Characterising the Physiological Responses of Chinook Salmon (Oncorhynchus tshawytscha) Subjected to Heat and Oxygen Stress
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chinook Salmon HSP90, SOD1, POLR2F and RPL18 Gene Discovery and Synteny Analysis
2.2. Sequence Alignments and Phylogenetic Analysis
2.3. Fish Rearing Prior to the Temperature/DO Trial
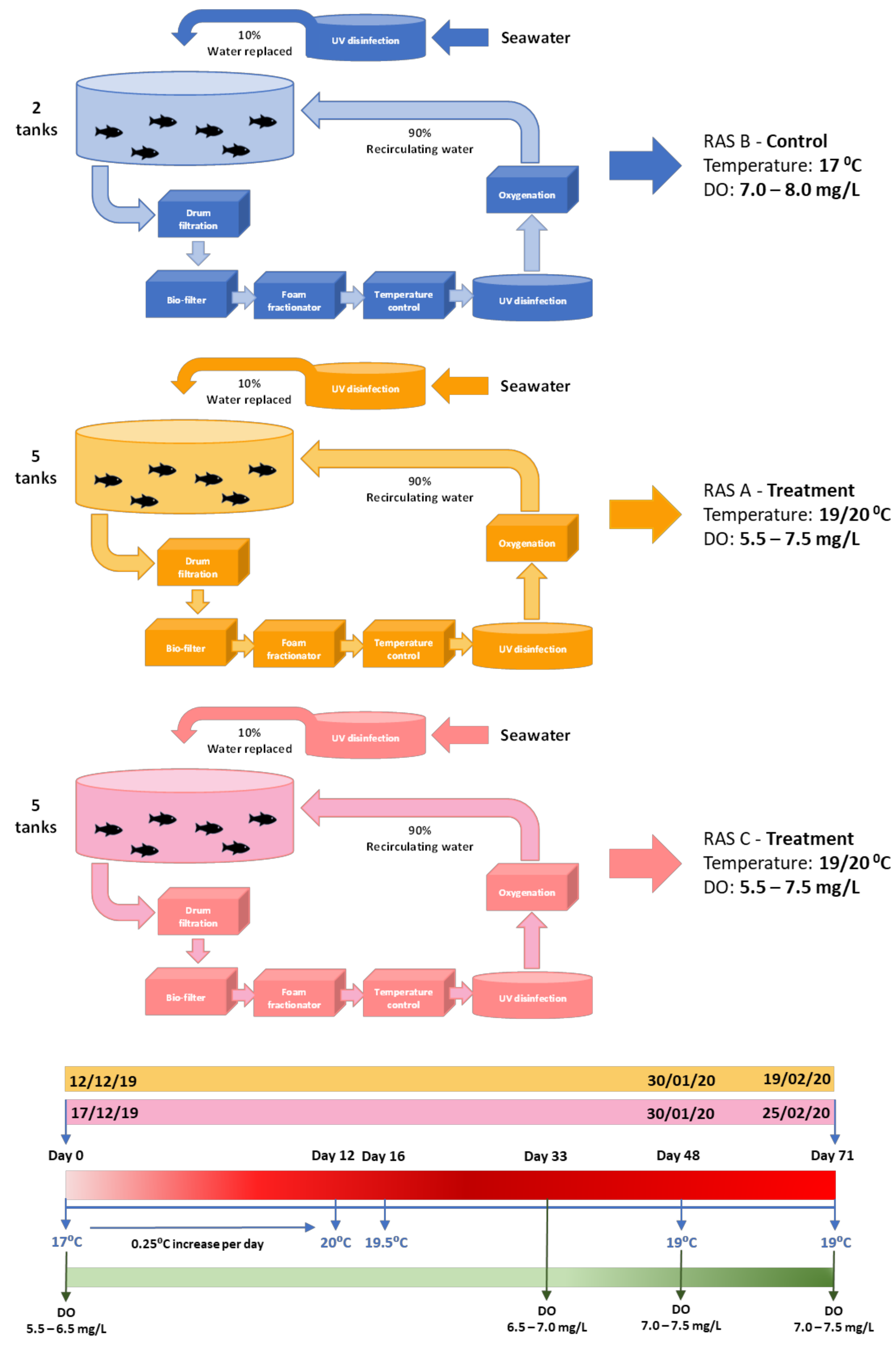
2.4. FRC Rearing Systems
2.5. Chinook Salmon Temperature/DO Trial
2.5.1. Fish Husbandry during the Trial
2.5.2. Assessment of Fish for the Trial
2.5.3. Trial Set-Up
2.5.4. Trial Adjustments
2.6. Fish Assessment and Sampling
2.7. RNA Extraction and cDNA Synthesis
2.8. Primer Design and Quantitative PCR (qPCR)
3. Results
3.1. Daily Temperature and DO of Each RAS System during the Trial
3.2. Chinook Salmon Performance Parameters
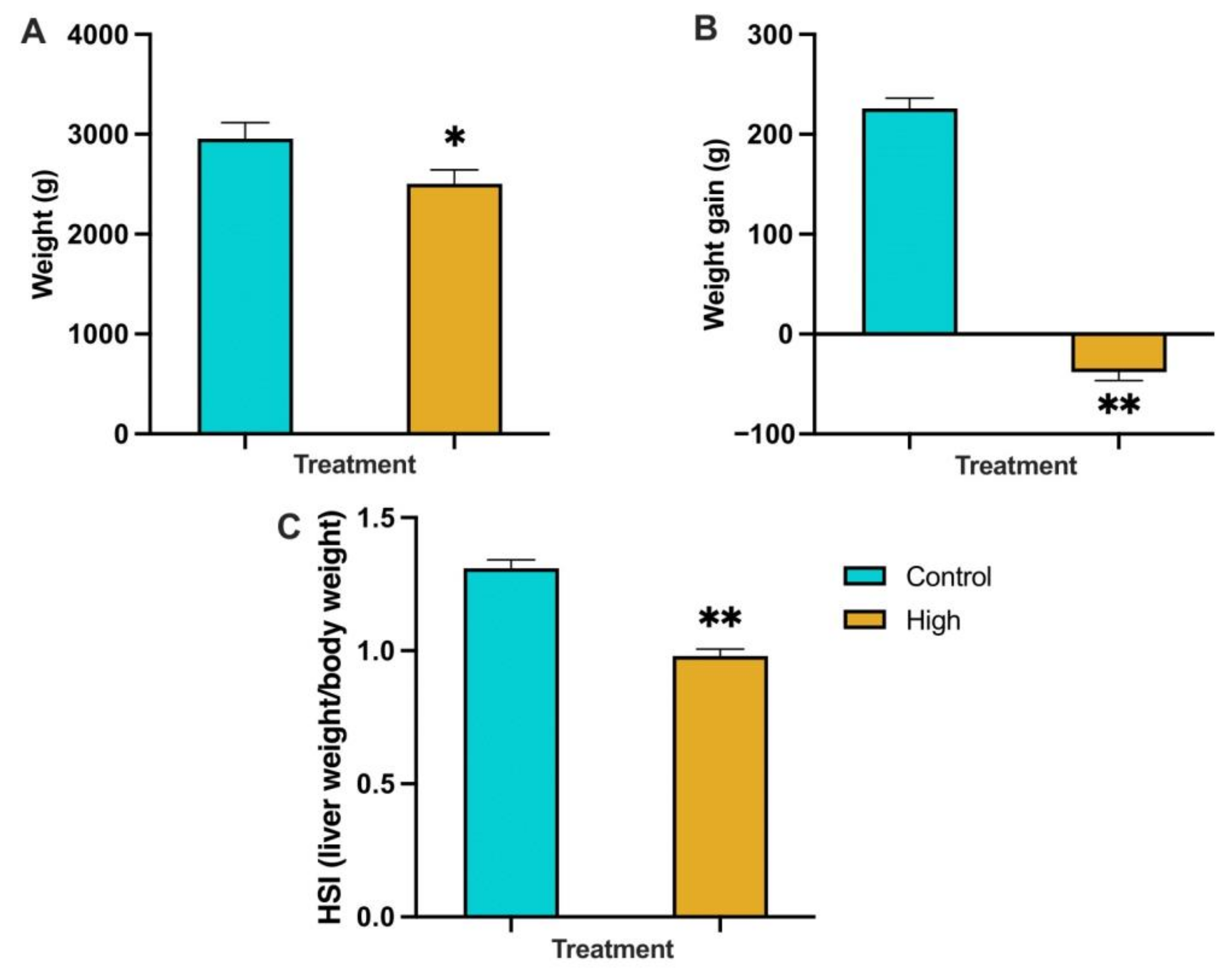
3.2.1. Fish Weight, Weight Gain and Hepatosomatic Index (HSI) at the End of the Trial (Sampled Population)
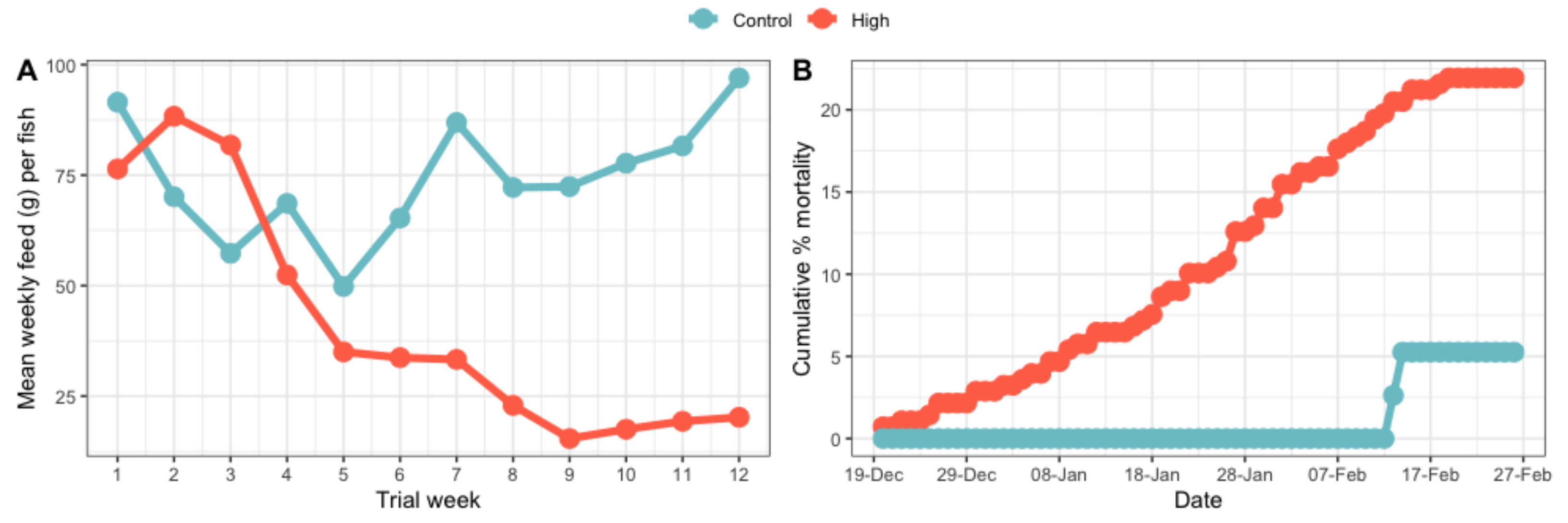
3.2.2. Feed Intake Per Fish and Fish Mortalities during the Trial (Whole Population)
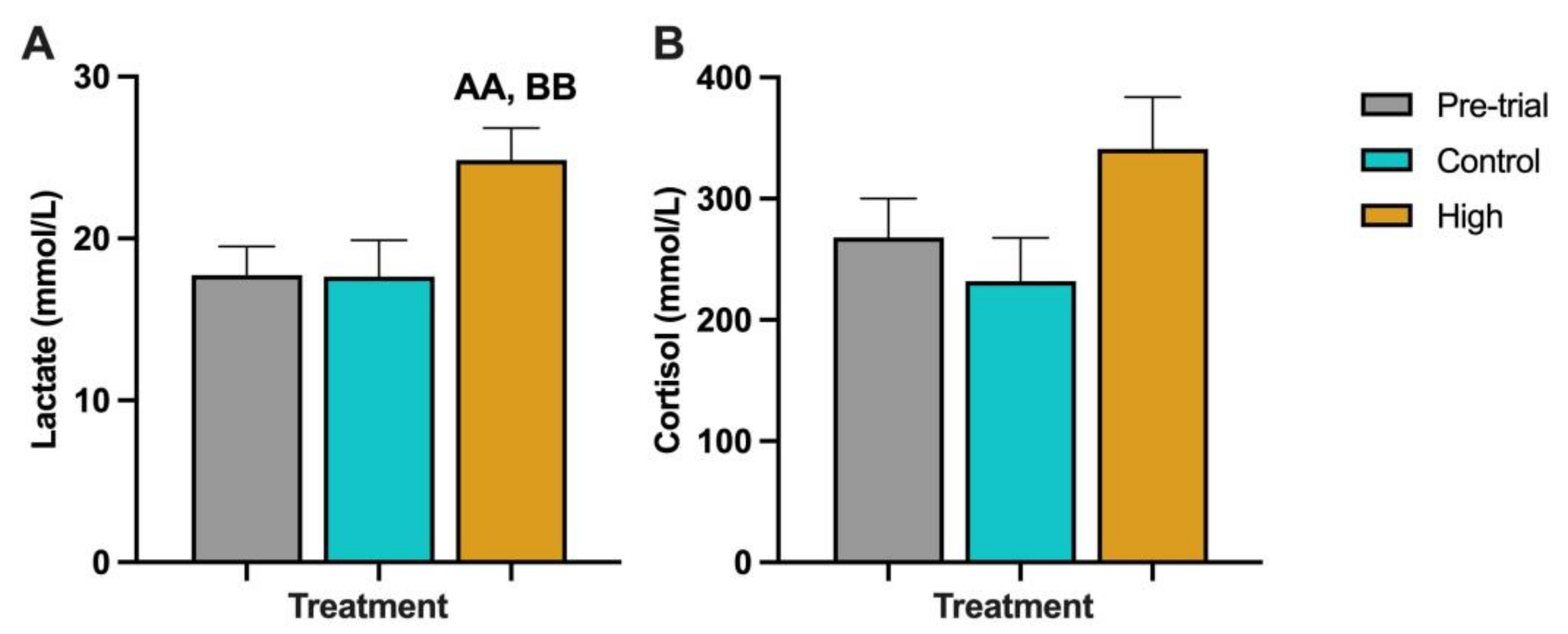
3.3. Chinook Salmon Haematology and Plasma Biochemistry
3.4. Characterisation of Chinook Salmon Housekeeping Genes
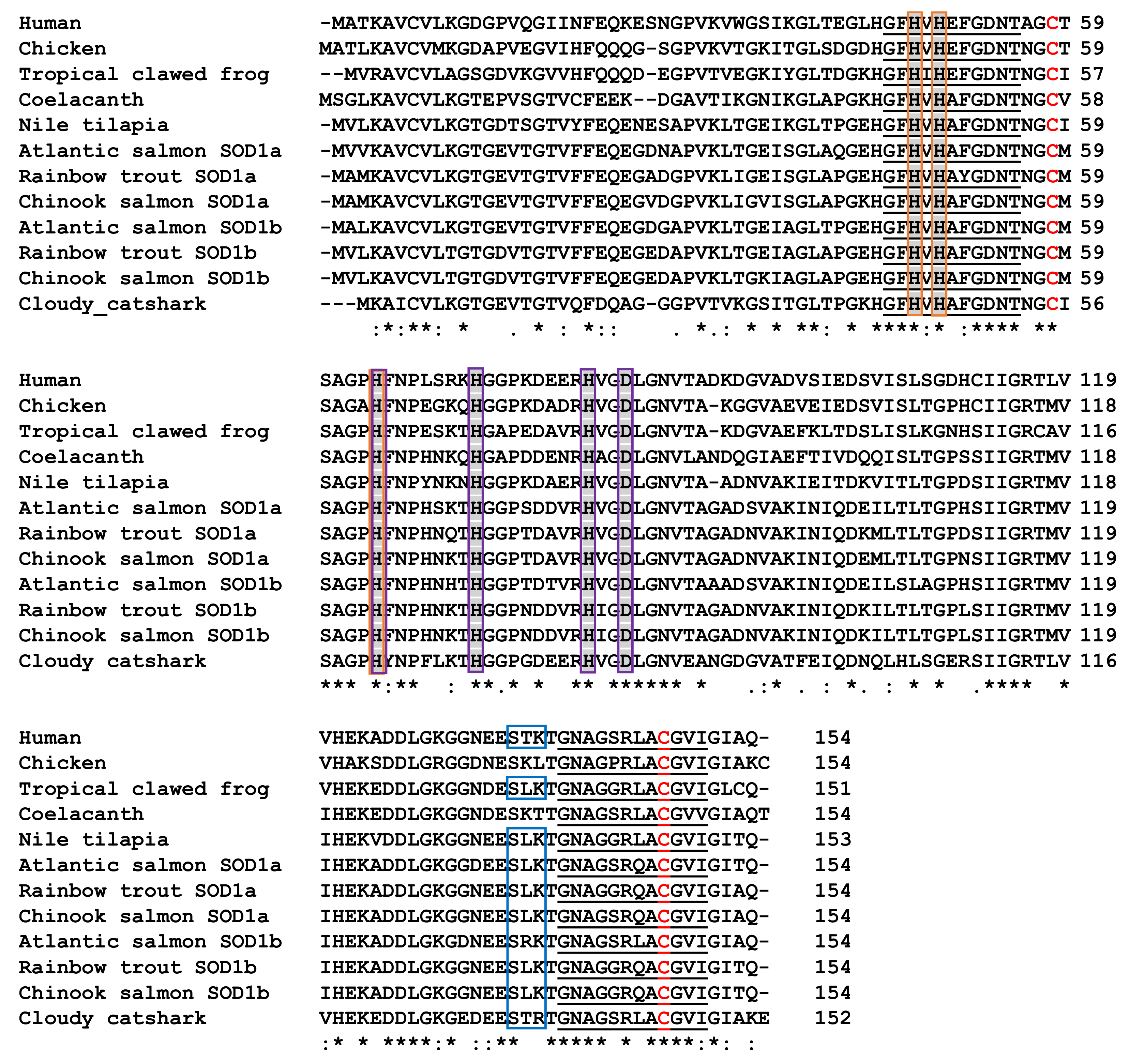
3.5. Characterisation of Chinook Salmon SOD1 Genes
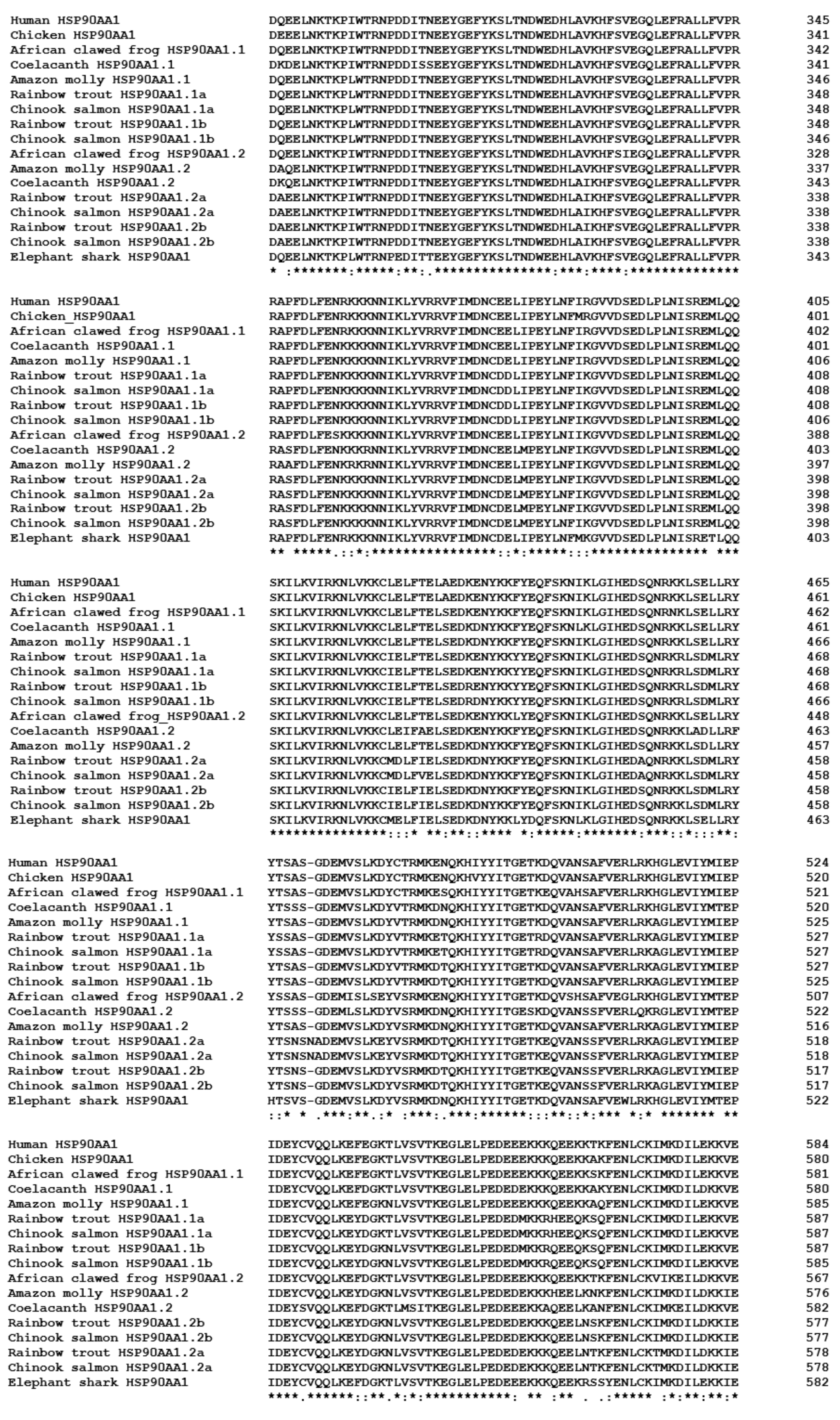
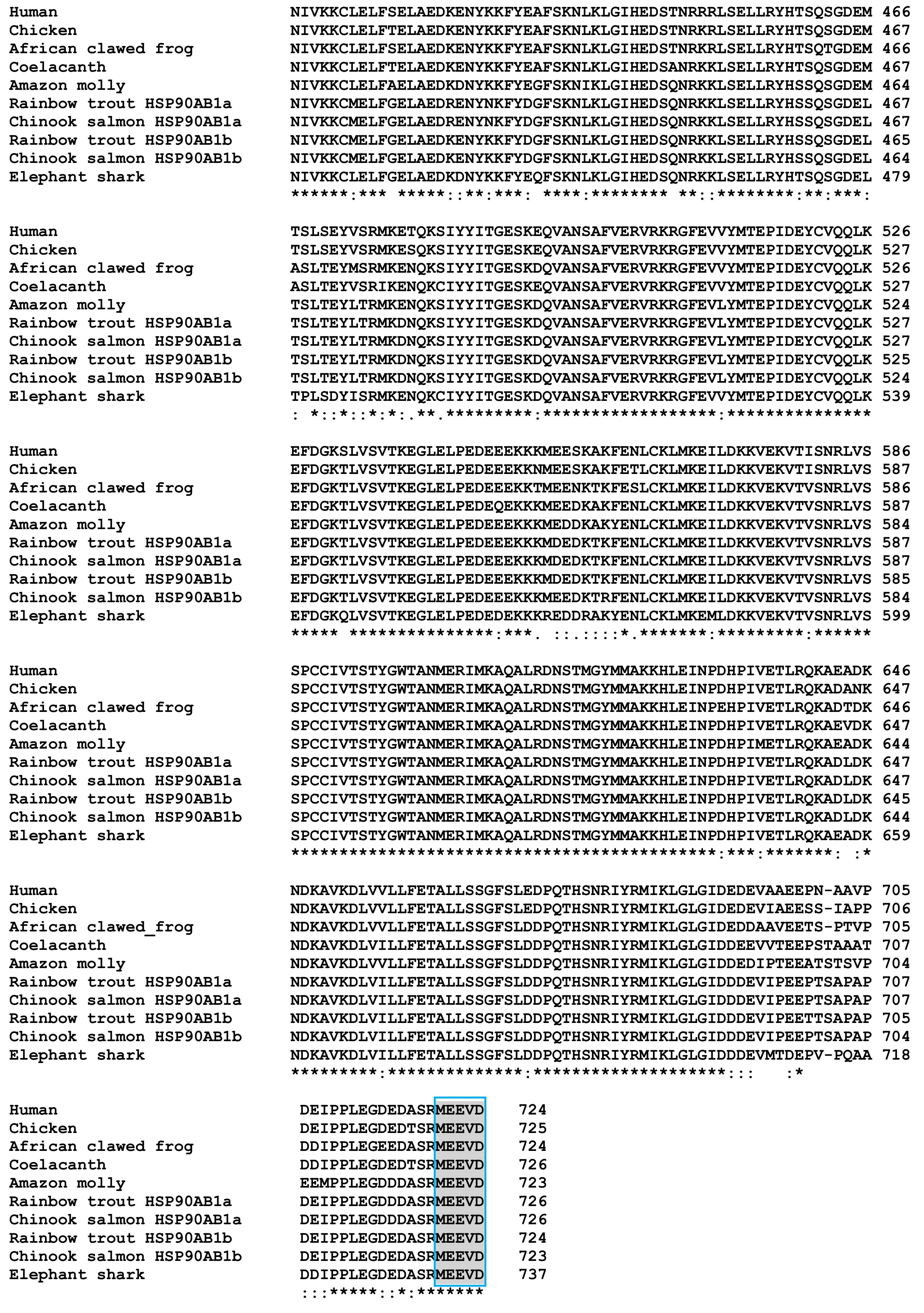
3.6. Characterisation of Chinook Salmon HSP90 Genes
3.7. Chinook Salmon SOD1 and HSP90 Expression during the Temperature/DO Trial
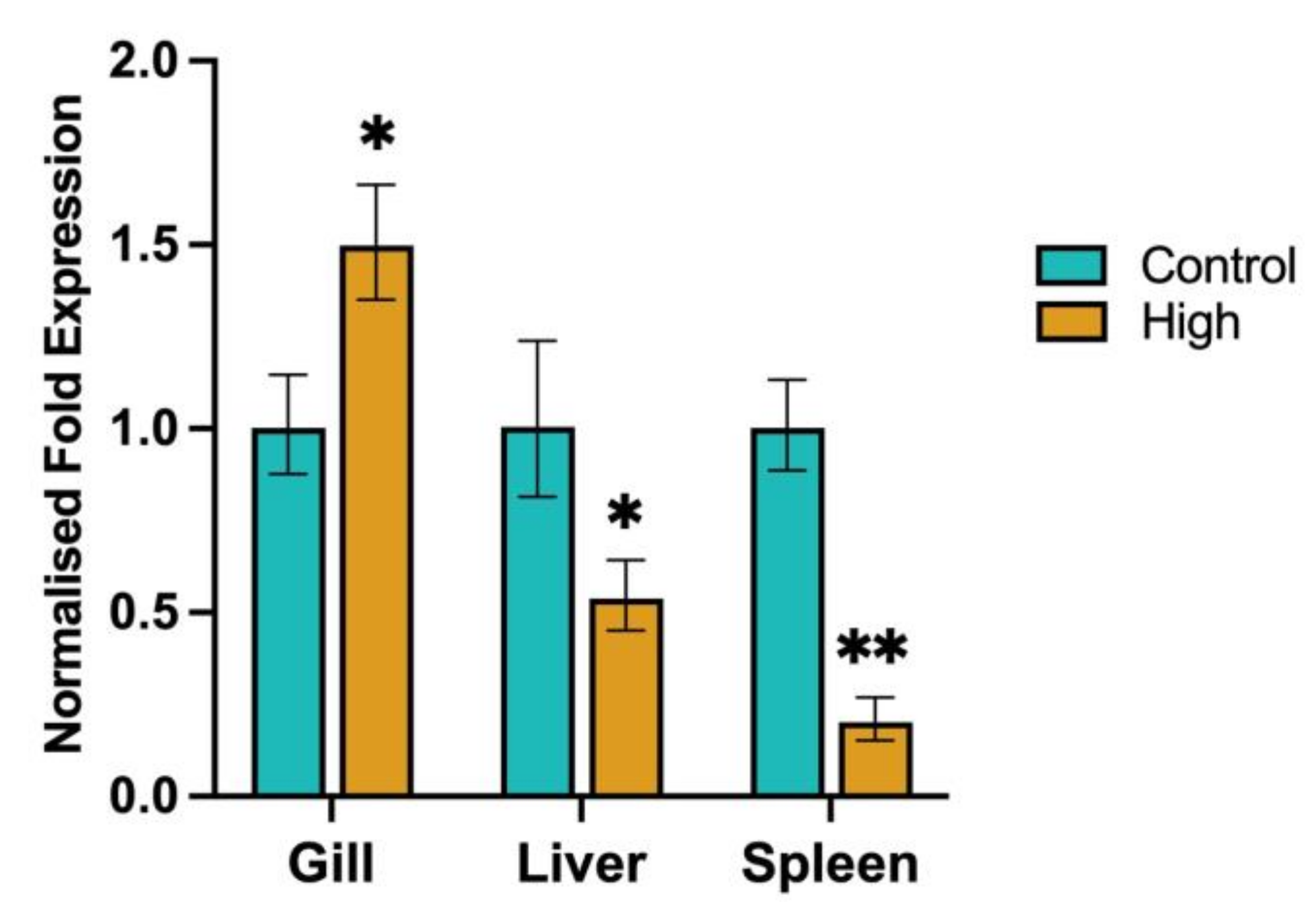
3.7.1. SOD1
3.7.2. HSP90
4. Discussion
4.1. General Health Parameters
4.1.1. Weight Change and Feed Intake
4.1.2. Mortality
4.2. Blood Health Parameters
4.3. Gene Expression Study
4.3.1. Chinook Salmon POLR2F and RPL18
4.3.2. Chinook Salmon SOD1 and HSP90
SOD1
HSP90
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olsvik, P.A.; Vikeså, V.; Lie, K.K. Transcriptional responses to temperature and low oxygen stress in Atlantic salmon studied with next-generation sequencing technology. BMC Genom. 2013, 14, 817. [Google Scholar] [CrossRef] [PubMed]
- Alfonso, S.; Gesto, M.; Sadoul, B. Temperature increase and its effects on fish stress physiology in the context of global warming. J. Fish Biol. 2020, 98, 1496–1508. [Google Scholar] [CrossRef] [PubMed]
- Beemelmanns, A.; Zanuzzo, F.S.; Xue, X. The transcriptomic responses of Atlantic salmon (Salmo salar) to high temperature stress alone, and in combination with moderate hypoxia. BMC Genom. 2021, 22, 261. [Google Scholar] [CrossRef] [PubMed]
- De Burgh-Day, C.O.; Spillman, C.M.; Smith, G.; Stevens, C.L. Forecasting extreme marine heat events in key aquaculture regions around New Zealand. J. South. Hemisph. Earth Syst. Sci. 2022, 72, 58–72. [Google Scholar] [CrossRef]
- Brosnahan, C. Diagnostic Investigation into Summer Mortality Events of Farmed Chinook salmon (Oncorhynchus tshawytscha) in New Zealand. Ph.D. Thesis, Massey University, Palmerston North, New Zealand, 2020. [Google Scholar]
- Chiswell, S.M.; Sutton, P.J. Relationships between long-term ocean warming, marine heat waves and primary production in the New Zealand region. N. Z. J. Mar. Freshw. Res. 2020, 54, 614–635. [Google Scholar] [CrossRef]
- NZKS. Annual Report FY19; New Zealand King Salmon Ltd.: Nelson, New Zealand, 2019. [Google Scholar]
- Broekhuizen, N.; Plew, D.R.; Pinkerton, M.H.; Gall, M.G. Sea temperature rise over the period 2002–2020 in Pelorus Sound, New Zealand—With possible implications for the aquaculture industry. N. Z. J. Mar. Freshw. Res. 2021, 55, 46–64. [Google Scholar] [CrossRef]
- Rosewarne, G.; Rodgers, B.; Lovell, G.; Ryder, J. FY22 Results and Equity Raising Presentation; New Zealand King Salmon Ltd.: Nelson, New Zealand, 2022. [Google Scholar]
- Huang, J.; Li, Y.; Liu, Z.; Kang, Y.; Wang, J. Transcriptomic responses to heat stress in rainbow trout Oncorhynchus mykiss head kidney. Fish Shellfish Immunol. 2018, 82, 32–40. [Google Scholar] [CrossRef]
- Hurst, T. Causes and consequences of winter mortality in fishes. J. Fish Biol. 2007, 71, 315–345. [Google Scholar] [CrossRef]
- Casillas, E.; Smith, L.S. Effect of stress on blood coagulation and haematology in rainbow trout (Salmo gairdneri). J. Fish Biol. 1977, 10, 481–491. [Google Scholar] [CrossRef]
- Caldwell, C.A.; Hinshaw, J. Physiological and haematological responses in rainbow trout subjected to supplemental dissolved oxygen in fish culture. Aquaculture 1994, 126, 183–193. [Google Scholar] [CrossRef]
- PagÉes, T.; Gomez, E.; Suner, O.; Viscor, G.; Tort, L. Effects of daily management stress on haematology and blood rheology of the gilthead seabream. J. Fish Biol. 1995, 46, 775–786. [Google Scholar] [CrossRef]
- Fazio, F.; Ferrantelli, V.; Fortino, G.; Arfuso, F.; Giangrosso, G.; Faggio, C. The influence of acute handling stress on some blood parameters in cultured sea bream (Sparus aurata Linnaeus, 1758). Ital. J. Food Saf. 2015, 4, 4174. [Google Scholar] [CrossRef] [PubMed]
- Narra, M.R.; Rajender, K.; Reddy, R.R.; Murty, U.S.; Begum, G. Insecticides induced stress response and recuperation in fish: Biomarkers in blood and tissues related to oxidative damage. Chemosphere 2017, 168, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Burgos-Aceves, M.A.; Lionetti, L.; Faggio, C. Multidisciplinary haematology as prognostic device in environmental and xenobiotic stress-induced response in fish. Sci. Total Environ. 2019, 670, 1170–1183. [Google Scholar] [CrossRef] [PubMed]
- Grzelak, A.K.; Davis, D.J.; Caraker, S.M.; Crim, M.J.; Spitsbergen, J.M.; Wiedmeyer, C.E. Stress leukogram induced by acute and chronic stress in zebrafish (Danio rerio). Comp. Med. 2017, 67, 263–269. [Google Scholar] [PubMed]
- Wendelaar Bonga, S.E. The stress response in fish. Physiol. Rev. 1997, 77, 591–625. [Google Scholar] [CrossRef] [PubMed]
- Palmisano, A.N.; Winton, J.R.; Dickhoff, W.W. Tissue-specific induction of Hsp90 mRNA and plasma cortisol response in chinook salmon following heat shock, seawater challenge, and handling challenge. Mar. Biotechnol. 2000, 2, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Calcagno, E.; Durando, P.; Valdés, M.E.; Franchioni, L.; de los Ángeles Bistoni, M. Effects of carbamazepine on cortisol levels and behavioral responses to stress in the fish Jenynsia multidentata. Physiol. Behav. 2016, 158, 68–75. [Google Scholar] [CrossRef]
- Guardiola, F.A.; Cuesta, A.; Esteban, M.Á. Using skin mucus to evaluate stress in gilthead seabream (Sparus aurata L.). Fish Shellfish. Immunol. 2016, 59, 323–330. [Google Scholar] [CrossRef]
- Schreck, C.B.; Tort, L.; Farrell, A.P.; Brauner, C.J. Biology of Stress in Fish; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Pottinger, T. Changes in blood cortisol, glucose and lactate in carp retained in anglers’ keepnets. J. Fish Biol. 1998, 53, 728–742. [Google Scholar] [CrossRef]
- Wells, R.M.; Pankhurst, N.W. Evaluation of simple instruments for the measurement of blood glucose and lactate, and plasma protein as stress indicators in fish. J. World Aquac. Soc. 1999, 30, 276–284. [Google Scholar] [CrossRef]
- Grutter, A.; Pankhurst, N. The effects of capture, handling, confinement and ectoparasite load on plasma levels of cortisol, glucose and lactate in the coral reef fish Hemigymnus melapterus. J. Fish Biol. 2000, 57, 391–401. [Google Scholar] [CrossRef]
- Pichavant, K.; Pichavant, K.; Maxime, V.; Thebault, M.; Ollivier, H.; Garnier, J.; Bousquet, B.; Diouris, M.; Boeuf, G.; Nonnotte, G. Effects of hypoxia and subsequent recovery on turbot Scophthalmus maximus: Hormonal changes and anaerobic metabolism. Mar. Ecol. Prog. Ser. 2002, 225, 275–285. [Google Scholar] [CrossRef]
- Lushchak, V.I.; Bagnyukova, T.V.; Lushchak, V.; Storey, J.M.; Storey, K.B. Hypoxia and recovery perturb free radical processes and antioxidant potential in common carp (Cyprinus carpio) tissues. Int. J. Biochem. Cell Biol. 2005, 37, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Pollock, M.; Clarke, L.; Dubé, M. The effects of hypoxia on fishes: From ecological relevance to physiological effects. Environ. Rev. 2007, 15, 1–14. [Google Scholar] [CrossRef]
- Abdel-Tawwab, M.; Monier, M.N.; Hoseinifar, S.H.; Faggio, C. Fish response to hypoxia stress: Growth, physiological, and immunological biomarkers. Fish Physiol. Biochem. 2019, 45, 997–1013. [Google Scholar] [CrossRef] [PubMed]
- Goikoetxea, A.; Sadoul, B.; Blondeau-Bidet, E.; Aerts, J.; Blanc, M.O.; Parrinello, H.; Barrachina, C.; Pratlong, M.; Geffroy, B. Genetic pathways underpinning hormonal stress responses in fish exposed to short- and long-term warm ocean temperatures. Ecol. Indic. 2021, 120, 106937. [Google Scholar] [CrossRef]
- Prunet, P.; Cairns, M.T.; Winberg, S.; Pottinger, T.G. Functional genomics of stress responses in fish. Rev. Fish. Sci. 2008, 16 (Suppl. 1), 157–166. [Google Scholar] [CrossRef]
- Currie, S.; Moyes, C.; Tufts, B. The effects of heat shock and acclimation temperature on hsp70 and hsp30 mRNA expression in rainbow trout: In vivo and in vitro comparisons. J. Fish Biol. 2000, 56, 398–408. [Google Scholar] [CrossRef]
- Kayhan, F.E.; Süsleyici Duman, B. Heat shock protein genes in fish. Turk. J. Fish. Aquat. Sci. 2010, 10, 287–293. [Google Scholar] [CrossRef]
- Ahmad, M.; Zuberi, A.; Ali, M.; Syed, A.; Khan, A.; Kamran, M. Effect of acclimated temperature on thermal tolerance, immune response and expression of HSP genes in Labeo rohita, Catla catla and their intergeneric hybrids. J. Therm. Biol. 2020, 89, 102570. [Google Scholar] [CrossRef] [PubMed]
- Dietz, T.J.; Somero, G.N. Species-and tissue-specific synthesis patterns for heat-shock proteins HSP70 and HSP90 in several marine teleost fishes. Physiol. Zool. 1993, 66, 863–880. [Google Scholar] [CrossRef]
- Parsell, D.; Lindquist, S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993, 27, 437–497. [Google Scholar] [CrossRef] [PubMed]
- Parsell, D.A.; Taulien, J.; Lindquist, S. The role of heat-shock proteins in thermotolerance. In Molecular Chaperones; Springer: Berlin/Heidelberg, Germany, 1993; pp. 23–30. [Google Scholar]
- Barua, D.; Heckathorn, S.A. The interactive effects of light and temperature on heat-shock protein accumulation in Solidago altissima (Asteraceae) in the field and laboratory. Am. J. Bot. 2006, 93, 102–109. [Google Scholar] [CrossRef]
- Welch, W.J. How cells respond to stress. Sci. Am. 1993, 268, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Liang, X.; Zhang, Y.; Li, Y.; Cao, X.; Gao, J. Cloning of three heat shock protein genes (HSP70, HSP90α and HSP90β) and their expressions in response to thermal stress in loach (Misgurnus anguillicaudatus) fed with different levels of vitamin C. Fish Shellfish Immunol. 2017, 66, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Veilleux, H.D.; Ryu, T.; Donelson, J.M.; Ravasi, T.; Munday, P.L. Molecular response to extreme summer temperatures differs between two genetically differentiated populations of a coral reef fish. Front. Mar. Sci. 2018, 5, 349. [Google Scholar] [CrossRef]
- Sales, C.F.; Lemos, F.S.; Morais, R.D.; Thomé, R.G.; Santos, H.B.; Pinheiro, A.P.; Bazzoli, N.; Rizzo, E. Thermal stress induces heat shock protein 70 and apoptosis during embryo development in a Neotropical freshwater fish. Reprod. Fertil. Dev. 2019, 31, 547–556. [Google Scholar] [CrossRef]
- De Alba, G.; López-Olmeda, J.F.; Sánchez-Vázquez, F.J. Rearing temperature conditions (constant vs. thermocycle) affect daily rhythms of thermal tolerance and sensing in zebrafish. J. Therm. Biol. 2021, 97, 102880. [Google Scholar] [CrossRef]
- Smith, T.; Tremblay, G.; Bradley, T. Characterization of the heat shock protein response of Atlantic salmon (Salmo salar). Fish Physiol. Biochem. 1999, 20, 279–292. [Google Scholar] [CrossRef]
- Lund, S.G.; Caissie, D.; Cunjak, R.A.; Vijayan, M.M.; Tufts, B.L. The effects of environmental heat stress on heat-shock mRNA and protein expression in Miramichi Atlantic salmon (Salmo salar) parr. Can. J. Fish. Aquat. Sci. 2002, 59, 1553–1562. [Google Scholar] [CrossRef]
- Gallant, M.J.; LeBlanc, S.; MacCormack, T.J.; Currie, S. Physiological responses to a short-term, environmentally realistic, acute heat stress in Atlantic salmon, Salmo salar. Facets 2017, 2, 330–341. [Google Scholar] [CrossRef]
- Akbarzadeh, A.; Günther, O.P.; Houde, A.L.; Li, S.; Ming, T.J.; Jeffries, K.M.; Hinch, S.G.; Miller, K.M. Developing specific molecular biomarkers for thermal stress in salmonids. BMC Genom. 2018, 19, 749. [Google Scholar] [CrossRef] [PubMed]
- Johnston, I.A. Expression of heat shock protein (Hsp90) paralogues is regulated by amino acids in skeletal muscle of Atlantic salmon. PLoS ONE 2013, 8, e74295. [Google Scholar]
- Ma, F.; Liu, Z.; Kang, Y.; Quan, J. Genome-wide identification of hsp90 gene in rainbow trout (Oncorhynchus mykiss) and their regulated expression in response to heat stress. DNA Cell Biol. 2020, 39, 428–440. [Google Scholar] [CrossRef] [PubMed]
- Palmisano, A.N.; Winton, J.R.; Dickhoff, W.W. Sequence features and phylogenetic analysis of the stress protein hsp90α in chinook salmon (Oncorhynchus tshawytscha), a poikilothermic vertebrate. Biochem. Biophys. Res. Commun. 1999, 258, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.J.P.; Shin, D.S.; Getzoff, E.D.; Tainer, J.A. The structural biochemistry of the superoxide dismutases. Biochim. Biophys. Acta 2010, 1804, 245–262. [Google Scholar] [CrossRef]
- Malek, R.L.; Sajadi, H.; Abraham, J.; Grundy, M.A.; Gerhard, G.S. The effects of temperature reduction on gene expression and oxidative stress in skeletal muscle from adult zebrafish. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2004, 138, 363–373. [Google Scholar] [CrossRef]
- Lushchak, V.I.; Bagnyukova, T.V. Temperature increase results in oxidative stress in goldfish tissues. 1. Indices of oxidative stress. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2006, 143, 30–35. [Google Scholar] [CrossRef]
- Bagnyukova, T.; Lushchak, O.; Storey, K.B.; Lushchak, V. Oxidative stress and antioxidant defense responses by goldfish tissues to acute change of temperature from 3 to 23 °C. J. Therm. Biol. 2007, 32, 227–234. [Google Scholar] [CrossRef]
- Vinagre, C.; Madeira, D.; Narciso, L.; Cabral, H.N.; Diniz, M. Effect of temperature on oxidative stress in fish: Lipid peroxidation and catalase activity in the muscle of juvenile seabass, Dicentrarchus labrax. Ecol. Indic. 2012, 23, 274–279. [Google Scholar] [CrossRef]
- Madeira, D.; Narciso, L.; Cabral, H.; Vinagre, C.; Diniz, M. Influence of temperature in thermal and oxidative stress responses in estuarine fish. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2013, 166, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-H.; Yang, F.F.; Liao, S.A.; Miao, Y.T.; Ye, C.X.; Wang, A.L.; Tan, J.W.; Chen, X.Y. High temperature induces apoptosis and oxidative stress in pufferfish (Takifugu obscurus) blood cells. J. Therm. Biol. 2015, 53, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.D.; Borges, V.D.; Rosa, C.E.; Colares, E.P.; Robaldo, R.B.; Martinez, P.E.; Bianchini, A. Effects of increasing temperature on antioxidant defense system and oxidative stress parameters in the Antarctic fish Notothenia coriiceps and Notothenia rossii. J. Therm. Biol. 2017, 68, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Pedrajas, J.; Peinado, J.; Lopez-Barea, J. Oxidative stress in fish exposed to model xenobiotics. Oxidatively modified forms of Cu, Zn-superoxide dismutase as potential biomarkers. Chem. -Biol. Interact. 1995, 98, 267–282. [Google Scholar] [CrossRef]
- Pandey, S.; Parvez, S.; Sayeed, I.; Haque, R.; Bin-Hafeez, B.; Raisuddin, S. Biomarkers of oxidative stress: A comparative study of river Yamuna fish Wallago attu (Bl. & Schn.). Sci. Total Environ. 2003, 309, 105–115. [Google Scholar] [PubMed]
- Pascual, P.; Pedrajas, J.; Toribio, F.; López-Barea, J.; Peinado, J. Effect of food deprivation on oxidative stress biomarkers in fish (Sparus aurata). Chem. -Biol. Interact. 2003, 145, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Olsvik, P.; Kristensen, T.; Waagbø, R.; Rosseland, B.; Tollefsen, K.E.; Baeverfjord, G.; Berntssen, M. mRNA expression of antioxidant enzymes (SOD, CAT and GSH-Px) and lipid peroxidative stress in liver of Atlantic salmon (Salmo salar) exposed to hyperoxic water during smoltification. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2005, 141, 314–323. [Google Scholar] [CrossRef]
- Sun, F.-H.; Zhou, Q.-X. Oxidative stress biomarkers of the polychaete Nereis diversicolor exposed to cadmium and petroleum hydrocarbons. Ecotoxicol. Environ. Saf. 2008, 70, 106–114. [Google Scholar] [CrossRef]
- Christensen, K.A.; Leong, J.S.; Sakhrani, D.; Biagi, C.A.; Minkley, D.R.; Withler, R.E.; Rondeau, E.B.; Koop, B.F.; Devlin, R.H. Chinook salmon (Oncorhynchus tshawytscha) genome and transcriptome. PLoS ONE 2018, 13, e0195461. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Burge, C.B.; Karlin, S. Finding the genes in genomic DNA. Curr. Opin. Struct. Biol. 1998, 8, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Salamov, A.A.; Solovyev, V.V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000, 10, 516–522. [Google Scholar] [CrossRef] [PubMed]
- Pearson, W.R.; Lipman, D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 1988, 85, 2444–2448. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools on the ExPASy Server; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Yates, A.; Akanni, W.; Amode, M.R.; Barrell, D.; Billis, K.; Carvalho-Silva, D.; Cummins, C.; Clapham, P.; Fitzgerald, S.; Gil, L. Ensembl 2016. Nucleic Acids Res. 2016, 44, D710–D716. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Page, R.D. Tree View: An application to display phylogenetic trees on personal computers. Bioinformatics 1996, 12, 357–358. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- De Castro, E.; Sigrist, C.J.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Symonds, J.E.; Walker, S.P.; Steiner, K.; Carter, C.G.; Bowman, J.P.; Nowak, B.F. Effects of feed ration and temperature on Chinook salmon (Oncorhynchus tshawytscha) microbiota in freshwater recirculating aquaculture systems. Aquaculture 2021, 543, 736965. [Google Scholar] [CrossRef]
- Lulijwa, R.; Young, T.; Symonds, J.E.; Walker, S.P.; Delorme, N.J.; Alfaro, A.C. Uncoupling thermotolerance and growth performance in chinook salmon: Blood biochemistry and immune capacity. Metabolites 2021, 11, 547. [Google Scholar] [CrossRef] [PubMed]
- Casanovas, P.; Walker, S.P.; Johnston, H.; Johnston, C.; Symonds, J.E. Comparative assessment of blood biochemistry and haematology normal ranges between Chinook salmon (Oncorhynchus tshawytscha) from seawater and freshwater farms. Aquaculture 2021, 537, 736464. [Google Scholar] [CrossRef]
- Ruijter, J.; Ramakers, C.; Hoogaars, W.; Karlen, Y.; Bakker, O.; Van den Hoff, M.; Moorman, A. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The ultimate qPCR experiment: Producing publication quality, reproducible data the first time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef]
- Baer, C.F.; Travis, J. Direct and correlated responses to artificial selection on acute thermal stress tolerance in a livebearing fish. Evolution 2000, 54, 238–244. [Google Scholar]
- Ashaf-Ud-Doulah, M.; Shahjahan, M.; Islam, S.M.; Al-Emran, M.; Rahman, M.S.; Hossain, M.A.R. Thermal stress causes nuclear and cellular abnormalities of peripheral erythrocytes in Indian major carp, rohu Labeo rohita. J. Therm. Biol. 2019, 86, 102450. [Google Scholar] [CrossRef] [PubMed]
- Petitjean, Q.; Jean, S.; Gandar, A.; Côte, J.; Laffaille, P.; Jacquin, L. Stress responses in fish: From molecular to evolutionary processes. Sci. Total Environ. 2019, 684, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Li, L.; Li, Q.; He, X.; Cui, Z. Transcriptomic characterization of temperature stress responses in larval zebrafish. PLoS ONE 2012, 7, e37209. [Google Scholar] [CrossRef] [PubMed]
- Handeland, S.O.; Imsland, A.K.; Stefansson, S.O. The effect of temperature and fish size on growth, feed intake, food conversion efficiency and stomach evacuation rate of Atlantic salmon post-smolts. Aquaculture 2008, 283, 36–42. [Google Scholar] [CrossRef]
- Smith, S.; Bernatchez, L.; Beheregaray, L.B. RNA-seq analysis reveals extensive transcriptional plasticity to temperature stress in a freshwater fish species. BMC Genom. 2013, 14, 375. [Google Scholar] [CrossRef] [PubMed]
- Tomalty, K.M.; Meek, M.H.; Stephens, M.R.; Rincón, G.; Fangue, N.A.; May, B.P.; Baerwald, M.R. Transcriptional response to acute thermal exposure in juvenile chinook salmon determined by RNAseq. G3 Genes Genomes Genet. 2015, 5, 1335–1349. [Google Scholar] [CrossRef] [PubMed]
- Poletto, J.B.; Cocherell, D.E.; Baird, S.E.; Nguyen, T.X.; Cabrera-Stagno, V.; Farrell, A.P.; Fangue, N.A. Unusual aerobic performance at high temperatures in juvenile Chinook salmon, Oncorhynchus tshawytscha. Conserv. Physiol. 2017, 5, cow067. [Google Scholar] [CrossRef] [PubMed]
- Meehl, G.A.; Tebaldi, C. More intense, more frequent, and longer lasting heat waves in the 21st century. Science 2004, 305, 994–997. [Google Scholar] [CrossRef]
- Somero, G.N. Adaptation of enzymes to temperature: Searching for basic “strategies”. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2004, 139, 321–333. [Google Scholar] [CrossRef]
- Somero, G. The physiology of climate change: How potentials for acclimatization and genetic adaptation will determine ‘winners’ and ‘losers’. J. Exp. Biol. 2010, 213, 912–920. [Google Scholar] [CrossRef]
- Evans, T.G.; Hammill, E.; Kaukinen, K.; Schulze, A.D.; Patterson, D.A.; English, K.K.; Curtis, J.M.; Miller, K.M. Transcriptomics of environmental acclimatization and survival in wild adult Pacific sockeye salmon (Oncorhynchus nerka) during spawning migration. Mol. Ecol. 2011, 20, 4472–4489. [Google Scholar] [CrossRef] [PubMed]
- Quinn, N.L.; McGowan, C.R.; Cooper, G.A.; Koop, B.F.; Davidson, W.S. Ribosomal genes and heat shock proteins as putative markers for chronic, sublethal heat stress in Arctic charr: Applications for aquaculture and wild fish. Physiol. Genom. 2011, 43, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Gracey, A.Y.; Troll, J.V.; Somero, G.N. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proc. Natl. Acad. Sci. USA 2001, 98, 1993–1998. [Google Scholar] [CrossRef] [PubMed]
- Martinovic, D.; Villeneuve, D.L.; Kahl, M.D.; Blake, L.S.; Brodin, J.D.; Ankley, G.T. Hypoxia alters gene expression in the gonads of zebrafish (Danio rerio). Aquat. Toxicol. 2009, 95, 258–272. [Google Scholar] [CrossRef] [PubMed]
- Kent, M.; Ojanguren, A.F. The effect of water temperature on routine swimming behaviour of new born guppies (Poecilia reticulata). Biol. Open 2015, 4, 547–552. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dell, A.I.; Pawar, S.; Savage, V.M. Systematic variation in the temperature dependence of physiological and ecological traits. Proc. Natl. Acad. Sci. USA 2011, 108, 10591–10596. [Google Scholar] [CrossRef] [PubMed]
- Madeira, D.; Araújo, J.E.; Vitorino, R.; Costa, P.M.; Capelo, J.L.; Vinagre, C.; Diniz, M.S. Molecular plasticity under ocean warming: Proteomics and fitness data provides clues for a better understanding of the thermal tolerance in fish. Front. Physiol. 2017, 8, 825. [Google Scholar] [CrossRef] [PubMed]
- Lepock, J.R. How do cells respond to their thermal environment? Int. J. Hyperth. 2005, 21, 681–687. [Google Scholar] [CrossRef]
- Boltaña, S.; Sanhueza, N.; Aguilar, A.; Gallardo-Escarate, C.; Arriagada, G.; Valdes, J.A.; Soto, D.; Quiñones, R.A. Influences of thermal environment on fish growth. Ecol. Evol. 2017, 7, 6814–6825. [Google Scholar] [CrossRef]
- Del Rio, A.M.; Davis, B.E.; Fangue, N.A.; Todgham, A.E. Combined effects of warming and hypoxia on early life stage Chinook salmon physiology and development. Conserv. Physiol. 2019, 7, coy078. [Google Scholar] [CrossRef]
- Blier, P.; Pelletier, D.; Dutil, J.D. Does aerobic capacity set a limit on fish growth rate? Rev. Fish. Sci. 1997, 5, 323–340. [Google Scholar] [CrossRef]
- Wade, N.M.; Clark, T.D.; Maynard, B.T.; Atherton, S.; Wilkinson, R.J.; Smullen, R.P.; Taylor, R.S. Effects of an unprecedented summer heatwave on the growth performance, flesh colour and plasma biochemistry of marine cage-farmed Atlantic salmon (Salmo salar). J. Therm. Biol. 2019, 80, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Gamperl, A.K.; Ajiboye, O.O.; Zanuzzo, F.S.; Sandrelli, R.M.; Ellen de Fátima, C.P.; Beemelmanns, A. The impacts of increasing temperature and moderate hypoxia on the production characteristics, cardiac morphology and haematology of Atlantic Salmon (Salmo salar). Aquaculture 2020, 519, 734874. [Google Scholar] [CrossRef]
- McCormick, S.; Shrimpton, J.; Carey, J.; O’dea, M.; Sloan, K.; Moriyama, S.; Björnsson, B.T. Repeated acute stress reduces growth rate of Atlantic salmon parr and alters plasma levels of growth hormone, insulin-like growth factor I and cortisol. Aquaculture 1998, 168, 221–235. [Google Scholar] [CrossRef]
- Bernier, N.J.; Craig, P.M. CRF-related peptides contribute to stress response and regulation of appetite in hypoxic rainbow trout. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2005, 289, R982–R990. [Google Scholar] [CrossRef] [PubMed]
- Bowen, L.; von Biela, V.R.; McCormick, S.D.; Regish, A.M.; Waters, S.C.; Durbin-Johnson, B.; Britton, M.; Settles, M.L.; Donnelly, D.S.; Laske, S.M. Transcriptomic response to elevated water temperatures in adult migrating Yukon River Chinook salmon (Oncorhynchus tshawytscha). Conserv. Physiol. 2020, 8, coaa084. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.C.; Bergersen, E.P. Influence of thermal and oxygen stratification on lake trout hooking mortality. N. Am. J. Fish. Manag. 1996, 16, 175–181. [Google Scholar] [CrossRef]
- Pastorino, P.; Vela Alonso, A.I.; Colussi, S.; Cavazza, G.; Menconi, V.; Mugetti, D.; Righetti, M.; Barbero, R.; Zuccaro, G.; Fernández-Garayzábal, J.F. A summer mortality outbreak of Lactococcosis by Lactococcus garvieae in a raceway system affecting farmed rainbow trout (Oncorhynchus mykiss) and brook trout (Salvelinus fontinalis). Animals 2019, 9, 1043. [Google Scholar] [CrossRef]
- Jeffries, K.M.; Hinch, S.G.; Sierocinski, T.; Pavlidis, P.; Miller, K.M. Transcriptomic responses to high water temperature in two species of Pacific salmon. Evol. Appl. 2014, 7, 286–300. [Google Scholar] [CrossRef]
- Matthews, W.J. Summer mortality of striped bass in reservoirs of the United States. Trans. Am. Fish. Soc. 1985, 114, 62–66. [Google Scholar] [CrossRef]
- Zale, A.V.; Wiechman, J.D.; Lochmiller, R.L.; Burroughs, J. Limnological conditions associated with summer mortality of striped bass in Keystone Reservoir, Oklahoma. Trans. Am. Fish. Soc. 1990, 119, 72–76. [Google Scholar] [CrossRef]
- Xiao, J.; Ford, S.E.; Yang, H.; Zhang, G.; Zhang, F.; Guo, X. Studies on mass summer mortality of cultured zhikong scallops (Chlamys farreri Jones et Preston) in China. Aquaculture 2005, 250, 602–615. [Google Scholar] [CrossRef]
- Choi, H.-S.; Myoung, J.I.; Park, M.; Cho, M.Y. A Study on the summer mortality of Korean rockfish Sebastes schlegelii in Korea. J. Fish Pathol. 2009, 22, 155–162. [Google Scholar]
- Fathi, M.; Dickson, C.; Dickson, M.; Leschen, W.; Baily, J.; Muir, F.; Ulrich, K.; Weidmann, M. Identification of tilapia lake virus in Egypt in Nile tilapia affected by ‘summer mortality’ syndrome. Aquaculture 2017, 473, 430–432. [Google Scholar] [CrossRef]
- Elsheshtawy, A.; Yehia, N.; Elkemary, M.; Soliman, H. Investigation of Nile tilapia summer mortality in Kafr El-Sheikh governorate, Egypt. Genet. Aquat. Org. 2019, 3, 17–25. [Google Scholar]
- Mommsen, T.P.; Vijayan, M.M.; Moon, T.W. Cortisol in teleosts: Dynamics, mechanisms of action, and metabolic regulation. Rev. Fish Biol. Fish. 1999, 9, 211–268. [Google Scholar] [CrossRef]
- Vijayan, M.M.; Aluru, N.; Leatherland, J.F. Stress response and the role of cortisol. Fish Dis. Disord. 2010, 2, 182–201. [Google Scholar]
- Wu, H.; Ohnuki, H.; Ota, S.; Murata, M.; Yoshiura, Y.; Endo, H. New approach for monitoring fish stress: A novel enzyme-functionalized label-free immunosensor system for detecting cortisol levels in fish. Biosens. Bioelectron. 2017, 93, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Maciuszek, M.; Rydz, L.; Świtakowska, I.; Verburg-van Kemenade, B.L.; Chadzińska, M. Effects of stress and cortisol on the polarization of carp macrophages. Fish Shellfish Immunol. 2019, 94, 27–37. [Google Scholar] [CrossRef]
- Ellis, T.; Yildiz, H.Y.; López-Olmeda, J.; Spedicato, M.T.; Tort, L.; Øverli, Ø.; Martins, C.I.M. Cortisol and finfish welfare. Fish Physiol. Biochem. 2012, 38, 163–188. [Google Scholar] [CrossRef]
- Schurr, A. Lactate, not pyruvate, is the end product of glucose metabolism via glycolysis. In Carbohydrate; Caliskan, M., Kavakli, I.H., Oz, G.C., Eds.; IntechOpen: London, UK, 2017. [Google Scholar]
- Omlin, T.; Weber, J.-M. Hypoxia stimulates lactate disposal in rainbow trout. J. Exp. Biol. 2010, 213, 3802–3809. [Google Scholar] [CrossRef] [PubMed]
- Mesa, M.G.; Weiland, L.; Wagner, P. Effects of acute thermal stress on the survival, predator avoidance, and physiology of juvenile fall chinook salmon. Northwest Sci. 2002, 76, 118–128. [Google Scholar]
- Romano, N.; Scapigliati, G.; Abelli, L. Water oxygen content affects distribution of T and B lymphocytes in lymphoid tissues of farmed Sea Bass (Dicentrarchus labrax). Fishes 2017, 2, 16. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Bjørløw, I.; Hagland, T.J.; Wergeland, H.I. Effect of seawater temperature on leucocyte populations in Atlantic salmon post-smolts. Vet. Immunol. Immunopathol. 2005, 106, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, O.D.R.; Rodríguez, M.H.; Ramirez, L.F.B. Thermal stress effect on tilapia, Oreochromis mossambicus (Pisces: Cichlidae), blood parameters. Mar. Freshw. Behav. Physiol. 2008, 41, 79–89. [Google Scholar] [CrossRef]
- Shahjahan, M.; Khatun, M.S.; Mun, M.M.; Islam, S.M.; Uddin, M.H.; Badruzzaman, M.; Khan, S. Nuclear and cellular abnormalities of erythrocytes in response to thermal stress in common carp, Cyprinus carpio. Front. Physiol. 2020, 11, 543. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.S.; Braasch, I.; Frickey, T.; Meyer, A.; Van de Peer, Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003, 13, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.R.; Kent, M.P.; Nome, T.; Davidson, W.S. The Atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef]
- Manchado, M.; Salas-Leiton, E.; Infante, C.; Ponce, M.; Asensio, E.; Crespo, A.; Cañavate, J.P. Molecular characterization, gene expression and transcriptional regulation of cytosolic HSP90 genes in the flatfish Senegalese sole (Solea senegalensis Kaup). Gene 2008, 416, 77–84. [Google Scholar] [CrossRef]
- Wang, P.F.; Zeng, S.; Xu, P.; Zhou, L.; Li, G.F. Two HSP90 genes in mandarin fish Siniperca chuatsi: Identification, characterization and their specific expression profiles during embryogenesis and under stresses. Fish Physiol. Biochem. 2016, 42, 1123–1136. [Google Scholar] [CrossRef]
- Acker, J.; de Graaff, M.; Cheynel, I.; Khazak, V.; Kedinger, C.; Vigneron, M. Interactions between the human RNA polymerase II subunits. J. Biol. Chem. 1997, 272, 16815–16821. [Google Scholar] [CrossRef] [PubMed]
- Kortner, T.M.; Valen, E.C.; Kortner, H.; Marjara, I.S.; Krogdahl, Å.; Bakke, A.M. Candidate reference genes for quantitative real-time PCR (qPCR) assays during development of a diet-related enteropathy in Atlantic salmon (Salmo salar L.) and the potential pitfalls of uncritical use of normalization software tools. Aquaculture 2011, 318, 355–363. [Google Scholar] [CrossRef]
- Kaarbo, M.; Crane, D.I.; Murrell, W.G. Isolation and characterisation of a chick cDNA encoding the RNA polymerase common subunit RPB6. DNA Seq. 2000, 11, 155–162. [Google Scholar] [CrossRef] [PubMed]
- McKune, K.; Woychik, N.A. Halobacterial S9 operon contains two genes encoding proteins homologous to subunits shared by eukaryotic RNA polymerases I, II, and III. J. Bacteriol. 1994, 176, 4754–4756. [Google Scholar] [CrossRef] [PubMed]
- Wool, I.G.; Chan, Y.L.; Glück, A. Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 1996, 73, 933–947. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yang, J.; Hou, Y.L.; Ding, X.; Peng, Z.S.; Hou, W.R. Cloning and sequence analysis of ribosomal protein L18gene (RPL18) from the Giant Panda (Ailuropoda melanoleuca). In Proceedings of the 2012 International Conference on Computer Science and Information Processing (CSIP), Xi’an, China, 24–26 August 2012; pp. 533–536. [Google Scholar]
- Barnard, G.F.; Staniunas, R.J.; Puder, M.; Steele, G.D., Jr.; Chen, L.B. Human ribosomal protein L37 has motifs predicting serine/threonine phosphorylation and a zinc-finger domain. Biochim. Biophys. Acta (BBA)—Gene Struct. Expr. 1994, 1218, 425–428. [Google Scholar] [CrossRef]
- Crespo, D.; Bogerd, J.; Sambroni, E.; LeGac, F.; Andersson, E.; Edvardsen, R.B.; Schulz, R.W. The initiation of puberty in Atlantic salmon brings about large changes in testicular gene expression that are modulated by the energy status. BMC Genom. 2019, 20, 475. [Google Scholar] [CrossRef]
- Chatzidimitriou, E.; Bisaccia, P.; Corrà, F.; Bonato, M.; Irato, P.; Manuto, L.; Toppo, S.; Bakiu, R.; Santovito, G. Copper/Zinc superoxide dismutase from the crocodile icefish Chionodraco hamatus: Antioxidant defense at constant sub-zero temperature. Antioxidants 2020, 9, 325. [Google Scholar] [CrossRef]
- Assfalg, M.; Banci, L.; Bertini, I.; Turano, P.; Vasos, P.R. Superoxide dismutase folding/unfolding pathway: Role of the metal ions in modulating structural and dynamical features. J. Mol. Biol. 2003, 330, 145–158. [Google Scholar] [CrossRef]
- Guyomard, R.; Boussaha, M.; Krieg, F.; Hervet, C.; Quillet, E. A synthetic rainbow trout linkage map provides new insights into the salmonid whole genome duplication and the conservation of synteny among teleosts. BMC Genet. 2012, 13, 15. [Google Scholar] [CrossRef]
- Shin, M.-K.; Park, H.R.; Yeo, W.J.; Han, K.N. Effects of thermal stress on the mRNA expression of SOD, HSP90, and HSP70 in the spotted sea bass (Lateolabrax maculatus). Ocean Sci. J. 2018, 53, 43–52. [Google Scholar] [CrossRef]
- Livingstone, D. Oxidative stress in aquatic organisms in relation to pollution and aquaculture. Rev. Med. Vet. 2003, 154, 427–430. [Google Scholar]
- Valavanidis, A.; Vlahogianni, T.; Dassenakis, M.; Scoullos, M. Molecular biomarkers of oxidative stress in aquatic organisms in relation to toxic environmental pollutants. Ecotoxicol. Environ. Saf. 2006, 64, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Regoli, F.; Giuliani, M.E. Oxidative pathways of chemical toxicity and oxidative stress biomarkers in marine organisms. Mar. Environ. Res. 2014, 93, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Birnie-Gauvin, K.; Costantini, D.; Cooke, S.J.; Willmore, W.G. A comparative and evolutionary approach to oxidative stress in fish: A review. Fish Fish. 2017, 18, 928–942. [Google Scholar] [CrossRef]
- Welker, T.L.; Congleton, J.L. Oxidative stress in juvenile chinook salmon, Oncorhynchus tshawytscha (Walbaum). Aquac. Res. 2004, 35, 881–887. [Google Scholar] [CrossRef]
- Mitchell, S.O.; Scholz, F.; Marcos, M.; Rodger, H. Sampling artefacts in gill histology of freshwater Atlantic Salmon (Salmo salar). Bull. Eur. Assoc. Fish Pathol. 2023, 43, 1–11. [Google Scholar] [CrossRef]
- Chen, B.; Piel, W.H.; Gui, L.; Bruford, E.; Monteiro, A. The HSP90 family of genes in the human genome: Insights into their divergence and evolution. Genomics 2005, 86, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Prodromou, C.; Roe, S.M.; O’Brien, R.; Ladbury, J.E.; Piper, P.W.; Pearl, L.H. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 1997, 90, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Scheufler, C.; Brinker, A.; Bourenkov, G.; Pegoraro, S.; Moroder, L.; Bartunik, H.; Hartl, F.U.; Moarefi, I. Structure of TPR domain–peptide complexes: Critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell 2000, 101, 199–210. [Google Scholar] [CrossRef]
- Lu, J.; Peatman, E.; Tang, H.; Lewis, J.; Liu, Z. Profiling of gene duplication patterns of sequenced teleost genomes: Evidence for rapid lineage-specific genome expansion mediated by recent tandem duplications. BMC Genom. 2012, 13, 246. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.Y.; Min, B.H.; Kim, N.N.; Cho, S.H.; Chang, Y.J. Expression of HSP90, HSP70 mRNA and change of plasma cortisol and glucose during water temperature rising in freshwater adapted black porgy, Acanthopagrus schlegeli. J. Aquac. 2006, 19, 315–322. [Google Scholar]
- Madeira, D.; Araújo, J.E.; Vitorino, R.; Capelo, J.L.; Vinagre, C.; Diniz, M.S. Ocean warming alters cellular metabolism and induces mortality in fish early life stages: A proteomic approach. Environ. Res. 2016, 148, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Von Biela, V.R.; Bowen, L.; McCormick, S.D.; Carey, M.P.; Donnelly, D.S.; Waters, S.; Regish, A.M.; Laske, S.M.; Brown, R.J.; Larson, S.; et al. Evidence of prevalent heat stress in Yukon River Chinook salmon. Can. J. Fish. Aquat. Sci. 2020, 77, 1878–1892. [Google Scholar] [CrossRef]
- Houde, A.L.S.; Akbarzadeh, A.; Günther, O.P.; Li, S.; Patterson, D.A.; Farrell, A.P.; Hinch, S.G.; Miller, K.M. Salmonid gene expression biomarkers indicative of physiological responses to changes in salinity and temperature, but not dissolved oxygen. J. Exp. Biol. 2019, 222, jeb198036. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Dorta, V.; Caballero, M.J.; Betancor, M.B.; Manchado, M.; Tort, L.; Torrecillas, S.; Zamorano, M.J.; Izquierdo, M.; Montero, D. Effects of thermal stress on the expression of glucocorticoid receptor complex linked genes in Senegalese sole (Solea senegalensis), Acute and adaptive stress responses. Gen. Comp. Endocrinol. 2017, 252, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Finerty, C.J. A Transcriptomics Approach to Examining the Effects of Prenatal and Thermal Stress on Developmental Plasticity in Chinook Salmon (Oncorhynchus tshawytscha). Master’s Thesis, Electronic Theses and Dissertations, University of Windsor, Windsor, UK, 2020. 8359. [Google Scholar]
- Meyer, E.; Aglyamova, G.; Matz, M. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol. Ecol. 2011, 20, 3599–3616. [Google Scholar] [CrossRef] [PubMed]
- Wellband, K.W.; Heath, J.W.; Heath, D.D. Environmental and genetic determinants of transcriptional plasticity in Chinook salmon. Heredity 2018, 120, 38–50. [Google Scholar] [CrossRef]
- Lewis, M.; Götting, M.; Anttila, K.; Kanerva, M.; Prokkola, J.M.; Seppänen, E.; Kolari, I.; Nikinmaa, M. Different relationship between hsp70 mRNA and hsp70 levels in the heat shock response of two salmonids with dissimilar temperature preference. Front. Physiol. 2016, 7, 511. [Google Scholar] [CrossRef]
- Ma, F.; Luo, L. Genome-wide identification of Hsp70/110 genes in rainbow trout and their regulated expression in response to heat stress. PeerJ 2020, 8, e10022. [Google Scholar] [CrossRef]
- Stenton-Dozey, J.M.; Heath, P.; Ren, J.S.; Zamora, L.N. New Zealand aquaculture industry: Research, opportunities and constraints for integrative multitrophic farming. N. Z. J. Mar. Freshw. Res. 2021, 55, 265–285. [Google Scholar] [CrossRef]




| Species | Genome Version | Accession Number |
|---|---|---|
| Human | GRCh38.p14 | GCA_000001405.29 |
| Chicken | GRCg6a | GCA_000002315.5 |
| Turkey | Turkey_5.1 | GCA_000146605.4 |
| Chinese softshelled turtle | PelSin_1.0 | GCA_000230535.1 |
| Green anole | AnoCar2.0 | GCA_000090745.2 |
| Xenopus laevis | Xenopus_laevis_v10.1 | GCA_017654675.1 |
| Xenopus tropicalis | UCB_Xtro_10.0 | GCA_000004195.4 |
| Coelacanth | LatCha1 | GCA_000225785.1 |
| Amazon molly | Poecilia_formosa-5.1.2 | GCA_000485575.1 |
| Rainbow trout | Omyk_2.0 | GCA_025558465.1 |
| Atlantic Salmon | Ssal_v3.1 | GCA_905237065.2 |
| Nile tilapia | O_niloticus_UMD_NMBU | GCA_001858045.3 |
| Elephant shark | Callorhinchus_milii-6.1.3 | GCA_000165045.2 |
| Gene | Primer Name | Sequence (5′-3′) | Product Size | Annealing Temperature |
|---|---|---|---|---|
| SOD1 | SOD1-F | GAGCTGACAATGTGGCTAAGA | 101 bp | 60 °C |
| SOD1-R | CAGCTTTCTCATGGATCACC | |||
| HSP90 | HSP90-F | CTTTGAGAACAAGAAGAAGAAGAAC | 92 bp | 65 °C |
| HSP90-R | CACACCCTTAATGAAGTTGAGGTAC | |||
| POLR2F | POLR2F-F | AGAAGAGGATCACAACCCAATAC | 162 bp | 61 °C |
| POLR2F-R | GGGATCTTCCTGCACTTTAGC | |||
| RPL18 | RPL18-F | GCTCTGAAGGTGACTGACGG | 206 bp | 61 °C |
| RPL18-R | TTGGAACGAATGTAGGGCTTGG |
| Blood Parameter | Pre-Trial (n = 16) | Control (n = 16) | High-Temp (n = 16) |
|---|---|---|---|
| Blood Haemoglobin (g/L) | 106.81 ± 3.41 | 101.18 ± 3.42 | 102.12 ± 3.44 |
| White blood cell count (109/L) | 25.00 ± 3.80 | 27.91 ± 3.78 | 25.78 ± 3.81 |
| Monocytes (%) | 1.25 ± 0.22 | 0.93 ± 0.16 | 0.87 ± 0.21 |
| Lymphocytes (%) | 92.19 ± 1.55 | 91.56 ± 1.32 | 81.94 ± 1.36 aa, b |
| Neutrophils (%) | 6.56 ± 0.81 | 7.50 ± 0.87 | 17.19 ± 1.10 aa, bb |
| Neutrophil/Lymphocyte ratio | 0.06 ± 0.01 | 0.08 ± 0.01 | 0.26 ± 0.04 aa, bb |
| Mean corpuscular haemoglobin (%) | 25.88 ± 0.73 | 28.70 ± 0.77 | 26.28 ± 0.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcoli, R.; Symonds, J.E.; Walker, S.P.; Battershill, C.N.; Bird, S. Characterising the Physiological Responses of Chinook Salmon (Oncorhynchus tshawytscha) Subjected to Heat and Oxygen Stress. Biology 2023, 12, 1342. https://doi.org/10.3390/biology12101342
Marcoli R, Symonds JE, Walker SP, Battershill CN, Bird S. Characterising the Physiological Responses of Chinook Salmon (Oncorhynchus tshawytscha) Subjected to Heat and Oxygen Stress. Biology. 2023; 12(10):1342. https://doi.org/10.3390/biology12101342
Chicago/Turabian StyleMarcoli, Roberta, Jane E. Symonds, Seumas P. Walker, Christopher N. Battershill, and Steve Bird. 2023. "Characterising the Physiological Responses of Chinook Salmon (Oncorhynchus tshawytscha) Subjected to Heat and Oxygen Stress" Biology 12, no. 10: 1342. https://doi.org/10.3390/biology12101342
APA StyleMarcoli, R., Symonds, J. E., Walker, S. P., Battershill, C. N., & Bird, S. (2023). Characterising the Physiological Responses of Chinook Salmon (Oncorhynchus tshawytscha) Subjected to Heat and Oxygen Stress. Biology, 12(10), 1342. https://doi.org/10.3390/biology12101342








