain OB3b. J. Bacteriol. 2010, 192, 6497–6498. [Google Scholar] [CrossRef]
- Chen, Y.; Crombie, A.; Rahman, M.T.; Dedysh, S.N.; Liesack, W.; Stott, M.B.; Alam, M.; Theisen, A.R.; Murrell, J.C.; Dunfield, P.F. Complete Genome Sequence of the Aerobic Facultative Methanotroph Methylocella silvestris BL2. J. Bacteriol. 2010, 192, 3840. [Google Scholar] [CrossRef]
- Boden, R.; Cunliffe, M.; Scanlan, J.; Moussard, H.; Kits, K.D.; Klotz, M.G.; Jetten, M.S.M.; Vuilleumier, S.; Han, J.; Peters, L.; et al. Complete genome sequence of the aerobic marine methanotroph Methylomonas methanica MC09. J. Bacteriol. 2011, 193, 7001–7002. [Google Scholar] [CrossRef]
- Svenning, M.M.; Hestnes, A.G.; Wartiainen, I.; Stein, L.Y.; Klotz, M.G.; Kalyuzhnaya, M.G.; Spang, A.; Bringel, F.; Vuilleumier, S.; Lajus, A.; et al. Genome Sequence of the Arctic Methanotroph Methylobacter tundripaludum SV96. J. Bacteriol. 2011, 193, 6418–6419. [Google Scholar] [CrossRef]
- Vuilleumier, S.; Khmelenina, V.N.; Bringel, F.; Reshetnikov, A.S.; Lajus, A.; Mangenot, S.; Rouy, Z.; Op den Camp, H.J.M.; Jetten, M.S.M.; Dispirito, A.A.; et al. Genome Sequence of the Haloalkaliphilic Methanotrophic Bacterium Methylomicrobium alcaliphilum 20Z. J. Bacteriol. 2012, 194, 551. [Google Scholar] [CrossRef]
- Hou, S.; Makarova, K.S.; Saw, J.H.; Senin, P.; Ly, B.V.; Zhou, Z.; Ren, Y.; Wang, J.; Galperin, M.Y.; Alam, M.; et al. Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol. Direct. 2008, 3, 1–25. [Google Scholar] [CrossRef]
- Knief, C. Diversity and Habitat Preferences of Cultivated and Uncultivated Aerobic Methanotrophic Bacteria Evaluated Based on pmoA as Molecular Marker occurrence and role of methane-oxidizing bacteria. Front. Microbiol. 2015, 6, 1346. [Google Scholar] [CrossRef]
- Lidstrom, M.E. Isolation and characterization of marine methanotrophs. Antonie Van Leeuwenhoek 1988, 54, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Lees, V.; Owens, N.J.P.; Murrell, J.C. Nitrogen metabolism in marine methanotrophs. Arch. Microbiol. 1991, 157, 60–65. [Google Scholar] [CrossRef]
- Khmelenina, V.N.; Kalyuzhnaya, M.G.; Starostina, N.G.; Suzina, N.E.; Trotsenko, Y.A. Isolation and characterization of halotolerant alkaliphilic methanotrophic bacteria from Tuva soda lakes. Curr. Microbiol. 1997, 35, 257–261. [Google Scholar] [CrossRef]
- Dedysh, S.N. Methanotrophic Bacteria of Acidic Sphagnum Peat Bogs. Microbiology 2002, 71, 638–650. [Google Scholar] [CrossRef]
- Stępniewska, Z.; Goraj, W.; Kuźniar, A.; Łopacka, N.; Małysza, M. Enrichment culture and identification of endophytic methanotrophs isolated from peatland plants. Folia Microbiol. 2017, 62, 381–391. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stepniewska, Z.; Pytlak, A.; Kuźniar, A. Methanotrophic activity in Carboniferous coalbed rocks. Int. J. Coal Geol. 2013, 106, 1–10. [Google Scholar] [CrossRef]
- Trembath-Reichert, E.; Morono, Y.; Ijiri, A.; Hoshino, T.; Dawson, K.S.; Inagaki, F.; Orphan, V.J. Methyl-compound use and slow growth characterize microbial life in 2-km-deep subseafloor coal and shale beds. Proc. Natl. Acad. Sci. USA 2017, 114, E9206–E9215. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, R.; Tian, K.; Zhao, S.; Shi, H.; Gong, W.; Lei, Q. Characteristics of the methanotroph used in coalbed methane emission reduction: Methane oxidation efficiency and coal wettability. Fuel 2023, 349, 128596. [Google Scholar] [CrossRef]
- Twing, K.I.; Brazelton, W.J.; Kubo, M.D.Y.; Hyer, A.J.; Cardace, D.; Hoehler, T.M.; McCollom, T.M.; Schrenk, M.O. Serpentinization-Influenced Groundwater Harbors Extremely Low Diversity Microbial Communities Adapted to High pH. Front. Microbiol. 2017, 8, 308. [Google Scholar] [CrossRef]
- Kraus, E.A.; Nothaft, D.; Stamps, B.W.; Rempfert, K.R.; Ellison, E.T.; Matter, J.M.; Templeton, A.S.; Boyd, E.S.; Spear, J.R. Molecular Evidence for an Active Microbial Methane Cycle in Subsurface Serpentinite-Hosted Groundwaters in the Samail Ophiolite, Oman. Appl. Environ. Microbiol. 2021, 87, e02068-20. [Google Scholar] [CrossRef] [PubMed]
- Nothaft, D.B.; Templeton, A.S.; Rhim, J.H.; Wang, D.T.; Labidi, J.; Miller, H.M.; Boyd, E.S.; Matter, J.M.; Ono, S.; Young, E.D.; et al. Geochemical, Biological, and Clumped Isotopologue Evidence for Substantial Microbial Methane Production under Carbon Limitation in Serpentinites of the Samail Ophiolite, Oman. J. Geophys. Res. Biogeosci. 2021, 126, e2020JG006025. [Google Scholar] [CrossRef]
- Head, I.M.; Jones, D.M.; Larter, S.R. Biological activity in the deep subsurface and the origin of heavy oil. Nature 2003, 426, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Stępniewska, Z.; Goraj, W.; Wolińska, A.; Szafranek-Nakonieczna, A.; Banach, A.; Górski, A. Methanotrophic activity of rocks surrounding Badenian salts in the “Wieliczka” Salt Mine. Carpathian J. Earth Environ. Sci. 2018, 13, 107–119. [Google Scholar] [CrossRef]
- Goraj, W.; Szafranek-Nakonieczna, A.; Grzadziel, J.; Polakowski, C.; Słowakiewicz, M.; Zheng, Y.; Gałazka, A.; Stępniewska, Z.; Pytlak, A. Microbial involvement in carbon transformation via ch4 and co2 in saline sedimentary pool. Biology 2021, 10, 792. [Google Scholar] [CrossRef] [PubMed]
- Goraj, W.; Stępniewska, Z. Biosynthesis of amino acids by methanotrophs in response to salinity stress. N. Biotechnol. 2016, 33, S198. [Google Scholar] [CrossRef]
- Ho, A.; De Roy, K.; Thas, O.; De Neve, J.; Hoefman, S.; Vandamme, P.; Heylen, K.; Boon, N. The more, the merrier: Heterotroph richness stimulates methanotrophic activity. ISME J. 2014, 8, 1945–1948. [Google Scholar] [CrossRef]
- Graham, D.W.; Chaudhary, J.A.; Hanson, R.S.; Arnold, R.G. Factors affecting competition between type I and type II methanotrophs in two-organism, continuous-flow reactors. Microb. Ecol. 1993, 25, 1–17. [Google Scholar] [CrossRef]
- Valverde-Pérez, B.; Xing, W.; Zachariae, A.A.; Skadborg, M.M.; Kjeldgaard, A.F.; Palomo, A.; Smets, B.F. Cultivation of methanotrophic bacteria in a novel bubble-free membrane bioreactor for microbial protein production. Bioresour. Technol. 2020, 310, 123388. [Google Scholar] [CrossRef]
- Scholten-Koerselman, I.; Houwaard, F.; Janssen, P.; Zehnder, A.J.B. Bacteroides xylanolyticus sp. nov., a xylanolytic bacterium from methane producing cattle manure. Antonie Van Leeuwenhoek 1986, 52, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Bąbel, M. Badenian evaporite basin of the northern Carpathian Foredeep as a drawdown salina basin. Acta Geol. Pol. 2004, 54, 313–337. [Google Scholar]
- Héry, M.; Singer, A.C.; Kumaresan, D.; Bodrossy, L.; Stralis-Pavese, N.; Prosser, J.I.; Thompson, I.P.; Murrell, J.C. Effect of earthworms on the community structure of active methanotrophic bacteria in a landfill cover soil. ISME J. 2008, 2, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Hatamoto, M.; Yamamoto, H.; Kindaichi, T.; Ozaki, N.; Ohashi, A. Biological oxidation of dissolved methane in effluents from anaerobic reactors using a down-flow hanging sponge reactor. Water Res. 2010, 44, 1409–1418. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, D.Y.; Lü Cker, S.; Vejmelkova, D.; Kostrikina, N.A.; Kleerebezem, R.; Rijpstra, I.C.; Sinninghe Damsté, J.S.; Le Paslier, D.; Muyzer, G.; Wagner, M.; et al. Nitrification expanded: Discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 2012, 6, 2245–2256. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2016. [Google Scholar]
- ES Wright. RDP v16 Modified Training Set for 16S rRNA Classification; ES Wright: Coventry, UK, 2019.
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 140. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R.; Kolde, M.R. Package “pheatmap”. R Packag. 2022, 1, 790. [Google Scholar]
- Dombrowski, H.J. Bacteria from paleozoic salt deposits. Ann. N. Y. Acad. Sci. 1963, 108, 453–460. [Google Scholar] [CrossRef]
- Tasch, P. Dead and viable fossil salt bacteria. Univ. Wichita Bull. 1963, 39, 2–7. [Google Scholar]
- Akhtar, N.; Ghauri, M.A.; Iqbal, A.; Anwar, M.A.; Akhtar, K. Biodiversity and phylogenetic analysis of culturable bacteria indigenous to Khewra salt mine of Pakistan and their industrial importance. Braz. J. Microbiol. 2008, 39, 143–150. [Google Scholar] [CrossRef]
- Roohi, A.; Ahmed, I.; Iqbal, M.; Jamil, M. Preliminary isolation and characterization of halotolerant and halophilic bacteria from salt mines of Karak, Pakistan. Pak. J. Bot. 2012, 44, 365–370. [Google Scholar]
- Lazar, C.S.; Braganca, J.M.; Cycil, L.M.; Hasan, F.; Dassarma, S.; Pecher, W.; Mcdonald, R.; Abdulsalam, M. Metagenomic Insights Into the Diversity of Halophilic Microorganisms Indigenous to the Karak Salt Mine, Pakistan. Front. Microbiol. 2020, 11, 1567. [Google Scholar] [CrossRef]
- Chauhan, M.; Garlapati, V.K. Production and Characterization of a Halo-, Solvent-, Thermo-tolerant Alkaline Lipase by Staphylococcus arlettae JPBW-1, Isolated from Rock Salt Mine. Appl. Biochem. Biotechnol. 2013, 171, 1429–1443. [Google Scholar] [CrossRef] [PubMed]
- Bachran, M.; Kluge, S.; Lopez-Fernandez, M.; Cherkouk, A. Microbial Diversity in an Arid, Naturally Saline Environment. Microb. Ecol. 2018, 78, 494–505. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-G.; Cui, X.-L.; Diger Pukall, R.; Li, H.-M.; Yang, Y.-L.; Xu, L.-H.; Wen, M.-L.; Peng, Q.; Jiang, C.-L. Salinicoccus kunmingensis sp. nov., a moderately halophilic bacterium isolated from a salt mine in Yunnan, south-west China. Int. J. Syst. Evol. Microbiol. 2007, 57, 2327–2332. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Wang, Z.G.; Wang, Y.X.; Schneegurt, M.A.; Li, Z.Y.; Lai, Y.H.; Zhang, S.Y.; Wen, M.L.; Cui, X.L. Comparative molecular analysis of the prokaryotic diversity of two salt mine soils in southwest China. J. Basic Microbiol. 2013, 53, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Pytlak, A.; Szafranek-Nakonieczna, A.; Sujak, A.; Grządziel, J.; Polakowski, C.; Kuźniar, A.; Proc, K.; Kubaczyński, A.; Goraj, W.; Gałązka, A.; et al. Stimulation of methanogenesis in bituminous coal from the upper Silesian coal basin. Int. J. Coal Geol. 2020, 231, 103609. [Google Scholar] [CrossRef]
- Pytlak, A.; Sujak, A.; Szafranek-Nakonieczna, A.; Grządziel, J.; Banach, A.; Goraj, W.; Gałązka, A.; Gruszecki, W.I.; Stępniewska, Z. Water-induced molecular changes of hard coals and lignites. Int. J. Coal Geol. 2020, 224, 103481. [Google Scholar] [CrossRef]
- Goonesekera, E.M.; Tsapekos, P.; Angelidaki, I.; Valverde-Pérez, B. Impact of recovered phosphorus supply on methanotrophic cultivation and microbial protein production. J. Environ. Manag. 2022, 322, 115820. [Google Scholar] [CrossRef]
- Bowman, J.P. Methylocystis. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley Online Labrary: Hoboken, NJ, USA, 2015; pp. 1–7. [Google Scholar] [CrossRef]
- Reed, D.W.; Fujita, Y.; Delwiche, M.E.; Blackwelder, D.B.; Sheridan, P.P.; Uchida, T.; Colwell, F.S. Microbial Communities from Methane Hydrate-Bearing Deep Marine Sediments in a Forearc Basin. Appl. Environ. Microbiol. 2002, 68, 3759–3770. [Google Scholar] [CrossRef] [PubMed]
- Reed, A.J.; Lutz, R.A.; Vetriani, C. Vertical distribution and diversity of bacteria and archaea in sulfide and methane-rich cold seep sediments located at the base of the Florida Escarpment. Extremophiles 2006, 10, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P. Methylomonas. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley Online Labrary: Hoboken, NJ, USA, 2016; pp. 1–11. [Google Scholar] [CrossRef]
- Auman, A.J.; Stolyar, S.; Costello, A.M.; Lidstrom, M.E. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl. Environ. Microbiol. 2000, 66, 5259–5266. [Google Scholar] [CrossRef] [PubMed]
- Auman, A.J.; Lidstrom, M.E. Analysis of sMMO-containing Type I methanotrophs in Lake Washington sediment. Environ. Microbiol. 2002, 4, 517–524. [Google Scholar] [CrossRef]
- Hutchens, E.; Radajewski, S.; Dumont, M.G.; McDonald, I.R.; Murrell, J.C. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 2004, 6, 111–120. [Google Scholar] [CrossRef]
- Lüke, C.; Krause, S.; Cavigiolo, S.; Greppi, D.; Lupotto, E.; Frenzel, P. Biogeography of wetland rice methanotrophs. Environ. Microbiol. 2010, 12, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Dianou, D.; Ueno, C.; Ogiso, T.; Kimura, M.; Asakawa, S. Diversity of Cultivable Methane-Oxidizing Bacteria in Microsites of a Rice Paddy Field: Investigation by Cultivation Method and Fluorescence in situ Hybridization (FISH). Microbes Environ. 2012, 27, 278–287. [Google Scholar] [CrossRef]
- Morris, S.A.; Radajewski, S.; Willison, T.W.; Murrell, J.C. Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 2002, 68, 1446–1453. [Google Scholar] [CrossRef]
- Jaatinen, K.; Tuittila, E.S.; Laine, J.; Yrjälä, K.; Fritze, H. Methane-oxidizing bacteria in a Finnish raised mire complex: Effects of site fertility and drainage. Microb. Ecol. 2005, 50, 429–439. [Google Scholar] [CrossRef]
- Chen, Y.; Dumont, M.G.; McNamara, N.P.; Chamberlain, P.M.; Bodrossy, L.; Stralis-Pavese, N.; Murrell, J.C. Diversity of the active methanotrophic community in acidic peatlands as assessed by mRNA and SIP-PLFA analyses. Environ. Microbiol. 2008, 10, 446–459. [Google Scholar] [CrossRef]
- Kip, N.; Ouyang, W.; van Winden, J.; Raghoebarsing, A.; van Niftrik, L.; Pol, A.; Pan, Y.; Bodrossy, L.; van Donselaar, E.G.; Reichart, G.J.; et al. Detection, isolation, and characterization of acidophilic methanotrophs from sphagnum mosses. Appl. Environ. Microbiol. 2011, 77, 5643–5654. [Google Scholar] [CrossRef] [PubMed]
- Bąkowska, A.; Karlicki, M.; Włodarczyk, A.; Matlakowska, R. Geomikrobiologia podziemnych środowisk kopalnianych monokliny przedsudeckiej. Biul. Państwowego Inst. Geol. 2017, 469, 201–216. [Google Scholar]
- Antony, C.P.; Shimpi, G.G.; Cockell, C.S.; Patole, M.S.; Shouche, Y.S. Molecular Characterization of Prokaryotic Communities Associated with Lonar Crater Basalts. Geomicrobiol. J. 2014, 31, 519–528. [Google Scholar] [CrossRef]
- Kalyuzhnaya, M.G. Methylomicrobium. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley Online Labrary: Hoboken, NJ, USA, 2016; pp. 1–10. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Berben, T.; Melton, E.D.; Overmars, L.; Vavourakis, C.D.; Muyzer, G. Microbial diversity and biogeochemical cycling in soda lakes. Extremophiles 2014, 18, 791–809. [Google Scholar] [CrossRef] [PubMed]
- Anderson, I.; Munk, C.; Lapidus, A.; Nolan, M.; Lucas, S.; Tice, H.; Del Rio, T.G.; Cheng, J.F.; Han, C.; Tapia, R.; et al. Genome sequence of the flexirubin-pigmented soil bacterium Niabella soli type strain (JS13-8T). Stand. Genom. Sci. 2012, 7, 210. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reddy, A.P.; Simmons, C.W.; D’haeseleer, P.; Khudyakov, J.; Burd, H.; Hadi, M.; Simmons, B.A.; Singer, S.W.; Thelen, M.P.; VanderGheynst, J.S. Discovery of Microorganisms and Enzymes Involved in High-Solids Decomposition of Rice Straw Using Metagenomic Analyses. PLoS ONE 2013, 8, e77985. [Google Scholar] [CrossRef] [PubMed]
- Eichorst, S.A.; Varanasi, P.; Stavila, V.; Zemla, M.; Auer, M.; Singh, S.; Simmons, B.A.; Singer, S.W. Community dynamics of cellulose-adapted thermophilic bacterial consortia. Environ. Microbiol. 2013, 15, 2573–2587. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.G.; Jeong, S.Y.; Cho, K.S. Functional rigidity of a methane biofilter during the temporal microbial succession. Appl. Microbiol. Biotechnol. 2014, 98, 3275–3286. [Google Scholar] [CrossRef]
- Spring, S.; Jäckel, U.; Wagner, M.; Kämpfer, P. Ottowia thiooxydans gen. nov., sp. nov., a novel facultatively anaerobic, N2O-producing bacterium isolated from activated sludge, and transfer of Aquaspirillum gracile to Hylemonella gracilis gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 99–106. [Google Scholar] [CrossRef]
- Li, Q.F.; Sun, L.N.; Kwon, S.W.; Chen, Q.; He, J.; Li, S.P.; Zhang, J. Xenophilus arseniciresistens sp. nov., an arsenite-resistant bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2014, 64, 1926–1931. [Google Scholar] [CrossRef]
- Lebert, B.M.; Sanford, S.A.; Reisinger, L.M.; Forsman, A.M.; Savage, A.E. Draft Genome Sequence of Xenophilus sp., a Novel Bacterium Isolated from the Skin of a Southern Leopard Frog (Rana sphenocephala) in Florida, USA. Am. Soc. Microbiol. 2017, 5, e01067-17. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Xu, Z.; Chen, C. Effect of mulching on labile soil organic matter pools, microbial community functional diversity and nitrogen transformations in two hardwood plantations of subtropical Australia. Appl. Soil Ecol. 2008, 40, 229–239. [Google Scholar] [CrossRef]
- Ho, A.; Lee, H.J.; Reumer, M.; Meima-Franke, M.; Raaijmakers, C.; Zweers, H.; de Boer, W.; Van der Putten, W.H.; Bodelier, P.L.E. Unexpected role of canonical aerobic methanotrophs in upland agricultural soils. Soil Biol. Biochem. 2019, 131, 1–8. [Google Scholar] [CrossRef]
- Patel, S.K.S.; Gupta, R.K.; Kumar, V.; Kondaveeti, S.; Kumar, A.; Das, D.; Kalia, V.C.; Lee, J.-K. Biomethanol Production from Methane by Immobilized Co-cultures of Methanotrophs. Indian J. Microbiol. 2020, 60, 318–324. [Google Scholar] [CrossRef]
- Singh, R.; Ryu, J.; Kim, S.W. Microbial consortia including methanotrophs: Some benefits of living together. J. Microbiol. 2019, 57, 939–952. [Google Scholar] [CrossRef]
- Tsapekos, P.; Zhu, X.; Pallis, E.; Angelidaki, I. Proteinaceous methanotrophs for feed additive using biowaste as carbon and nutrients source. Bioresour. Technol. 2020, 313, 123646. [Google Scholar] [CrossRef]
- Zha, X.; Tsapekos, P.; Zhu, X.; Khoshnevisan, B.; Lu, X.; Angelidaki, I. Bioconversion of wastewater to single cell protein by methanotrophic bacteria. Bioresour. Technol. 2021, 320, 124351. [Google Scholar] [CrossRef]
- Helm, J.; Wendlandt, K.D.; Rogge, G.; Kappelmeyer, U. Characterizing a stable methane-utilizing mixed culture used in the synthesis of a high-quality biopolymer in an open system. J. Appl. Microbiol. 2006, 101, 387–395. [Google Scholar] [CrossRef]
- Chidambarampadmavathy, K.; Karthikeyan, O.P.; Heimann, K. Biopolymers made from methane in bioreactors. Eng. Life Sci. 2015, 15, 689–699. [Google Scholar] [CrossRef]
- Fergala, A.; Alsayed, A.; Khattab, S.; Ramirez, M.; Eldyasti, A. Development of Methane-Utilizing Mixed Cultures for the Production of Polyhydroxyalkanoates (PHAs) from Anaerobic Digester Sludge. Environ. Sci. Technol. 2018, 52, 12376–12387. [Google Scholar] [CrossRef]
- Cantera, S.; Phandanouvong-Lozano, V.; Pascual, C.; García-Encina, P.A.; Lebrero, R.; Hay, A.; Muñoz, R. A systematic comparison of ectoine production from upgraded biogas using Methylomicrobium alcaliphilum and a mixed haloalkaliphilic consortium. Waste Manag. 2020, 102, 773–781. [Google Scholar] [CrossRef]
- Bothe, H.; Møller Jensen, K.; Mergel, A.; Larsen, J.; Jørgensen, C.; Bothe, H.; Jørgensen, L. Heterotrophic bacteria growing in association with Methylococcus capsulatus (Bath) in a single cell protein production process. Appl. Microbiol. Biotechnol. 2002, 59, 33–39. [Google Scholar] [CrossRef]
- Tsapekos, P.; Khoshnevisan, B.; Zhu, X.; Zha, X.; Angelidaki, I. Methane oxidising bacteria to upcycle effluent streams from anaerobic digestion of municipal biowaste. J. Environ. Manag. 2019, 251, 109590. [Google Scholar] [CrossRef]
- Lau, E.; Iv, E.J.N.; Dillard, Z.W.; Dague, R.D.; Semple, A.L.; Wentzell, W.L. High Throughput Sequencing to Detect Differences in Methanotrophic Methylococcaceae and Methylocystaceae in Surface Peat, Forest Soil, and Sphagnum Moss in Cranesville Swamp Preserve, West Virginia, USA. Microorganisms 2015, 3, 113–136. [Google Scholar] [CrossRef]
- Sengupta, A.; Dick, W.A. Methanotrophic bacterial diversity in two diverse soils under varying land-use practices as determined by high-throughput sequencing of the pmoA gene. Appl. Soil Ecol. 2017, 119, 35–45. [Google Scholar] [CrossRef]
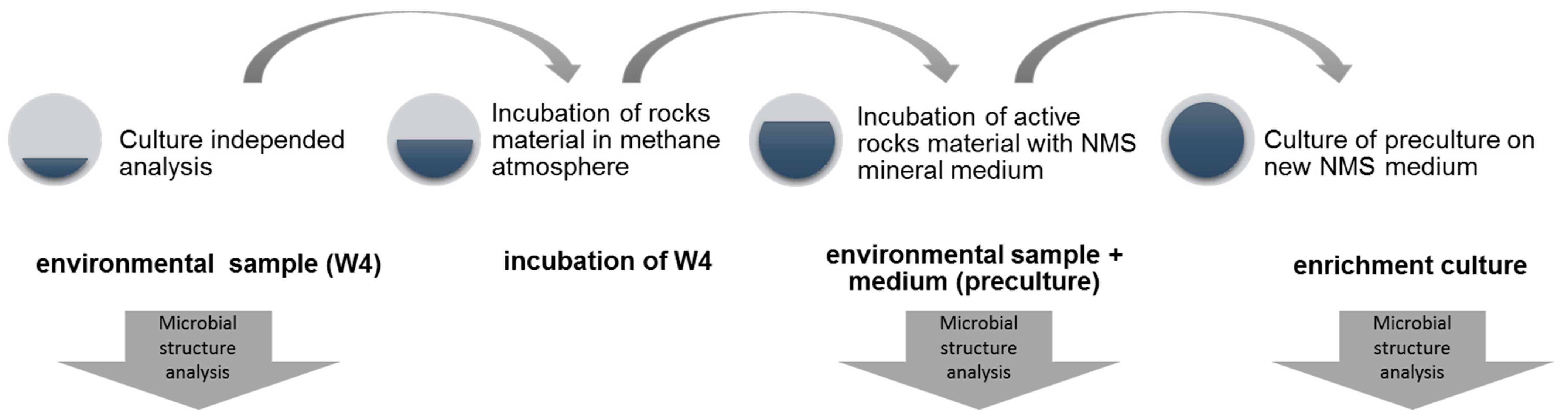
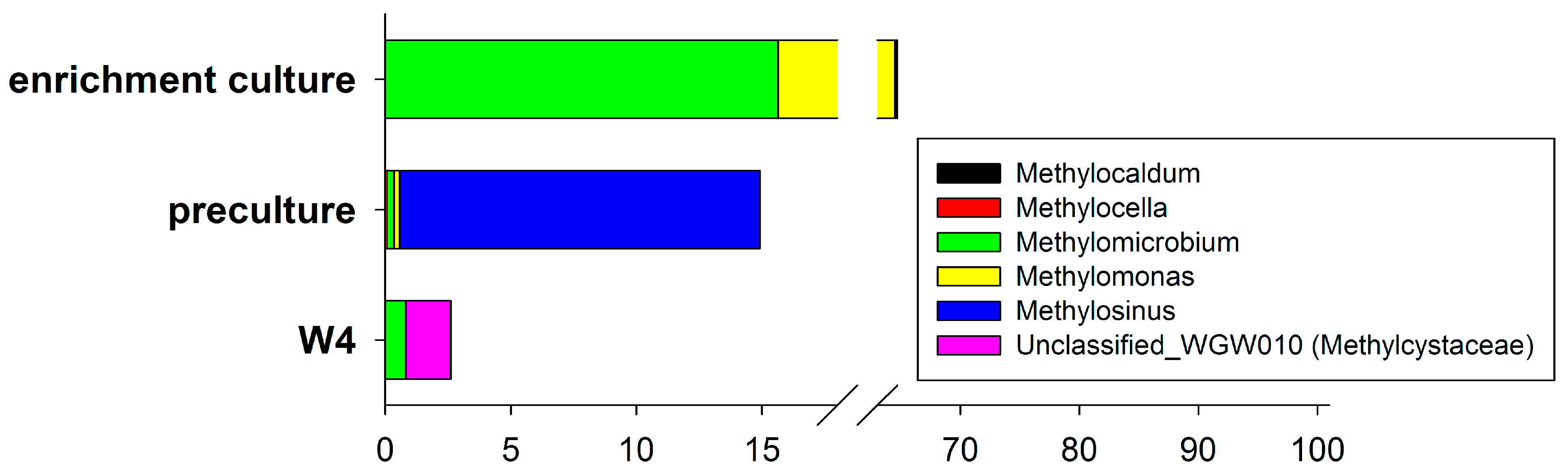
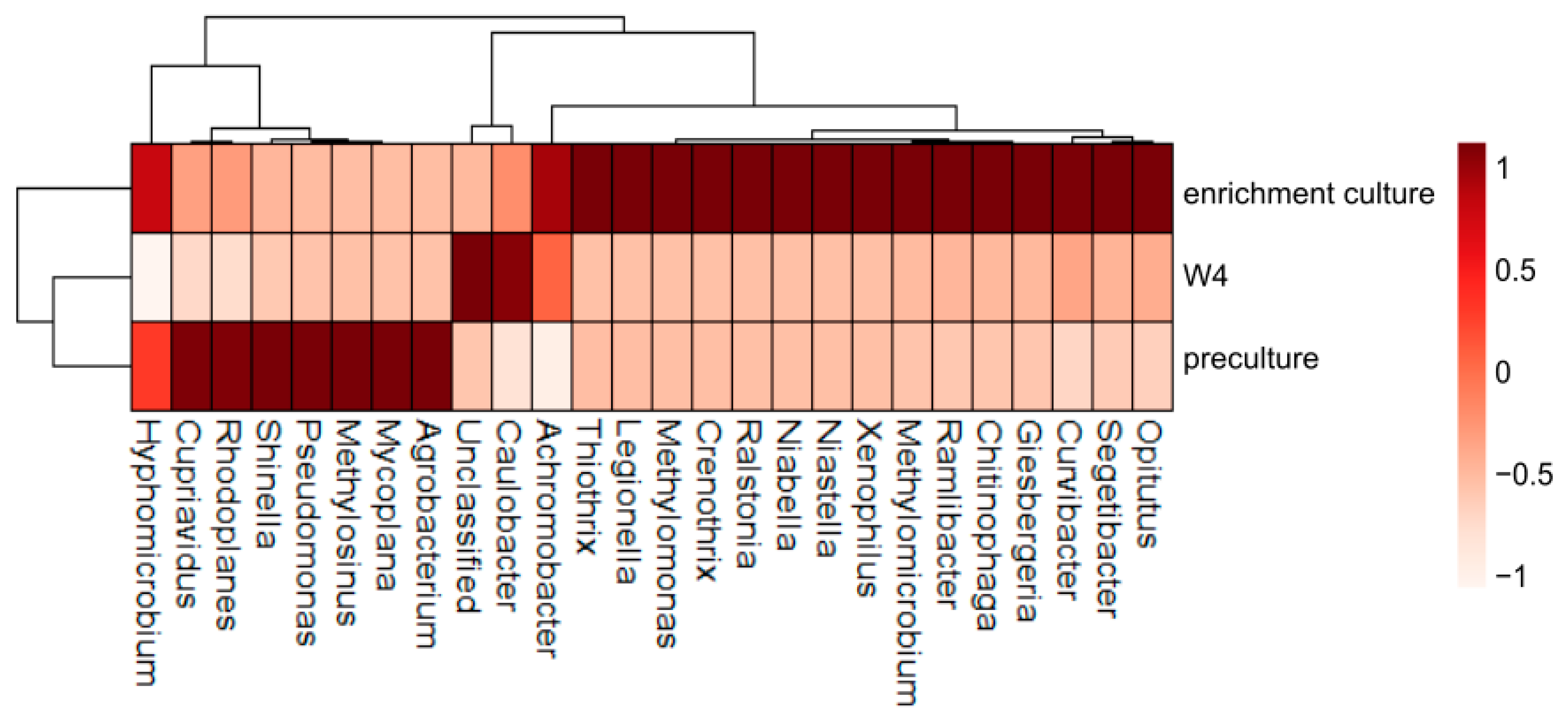
| Sample Name | Research Material | Medium | Substrate Addition (CH4) |
|---|---|---|---|
| W4 | crushed rocks surrounding Wieliczka salt deposits | no medium | no CH4 |
| W4 incubation | crushed rocks surrounding Wieliczka salt deposits | destiled water | approx. 10% CH4 |
| preculture | W4 incubation | mineral medium (NMS) | approx. 10% CH4 |
| enrichment culture | preculture | mineral medium (NMS) | approx. 10% CH4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goraj, W.; Pytlak, A.; Grządziel, J.; Gałązka, A.; Stępniewska, Z.; Szafranek-Nakonieczna, A. Dynamics of Methane-Consuming Biomes from Wieliczka Formation: Environmental and Enrichment Studies. Biology 2023, 12, 1420. https://doi.org/10.3390/biology12111420
Goraj W, Pytlak A, Grządziel J, Gałązka A, Stępniewska Z, Szafranek-Nakonieczna A. Dynamics of Methane-Consuming Biomes from Wieliczka Formation: Environmental and Enrichment Studies. Biology. 2023; 12(11):1420. https://doi.org/10.3390/biology12111420
Chicago/Turabian StyleGoraj, Weronika, Anna Pytlak, Jarosław Grządziel, Anna Gałązka, Zofia Stępniewska, and Anna Szafranek-Nakonieczna. 2023. "Dynamics of Methane-Consuming Biomes from Wieliczka Formation: Environmental and Enrichment Studies" Biology 12, no. 11: 1420. https://doi.org/10.3390/biology12111420
APA StyleGoraj, W., Pytlak, A., Grządziel, J., Gałązka, A., Stępniewska, Z., & Szafranek-Nakonieczna, A. (2023). Dynamics of Methane-Consuming Biomes from Wieliczka Formation: Environmental and Enrichment Studies. Biology, 12(11), 1420. https://doi.org/10.3390/biology12111420








