Monooxygenases and Antibiotic Resistance: A Focus on Carbapenems
Abstract
Simple Summary
Abstract
1. Introduction
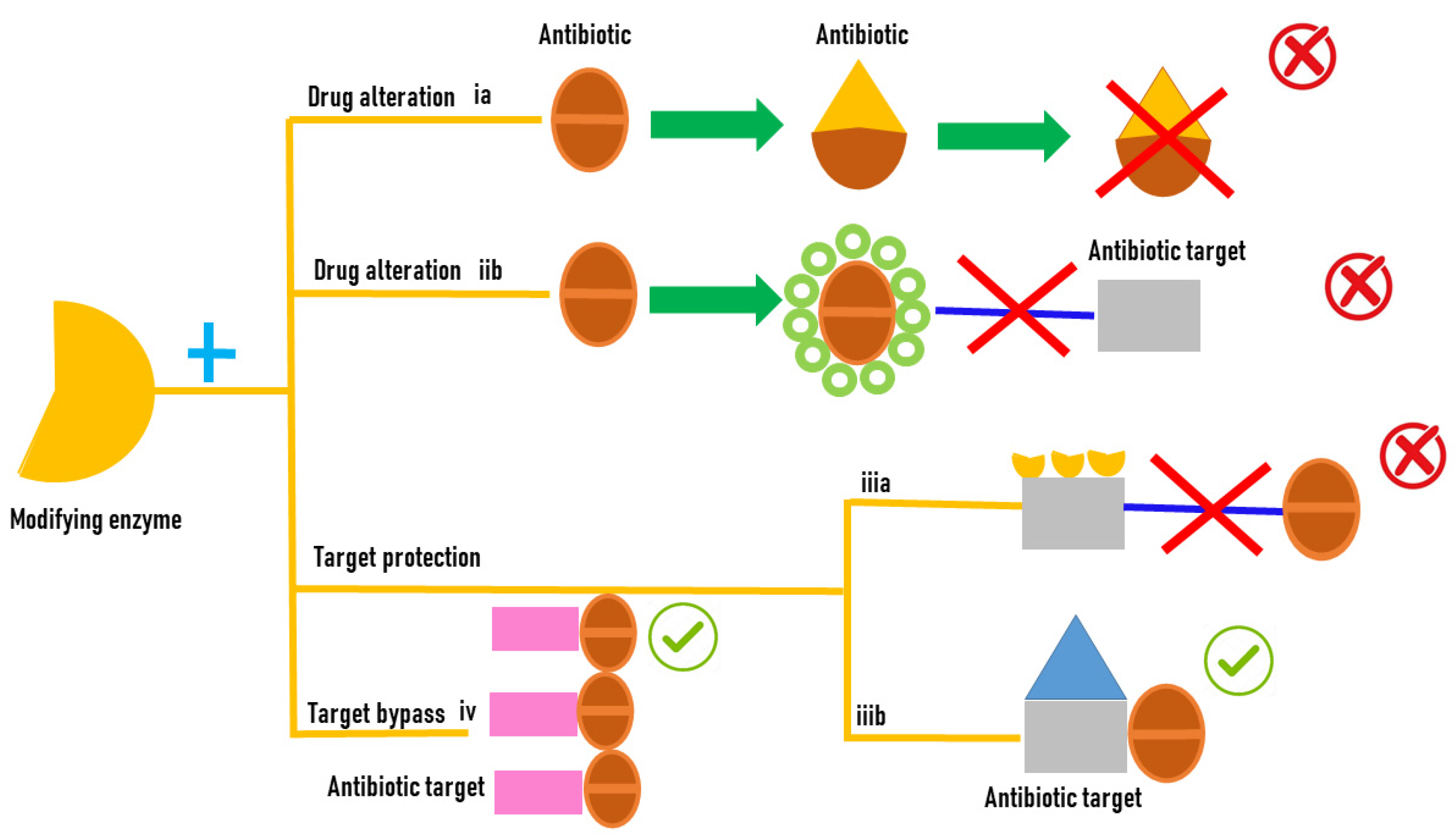
2. Antibiotic Resistance Mechanisms
2.1. Modification of Antibiotic Target
2.1.1. Decreased Permeability
2.1.2. Overexpression of Drug Efflux Pumps
2.1.3. Hydrolysis of Antibiotic Molecules
2.2. Antibiotic Resistance Mediated by Bacterial Enzymes
2.2.1. Flavin-Dependent Monooxygenases (FMOs)
2.2.2. Baeyer–Villiger Monooxygenases (BVMOs)
2.3. FMOs and Antibiotic Resistance
2.3.1. Tetracyclines
2.3.2. Rifamycins
2.3.3. Sulfonamides
3. Carbapenems
3.1. Mechanism of Action
3.2. Carbapenem Resistance in ESKAPE Pathogens
3.3. Incidence of Carbapenem Resistance
4. BVMO and β-Lactams Resistance
4.1. Carbapenems
4.2. β-Lactams
5. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Popa, S.L.; Pop, C.; Dita, M.O.; Brata, V.D.; Bolchis, R.; Czako, Z.; Saadani, M.M.; Ismaiel, A.; Dumitrascu, D.I.; Grad, S. Deep Learning and Antibiotic Resistance. Antibiotics 2022, 11, 1674. [Google Scholar] [CrossRef]
- Mohsin, S.; Amin, M.N. Superbugs: A Constraint to Achieving the Sustainable Development Goals. Bull. Natl. Res. Cent. 2023, 47, 1–13. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.M.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K. Antibiotic Resistance in Microbes: History, Mechanisms, Therapeutic Strategies and Future Prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Giacometti, F.; Shirzad-Aski, H.; Ferreira, S. Antimicrobials and food-related stresses as selective factors for antibiotic resistance along the farm to fork continuum. Antibiotics 2021, 10, 671. [Google Scholar] [CrossRef]
- McCracken, C.M.; Tucker, K.J.; Tallman, G.B.; Holmer, H.K.; Noble, B.N.; McGregor, J.C. General Perceptions and Knowledge of Antibiotic Resistance and Antibiotic Use Behavior: A Cross-Sectional Survey of US Adults. Antibiotics 2023, 12, 672. [Google Scholar] [CrossRef]
- Kamoshida, G.; Akaji, T.; Takemoto, N.; Suzuki, Y.; Sato, Y.; Kai, D.; Hibino, T.; Yamaguchi, D.; Kikuchi-Ueda, T.; Nishida, S. Lipopolysaccharide-Deficient Acinetobacter Baumannii Due to Colistin Resistance Is Killed by Neutrophil-Produced Lysozyme. Front. Microbiol. 2020, 11, 573. [Google Scholar] [CrossRef]
- Choi, U.; Lee, C.-R. Distinct Roles of Outer Membrane Porins in Antibiotic Resistance and Membrane Integrity in Escherichia Coli. Front. Microbiol. 2019, 10, 953. [Google Scholar]
- Rashid Mahmood, A.; Mansour Hussein, N. Study of Antibiotic Resistant Genes in Pseudomonas Aeroginosa Isolated from Burns and Wounds. Arch. Razi Inst. 2022, 77, 403–411. [Google Scholar]
- Nishino, K.; Yamasaki, S.; Nakashima, R.; Zwama, M.; Hayashi-Nishino, M. Function and Inhibitory Mechanisms of Multidrug Efflux Pumps. Front. Microbiol. 2021, 12, 737288. [Google Scholar] [CrossRef]
- Shen, J.; Wang, Y.; Schwarz, S. Presence and Dissemination of the Multiresistance Gene Cfr in Gram-Positive and Gram-Negative Bacteria. J. Antimicrob. Chemother. 2013, 68, 1697–1706. [Google Scholar] [CrossRef]
- Garcia, M.M. Carbapenemases: A Real Threat. APUA Newsl. 2013, 31, 4–6. [Google Scholar]
- Ortiz de la Rosa, J.M.; Nordmann, P.; Poirel, L. Antioxidant Molecules as a Source of Mitigation of Antibiotic Resistance Gene Dissemination. Antimicrob. Agents Chemother. 2021, 65, 10–1128. [Google Scholar] [CrossRef]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular Mechanisms of Antibiotic Resistance Revisited. Nat. Rev. Microbiol. 2023, 21, 280–295. [Google Scholar] [CrossRef]
- Egorov, A.M.; Ulyashova, M.M.; Rubtsova, M.Y. Bacterial Enzymes and Antibiotic Resistance. Acta Naturae (Aнглoязычная Bepcuя) 2018, 10, 33–48. [Google Scholar] [CrossRef]
- Babic, M.; Hujer, A.M.; Bonomo, R.A. What’s New in Antibiotic Resistance? Focus on Beta-Lactamases. Drug Resist. Updates 2006, 9, 142–156. [Google Scholar] [CrossRef]
- Souque, C.; Escudero, J.A.; MacLean, R.C. Integron Activity Accelerates the Evolution of Antibiotic Resistance. eLife 2021, 10, e62474. [Google Scholar] [CrossRef]
- Golkar, T.; Zieliński, M.; Berghuis, A.M. Look and Outlook on Enzyme-Mediated Macrolide Resistance. Front. Microbiol. 2018, 9, 1942. [Google Scholar] [CrossRef]
- Chaiyen, P.; Fraaije, M.W.; Mattevi, A. The Enigmatic Reaction of Flavins with Oxygen. Trends Biochem. Sci. 2012, 37, 373–380. [Google Scholar] [CrossRef]
- Mascotti, M.L.; Ayub, M.J.; Furnham, N.; Thornton, J.M.; Laskowski, R.A. Chopping and Changing: The Evolution of the Flavin-Dependent Monooxygenases. J. Mol. Biol. 2016, 428, 3131–3146. [Google Scholar] [CrossRef]
- Palfey, B.A.; McDonald, C.A. Control of Catalysis in Flavin-Dependent Monooxygenases. Arch. Biochem. Biophys. 2010, 493, 26–36. [Google Scholar] [CrossRef]
- Massey, V. Activation of Molecular Oxygen by Flavins and Flavoproteins. J. Biol. Chem. 1994, 269, 22459–22462. [Google Scholar] [CrossRef] [PubMed]
- Huijbers, M.M.E.; Montersino, S.; Westphal, A.H.; Tischler, D.; Van Berkel, W.J.H. Flavin Dependent Monooxygenases. Arch. Biochem. Biophys. 2014, 544, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Leisch, H.; Morley, K.; Lau, P.C.K. Baeyer−Villiger Monooxygenases: More than Just Green Chemistry. Chem. Rev. 2011, 111, 4165–4222. [Google Scholar] [CrossRef] [PubMed]
- Fraaije, M.W.; Kamerbeek, N.M.; van Berkel, W.J.H.; Janssen, D.B. Identification of a Baeyer–Villiger Monooxygenase Sequence Motif. FEBS Lett. 2002, 518, 43–47. [Google Scholar] [CrossRef]
- Kamerbeek, N.M.; Janssen, D.B.; van Berkel, W.J.H.; Fraaije, M.W. Baeyer–Villiger Monooxygenases, an Emerging Family of Flavin-dependent Biocatalysts. Adv. Synth. Catal. 2003, 345, 667–678. [Google Scholar] [CrossRef]
- Fraaije, M.W.; Kamerbeek, N.M.; Heidekamp, A.J.; Fortin, R.; Janssen, D.B. The Prodrug Activator EtaA from Mycobacterium Tuberculosis Is a Baeyer-Villiger Monooxygenase. J. Biol. Chem. 2004, 279, 3354–3360. [Google Scholar] [CrossRef]
- Nomura, T.; Kushiro, T.; Yokota, T.; Kamiya, Y.; Bishop, G.J.; Yamaguchi, S. The Last Reaction Producing Brassinolide Is Catalyzed by Cytochrome P-450s, CYP85A3 in Tomato and CYP85A2 in Arabidopsis. J. Biol. Chem. 2005, 280, 17873–17879. [Google Scholar] [CrossRef]
- Speer, B.S.; Bedzyk, L.; Salyers, A. Evidence That a Novel Tetracycline Resistance Gene Found on Two Bacteroides Transposons Encodes an NADP-Requiring Oxidoreductase. J. Bacteriol. 1991, 173, 176–183. [Google Scholar] [CrossRef]
- Yang, W.; Moore, I.F.; Koteva, K.P.; Bareich, D.C.; Hughes, D.W.; Wright, G.D. TetX Is a Flavin-Dependent Monooxygenase Conferring Resistance to Tetracycline Antibiotics. J. Biol. Chem. 2004, 279, 52346–52352. [Google Scholar] [CrossRef]
- Volkers, G.; Palm, G.J.; Weiss, M.S.; Wright, G.D.; Hinrichs, W. Structural Basis for a New Tetracycline Resistance Mechanism Relying on the TetX Monooxygenase. FEBS Lett. 2011, 585, 1061–1066. [Google Scholar] [CrossRef]
- Rudra, P.; Hurst-Hess, K.; Lappierre, P.; Ghosh, P. High Levels of Intrinsic Tetracycline Resistance in Mycobacterium Abscessus Are Conferred by a Tetracycline-Modifying Monooxygenase. Antimicrob. Agents Chemother. 2018, 62, e00119-18. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Gasparrini, A.J.; Reck, M.R.; Symister, C.T.; Elliott, J.L.; Vogel, J.P.; Wencewicz, T.A.; Dantas, G.; Tolia, N.H. Plasticity, Dynamics, and Inhibition of Emerging Tetracycline Resistance Enzymes. Nat. Chem. Biol. 2017, 13, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Koteva, K.; Cox, G.; Kelso, J.K.; Surette, M.D.; Zubyk, H.L.; Ejim, L.; Stogios, P.; Savchenko, A.; Sørensen, D.; Wright, G.D. Rox, a Rifamycin Resistance Enzyme with an Unprecedented Mechanism of Action. Cell Chem. Biol. 2018, 25, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-Y.; Huang, C.-H.; Lu, P.-L.; Ko, W.-C.; Chen, Y.-H.; Hsueh, P.-R. Role of Rifampin for the Treatment of Bacterial Infections Other than Mycobacteriosis. J. Infect. 2017, 75, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Abdelwahab, H.; Martin Del Campo, J.S.; Dai, Y.; Adly, C.; El-Sohaimy, S.; Sobrado, P. Mechanism of Rifampicin Inactivation in Nocardia Farcinica. PLoS ONE 2016, 11, e0162578. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.J.; Quan, S.; Gowan, B.; Dabbs, E.R. Monooxygenase-like Sequence of a Rhodococcus Equi Gene Conferring Increased Resistance to Rifampin by Inactivating This Antibiotic. Antimicrob. Agents Chemother. 1997, 41, 218–221. [Google Scholar] [CrossRef]
- Hoshino, Y.; Fujii, S.; Shinonaga, H.; Arai, K.; Saito, F.; Fukai, T.; Satoh, H.; Miyazaki, Y.; Ishikawa, J. Monooxygenation of Rifampicin Catalyzed by the Rox Gene Product of Nocardia Farcinica: Structure Elucidation, Gene Identification and Role in Drug Resistance. J. Antibiot. 2010, 63, 23–28. [Google Scholar] [CrossRef]
- Liu, L.-K.; Dai, Y.; Abdelwahab, H.; Sobrado, P.; Tanner, J.J. Structural Evidence for Rifampicin Monooxygenase Inactivating Rifampicin by Cleaving Its Ansa-Bridge. Biochemistry 2018, 57, 2065–2068. [Google Scholar] [CrossRef]
- Baunach, M.; Ding, L.; Willing, K.; Hertweck, C. Bacterial Synthesis of Unusual Sulfonamide and Sulfone Antibiotics by Flavoenzyme-Mediated Sulfur Dioxide Capture. Angew. Chem. Int. Ed. 2015, 54, 13279–13283. [Google Scholar] [CrossRef]
- Ricken, B.; Kolvenbach, B.A.; Bergesch, C.; Benndorf, D.; Kroll, K.; Strnad, H.; Vlček, Č.; Adaixo, R.; Hammes, F.; Shahgaldian, P. FMNH2-Dependent Monooxygenases Initiate Catabolism of Sulfonamides in Microbacterium Sp. Strain BR1 Subsisting on Sulfonamide Antibiotics. Sci. Rep. 2017, 7, 15783. [Google Scholar] [CrossRef]
- Kim, D.-W.; Thawng, C.N.; Lee, K.; Wellington, E.M.H.; Cha, C.-J. A Novel Sulfonamide Resistance Mechanism by Two-Component Flavin-Dependent Monooxygenase System in Sulfonamide-Degrading Actinobacteria. Environ. Int. 2019, 127, 206–215. [Google Scholar] [CrossRef] [PubMed]
- JACOBS, R.F. Imipenem-Cilastatin: The First Thienamycin Antibiotic. Pediatr. Infect. Dis. J. 1986, 5, 444–448. [Google Scholar] [CrossRef]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, Present, and Future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef]
- Bradley, J.S. Meropenem: A New, Extremely Broad Spectrum Beta-Lactam Antibiotic for Serious Infections in Pediatrics. Pediatr. Infect Dis. J. 1997, 16, 263–268. [Google Scholar] [CrossRef]
- El-Gamal, M.I.; Brahim, I.; Hisham, N.; Aladdin, R.; Mohammed, H.; Bahaaeldin, A. Recent Updates of Carbapenem Antibiotics. Eur. J. Med. Chem. 2017, 131, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Papp-Wallace, K.M.; Bonomo, R.A. New β-Lactamase Inhibitors in the Clinic. Infect. Dis. Clin. 2016, 30, 441–464. [Google Scholar] [CrossRef]
- Watkins, R.R.; Bonomo, R.A. Increasing Prevalence of Carbapenem-Resistant Enterobacteriaceae and Strategies to Avert a Looming Crisis. Expert Rev. Anti Infect. Ther. 2013, 11, 543–545. [Google Scholar] [CrossRef]
- Hoskins, J.; Matsushima, P.; Mullen, D.L.; Tang, J.; Zhao, G.; Meier, T.I.; Nicas, T.I.; Jaskunas, S.R. Gene Disruption Studies of Penicillin-Binding Proteins 1a, 1b, and 2a in Streptococcus Pneumoniae. J. Bacteriol. 1999, 181, 6552–6555. [Google Scholar] [CrossRef]
- Elshamy, A.A.; Aboshanab, K.M. A Review on Bacterial Resistance to Carbapenems: Epidemiology, Detection and Treatment Options. Future Sci. OA 2020, 6, FSO438. [Google Scholar] [CrossRef]
- Davies, T.A.; Shang, W.; Bush, K.; Flamm, R.K. Affinity of Doripenem and Comparators to Penicillin-Binding Proteins in Escherichia Coli and Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2008, 52, 1510–1512. [Google Scholar] [CrossRef]
- Aurilio, C.; Sansone, P.; Barbarisi, M.; Pota, V.; Giaccari, L.G.; Coppolino, F.; Barbarisi, A.; Passavanti, M.B.; Pace, M.C. Mechanisms of Action of Carbapenem Resistance. Antibiotics 2022, 11, 421. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Almugadam, B.S.; Ali, N.; Ahmed, A.B.; Ahmed, E.; Wang, L. Prevalence and Antibiotics Susceptibility Patterns of Carbapenem Resistant Enterobacteriaceae. J. Bacteriol. Mycol. Open Access 2018, 6, 187–190. [Google Scholar] [CrossRef][Green Version]
- Doumith, M.; Ellington, M.J.; Livermore, D.M.; Woodford, N. Molecular Mechanisms Disrupting Porin Expression in Ertapenem-Resistant Klebsiella and Enterobacter Spp. Clinical Isolates from the UK. J. Antimicrob. Chemother. 2009, 63, 659–667. [Google Scholar] [CrossRef]
- Huang, W.; Qiao, F.; Zhang, Y.; Huang, J.; Deng, Y.; Li, J.; Zong, Z. In-Hospital Medical Costs of Infections Caused by Carbapenem-Resistant Klebsiella Pneumoniae. Clin. Infect. Dis. 2018, 67, S225–S230. [Google Scholar] [CrossRef]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef]
- Cuzon, G.; Naas, T.; Guibert, M.; Nordmann, P. In Vivo Selection of Imipenem-Resistant Klebsiella Pneumoniae Producing Extended-Spectrum β-Lactamase CTX-M-15 and Plasmid-Encoded DHA-1 Cephalosporinase. Int. J. Antimicrob. Agents 2010, 35, 265–268. [Google Scholar] [CrossRef][Green Version]
- uz Zaman, T.; Aldrees, M.; Al Johani, S.M.; Alrodayyan, M.; Aldughashem, F.A.; Balkhy, H.H. Multi-Drug Carbapenem-Resistant Klebsiella Pneumoniae Infection Carrying the OXA-48 Gene and Showing Variations in Outer Membrane Protein 36 Causing an Outbreak in a Tertiary Care Hospital in Riyadh, Saudi Arabia. Int. J. Infect. Dis. 2014, 28, 186–192. [Google Scholar] [CrossRef]
- Munoz-Price, L.S.; Poirel, L.; Bonomo, R.A.; Schwaber, M.J.; Daikos, G.L.; Cormican, M.; Cornaglia, G.; Garau, J.; Gniadkowski, M.; Hayden, M.K. Clinical Epidemiology of the Global Expansion of Klebsiella Pneumoniae Carbapenemases. Lancet Infect. Dis. 2013, 13, 785–796. [Google Scholar] [CrossRef]
- Baroud, Á.; Dandache, I.; Araj, G.F.; Wakim, R.; Kanj, S.; Kanafani, Z.; Khairallah, M.; Sabra, A.; Shehab, M.; Dbaibo, G. Underlying Mechanisms of Carbapenem Resistance in Extended-Spectrum β-Lactamase-Producing Klebsiella Pneumoniae and Escherichia Coli Isolates at a Tertiary Care Centre in Lebanon: Role of OXA-48 and NDM-1 Carbapenemases. Int. J. Antimicrob. Agents 2013, 41, 75–79. [Google Scholar] [CrossRef]
- Tian, X.; Zheng, X.; Sun, Y.; Fang, R.; Zhang, S.; Zhang, X.; Lin, J.; Cao, J.; Zhou, T. Molecular Mechanisms and Epidemiology of Carbapenem-Resistant Escherichia Coli Isolated from Chinese Patients during 2002–2017. Infect. Drug Resist. 2020, 13, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Govindaswamy, A.; Bajpai, V.; Khurana, S.; Aravinda, A.; Batra, P.; Malhotra, R.; Mathur, P. Prevalence and Characterization of Beta-Lactamase-Producing Escherichia Coli Isolates from a Tertiary Care Hospital in India. J. Lab. Physicians 2019, 11, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Lima, W.G.; Alves, G.C.S.; Sanches, C.; Fernandes, S.O.A.; de Paiva, M.C. Carbapenem-Resistant Acinetobacter Baumannii in Patients with Burn Injury: A Systematic Review and Meta-Analysis. Burns 2019, 45, 1495–1508. [Google Scholar] [CrossRef] [PubMed]
- Almasaudi, S.B. Acinetobacter Spp. as Nosocomial Pathogens: Epidemiology and Resistance Features. Saudi J. Biol. Sci. 2018, 25, 586–596. [Google Scholar] [CrossRef]
- Wong, D.; Nielsen, T.B.; Bonomo, R.A.; Pantapalangkoor, P.; Luna, B.; Spellberg, B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 2017, 30, 409–447. [Google Scholar] [CrossRef]
- Zhu, L.-J.; Pan, Y.; Gao, C.-Y.; Hou, P.-F. Distribution of Carbapenemases and Efflux Pump in Carbapenem-Resistance Acinetobacter Baumannii. Ann. Clin. Lab Sci. 2020, 50, 241–246. [Google Scholar]
- Bansal, G.; Allen-McFarlane, R.; Eribo, B. Antibiotic Susceptibility, Clonality, and Molecular Characterization of Carbapenem-Resistant Clinical Isolates of Acinetobacter Baumannii from Washington DC. Int. J. Microbiol. 2020, 2020, 2120159. [Google Scholar] [CrossRef]
- Reynolds, D.; Kollef, M. The Epidemiology and Pathogenesis and Treatment of Pseudomonas Aeruginosa Infections: An Update. Drugs 2021, 81, 2117–2131. [Google Scholar] [CrossRef]
- Perez, F.; Villegas, M.V. The Role of Surveillance Systems in Confronting the Global Crisis of Antibiotic-Resistant Bacteria. Curr. Opin. Infect. Dis. 2015, 28, 375. [Google Scholar] [CrossRef]
- Feil, E.J. Enterobacteriaceae: Joining the Dots with Pan-European Epidemiology. Lancet Infect. Dis. 2017, 17, 118–119. [Google Scholar] [CrossRef]
- Xu, Y.; Gu, B.; Huang, M.; Liu, H.; Xu, T.; Xia, W.; Wang, T. Epidemiology of Carbapenem Resistant Enterobacteriaceae (CRE) during 2000–2012 in Asia. J. Thorac. Dis. 2015, 7, 376. [Google Scholar] [PubMed]
- Tesfa, T.; Mitiku, H.; Edae, M.; Assefa, N. Prevalence and Incidence of Carbapenem-Resistant K. Pneumoniae Colonization: Systematic Review and Meta-Analysis. Syst. Rev. 2022, 11, 240. [Google Scholar] [CrossRef] [PubMed]
- Agyeman, A.A.; Bergen, P.J.; Rao, G.G.; Nation, R.L.; Landersdorfer, C.B. A Systematic Review and Meta-Analysis of Treatment Outcomes Following Antibiotic Therapy among Patients with Carbapenem-Resistant Klebsiella Pneumoniae Infections. Int. J. Antimicrob. Agents 2020, 55, 105833. [Google Scholar] [CrossRef]
- Nordmann, P.; Cuzon, G.; Naas, T. The Real Threat of Klebsiella Pneumoniae Carbapenemase-Producing Bacteria. Lancet Infect. Dis. 2009, 9, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Ayobami, O.; Willrich, N.; Harder, T.; Okeke, I.N.; Eckmanns, T.; Markwart, R. The Incidence and Prevalence of Hospital-Acquired (Carbapenem-Resistant) Acinetobacter Baumannii in Europe, Eastern Mediterranean and Africa: A Systematic Review and Meta-Analysis. Emerg. Microbes Infect. 2019, 8, 1747–1759. [Google Scholar] [CrossRef]
- Al-Rashed, N.; Bindayna, K.M.; Shahid, M.; Saeed, N.K.; Darwish, A.; Joji, R.M.; Al-Mahmeed, A. Prevalence of Carbapenemases in Carbapenem-Resistant Acinetobacter Baumannii Isolates from the Kingdom of Bahrain. Antibiotics 2023, 12, 1198. [Google Scholar] [CrossRef]
- Tenover, F.C.; Nicolau, D.P.; Gill, C.M. Carbapenemase-Producing Pseudomonas Aeruginosa–an Emerging Challenge. Emerg. Microbes Infect. 2022, 11, 811–814. [Google Scholar] [CrossRef]
- Woodworth, K.R.; Walters, M.S.; Weiner, L.M.; Edwards, J.; Brown, A.C.; Huang, J.Y.; Malik, S.; Slayton, R.B.; Paul, P.; Capers, C. Vital Signs: Containment of Novel Multidrug-Resistant Organisms and Resistance Mechanisms—United States, 2006–2017. Morb. Mortal. Wkly. Rep. 2018, 67, 396. [Google Scholar] [CrossRef]
- Minerdi, D.; Zgrablic, I.; Castrignanò, S.; Catucci, G.; Medana, C.; Terlizzi, M.E.; Gribaudo, G.; Gilardi, G.; Sadeghi, S.J. Escherichia Coli Overexpressing a Baeyer-Villiger Monooxygenase from Acinetobacter Radioresistens Becomes Resistant to Imipenem. Antimicrob. Agents Chemother. 2016, 60, 64–74. [Google Scholar] [CrossRef]
- Gao, J.; Zhao, X.; Bao, Y.; Ma, R.; Zhou, Y.; Li, X.; Chai, T.; Cai, Y. Antibiotic Resistance and OXA-Type Carbapenemases-Encoding Genes in Airborne Acinetobacter Baumannii Isolated from Burn Wards. Burns 2014, 40, 295–299. [Google Scholar] [CrossRef]
- Opazo, A.; Domínguez, M.; Bello, H.; Amyes, S.G.B.; González-Rocha, G. OXA-Type Carbapenemases in Acinetobacter Baumannii in South America. J. Infect. Dev. Ctries. 2012, 6, 311–316. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Poirel, L.; Figueiredo, S.; Cattoir, V.; Carattoli, A.; Nordmann, P. Acinetobacter Radioresistens as a Silent Source of Carbapenem Resistance for Acinetobacter spp. Antimicrob. Agents Chemother. 2008, 52, 1252–1256. [Google Scholar] [CrossRef] [PubMed]
- Seifert, H.; Dijkshoorn, L.; Gerner-Smidt, P.; Pelzer, N.; Tjernberg, I.; Vaneechoutte, M. Distribution of Acinetobacter Species on Human Skin: Comparison of Phenotypic and Genotypic Identification Methods. J. Clin. Microbiol. 1997, 35, 2819–2825. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.C.; Xu, Q.; Wu, B.; Godzik, A. Crystal Structure of a Baeyer–Villiger Flavin-Containing Monooxygenase from Staphylococcus Aureus MRSA Strain MU50. Proteins 2018, 86, 269. [Google Scholar] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minerdi, D.; Loqui, D.; Sabbatini, P. Monooxygenases and Antibiotic Resistance: A Focus on Carbapenems. Biology 2023, 12, 1316. https://doi.org/10.3390/biology12101316
Minerdi D, Loqui D, Sabbatini P. Monooxygenases and Antibiotic Resistance: A Focus on Carbapenems. Biology. 2023; 12(10):1316. https://doi.org/10.3390/biology12101316
Chicago/Turabian StyleMinerdi, Daniela, Davide Loqui, and Paolo Sabbatini. 2023. "Monooxygenases and Antibiotic Resistance: A Focus on Carbapenems" Biology 12, no. 10: 1316. https://doi.org/10.3390/biology12101316
APA StyleMinerdi, D., Loqui, D., & Sabbatini, P. (2023). Monooxygenases and Antibiotic Resistance: A Focus on Carbapenems. Biology, 12(10), 1316. https://doi.org/10.3390/biology12101316






