Genome-Wide Analysis of Q-Type C2H2 ZFP Genes in Response to Biotic and Abiotic Stresses in Sugar Beet
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Acquisition of the Sequences of the Q-Type ZFP Transcription Factor Family in Sugar Beet
2.2. Chromosomal Distribution, Protein Characterization, and Amino Acid Properties
2.3. Multiple Sequence Alignment and Phylogenetic Tree Construction
2.4. Analysis of Members of the C2H2 ZFP Gene Family
2.5. Promoter Analysis of the BvZFP Genes in Sugar Beet
2.6. Collinearity Analysis of Arabidopsis and Sugar Beet BvZFP Genes
2.7. BvZFP Genes Expression Network Analysis
3. Results
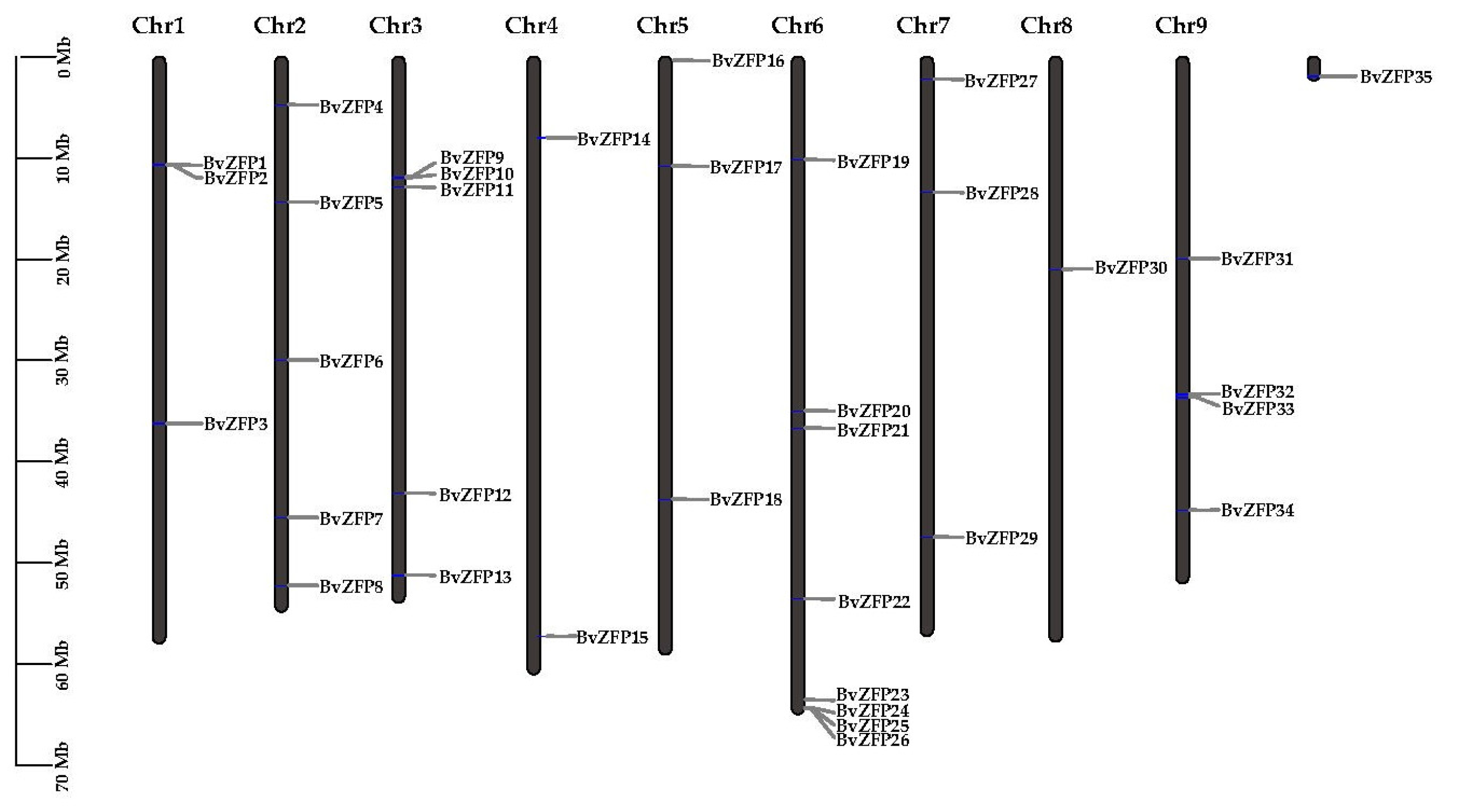
3.1. Identification and Chromosomal Localization of the C2H2 Q-Type ZFP Subclass in Sugar Beet
3.2. Characterization of the Sugar Beet C2H2 Q-Type ZFP Subclass
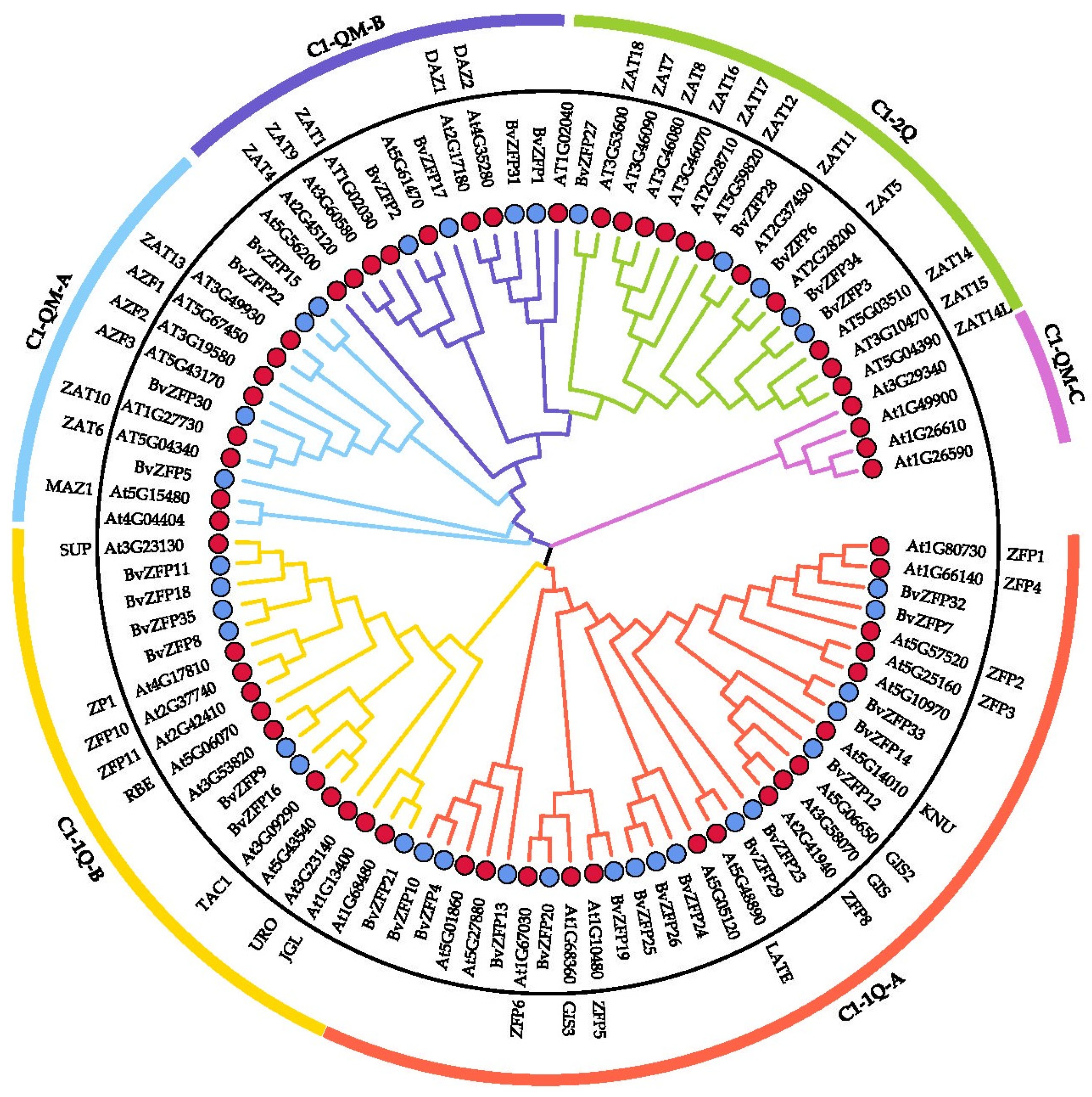
3.3. Phylogenetic Analysis of Q-Type BvZFP Genes
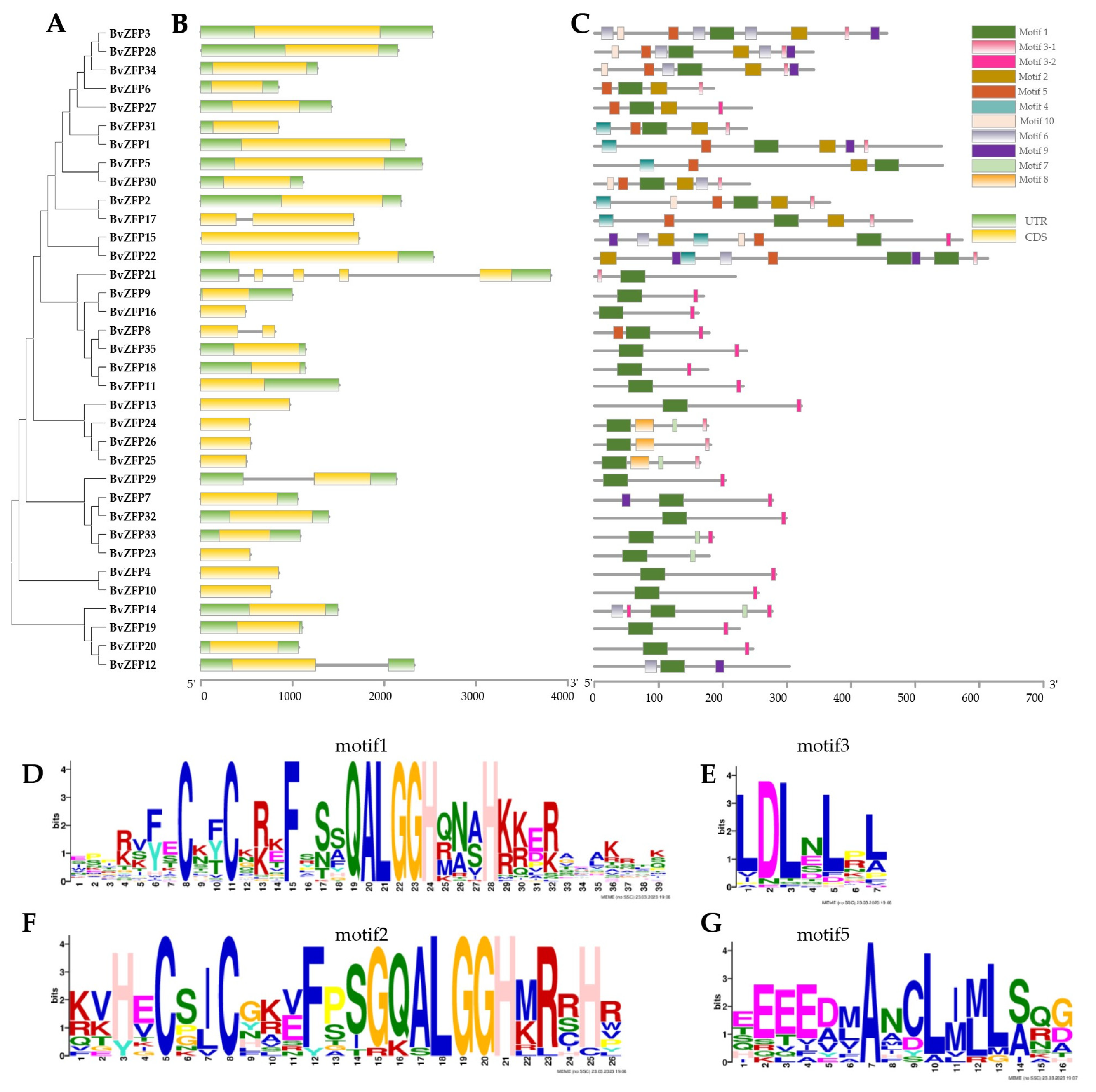
3.4. Gene Structure and Conservative Motif Analysis of C2H2 Q-Type BvZFPs
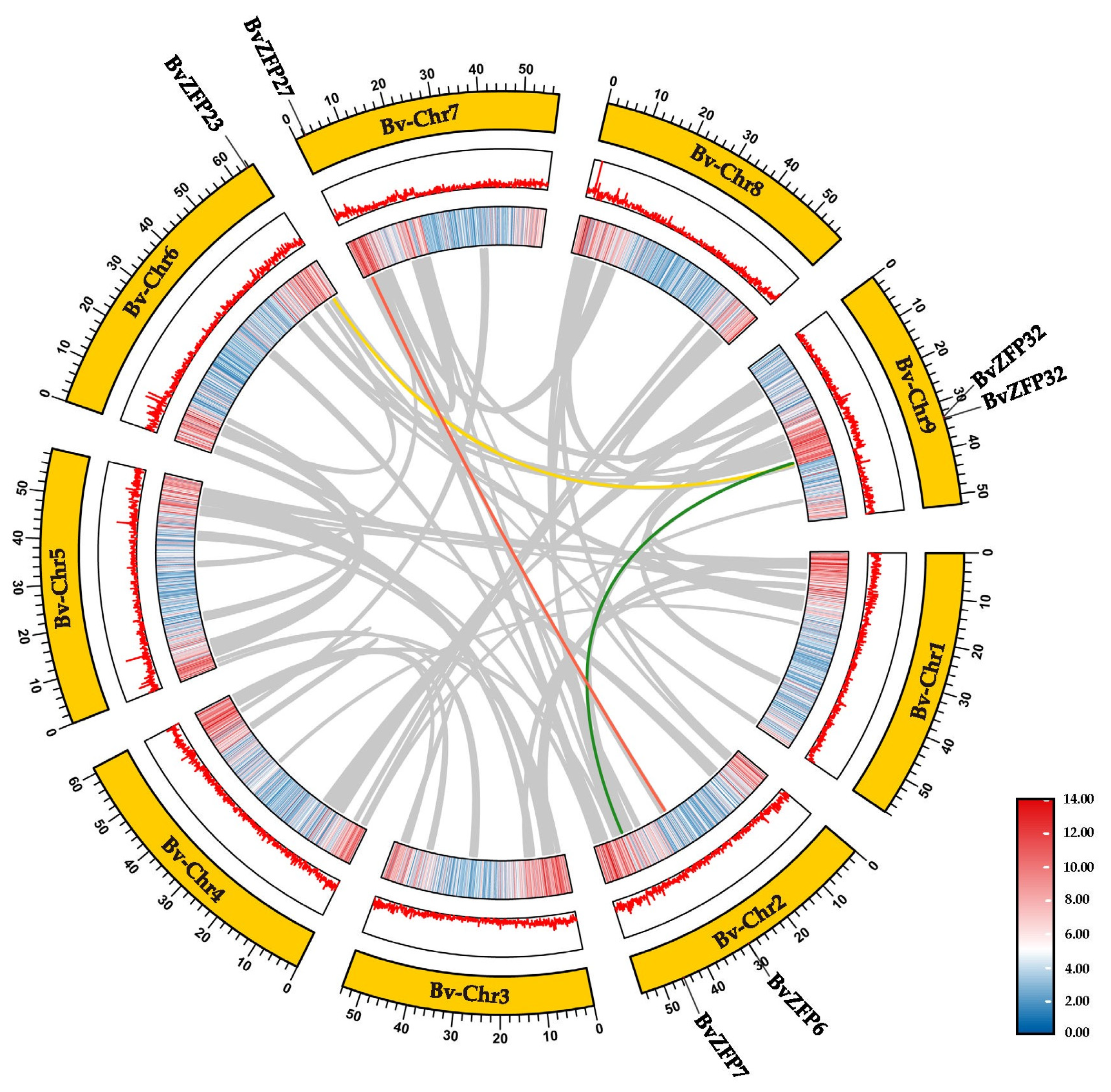
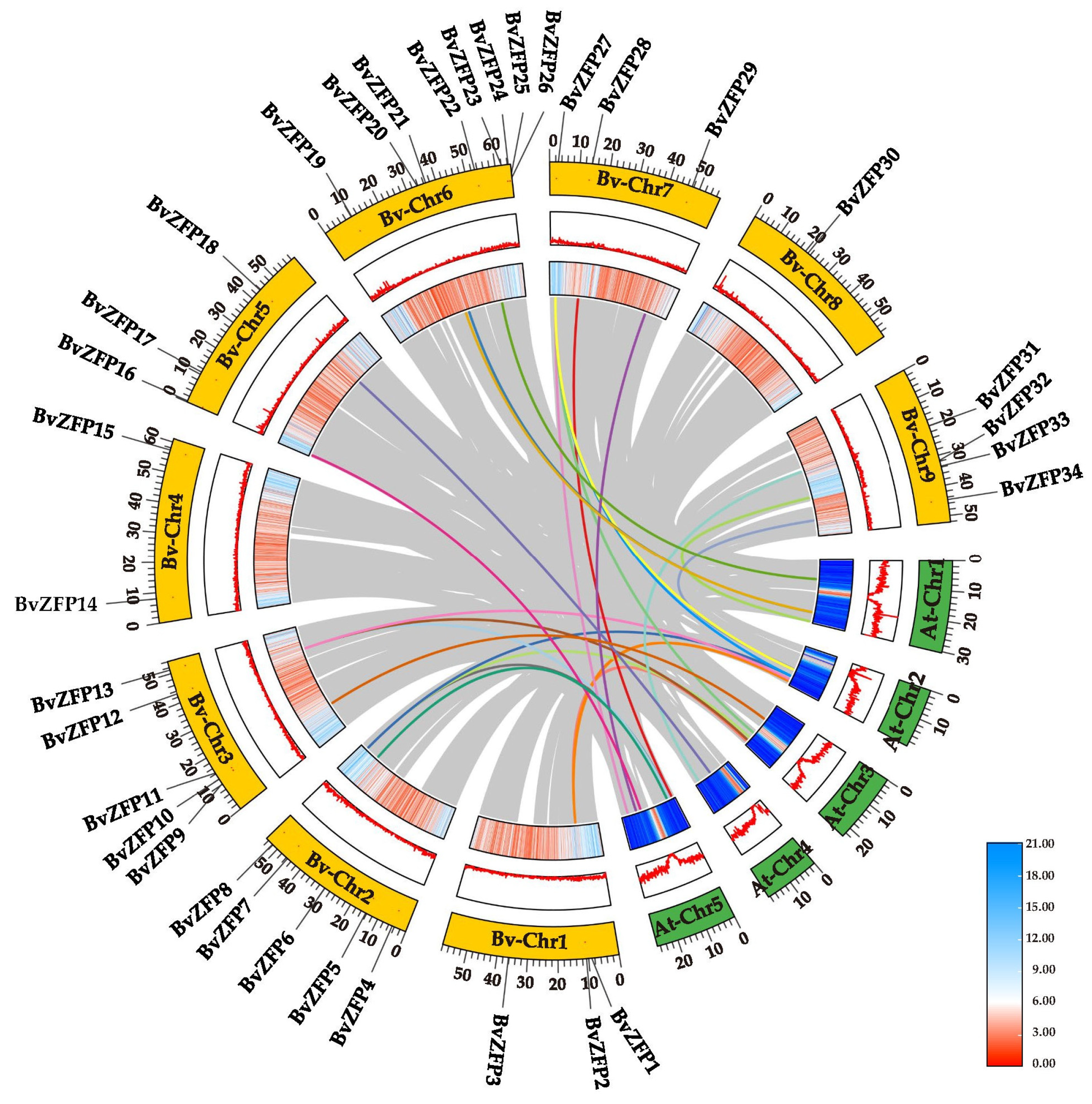
3.5. Genomic Collinearity Analysis of C2H2 Q-Type BvZFPs between Sugar Beet and Arabidopsis
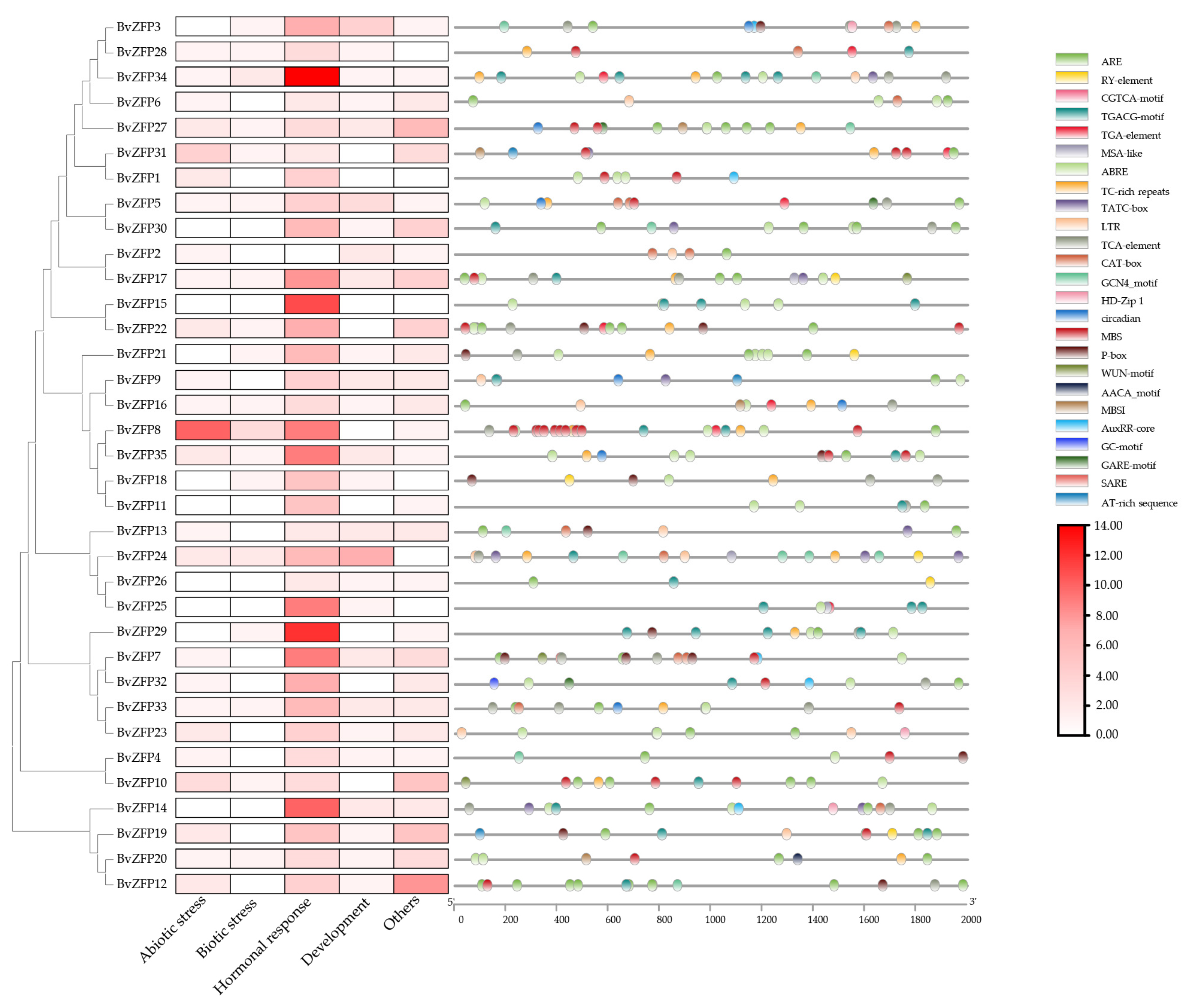
3.6. Promoter Analysis of the Q-Type BVZFP Genes in Sugar Beet
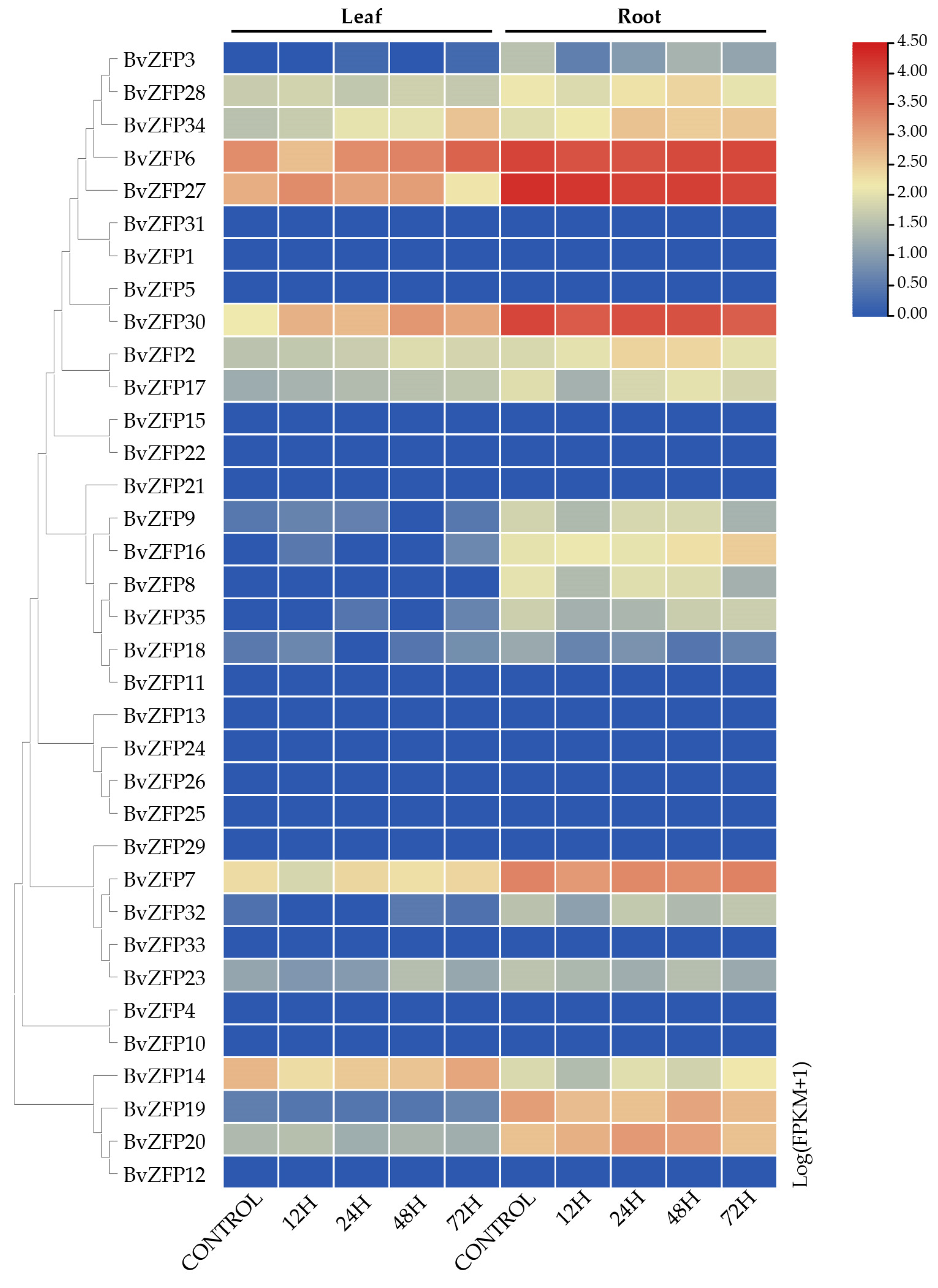
3.7. Expression Profiles Analysis of Q-Type BvZFPs in Different Tissue
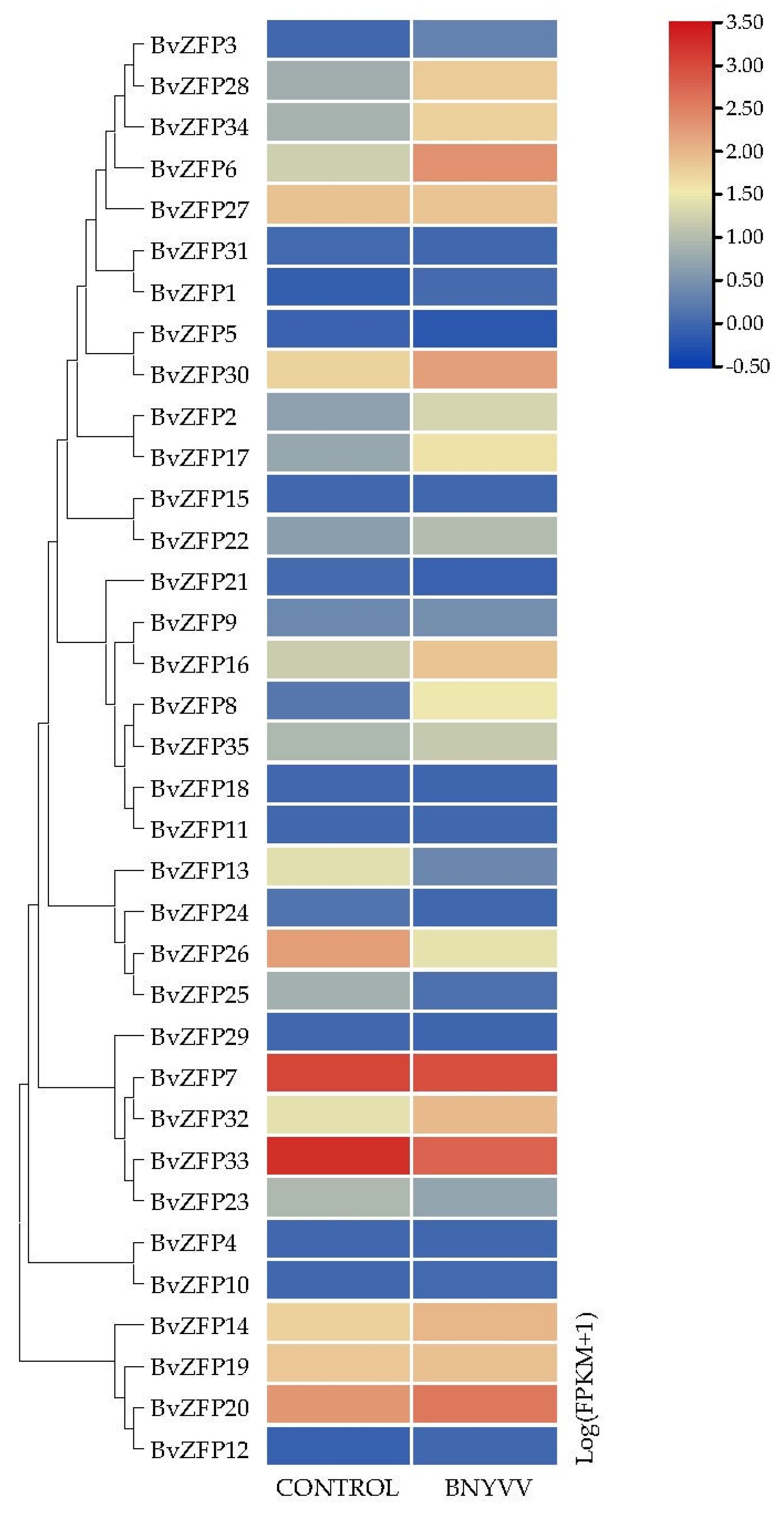
3.8. Responses of Q-Type BvZFP Genes under Salt Treatment and Viral Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weidemüller, P.; Kholmatov, M.; Petsalaki, E.; Zaugg, J.B. Transcription factors: Bridge between cell signaling and gene regulation. Proteomics 2021, 21, 23–24. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family—An update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Viana, V.E.; Busanello, C.; da Maia, L.C.; Pegoraro, C.; Costa de Oliveira, A. Activation of rice WRKY transcription factors: An army of stress fighting soldiers? Curr. Opin. Plant Biol. 2018, 45, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Constabel, C.P. MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef]
- Wang, K.; Ding, Y.; Cai, C.; Chen, Z.; Zhu, C. The role of C2H2 zinc finger proteins in plant responses to abiotic stresses. Physiol. Plant. 2019, 165, 690–700. [Google Scholar] [CrossRef]
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef]
- Yuan, S.; Li, X.; Li, R.; Wang, L.; Zhang, C.; Chen, L.; Hao, Q.; Zhang, X.; Chen, H.; Shan, Z.; et al. Genome-Wide identification and classification of soybean C2H2 zinc finger proteins and their expression analysis in legume-rhizobium symbiosis. Front. Microbiol. 2018, 9, 126. [Google Scholar] [CrossRef]
- Lyu, T.; Liu, W.; Hu, Z.; Xiang, X.; Liu, T.; Xiong, X.; Cao, J. Molecular characterization and expression analysis reveal the roles of Cys2/His2 zinc-finger transcription factors during flower development of Brassica rapa subsp. chinensis. Plant Mol. Biol. 2020, 102, 123–141. [Google Scholar] [CrossRef]
- Jiao, Z.; Wang, L.; Du, H.; Wang, Y.; Liu, J.; Huang, J.; Huang, W.; Ge, L. Genome-wide study of C2H2 zinc finger gene family in Medicago truncatula. BMC Plant Biol. 2020, 20, 401. [Google Scholar] [CrossRef]
- Li, Y.; Sun, A.; Wu, Q.; Zou, X.; Chen, F.; Cai, R.; Xie, H.; Zhang, M.; Guo, X. Comprehensive genomic survey, structural classification and expression analysis of C2H2-type zinc finger factor in wheat (Triticum aestivum L.). BMC Plant Biol. 2021, 21, 380. [Google Scholar] [CrossRef]
- Agarwal, P.; Arora, R.; Ray, S.; Singh, A.K.; Singh, V.P.; Takatsuji, H.; Kapoor, S.; Tyagi, A.K. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 2007, 65, 467–485. [Google Scholar] [CrossRef]
- Li, X.; Cao, X.; Li, J.; Niu, Q.; Mo, Y.; Xiao, L. Genome-wide characterization of C2H2 zinc-finger gene family provides insight into the mechanisms and evolution of the dehydration-rehydration responses in Physcomitrium and Arabidopsis. Front. Plant Sci. 2022, 13, 953459. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, Y.; Cai, Q.; Li, X.; Sun, Y.; Yu, T.; Yang, J.; Zhang, J. Analysis of the C2H2 gene family in maize (Zea mays L.) under cold stress: Identification and expression. Life 2022, 13, 122. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Chen, J.; Liu, M.; Zhang, H.; Liu, D.; Chen, S. Genome-wide analysis of C2H2 zinc finger gene family and its response to cold and drought stress in sorghum [Sorghum bicolor (L.) Moench]. Int. J. Mol. Sci. 2022, 23, 5571. [Google Scholar] [CrossRef]
- Yin, J.; Wang, L.; Zhao, J.; Li, Y.; Huang, R.; Jiang, X.; Zhou, X.; Zhu, X.; He, Y.; He, Y.; et al. Genome-wide characterization of the C2H2 zinc-finger genes in Cucumis sativus and functional analyses of four CsZFPs in response to stresses. BMC Plant Biol. 2020, 20, 359. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Z.; Xu, X.; Zhang, H.; Li, C. Genome-wide analysis of C2H2 zinc-finger family transcription factors and their responses to abiotic stresses in poplar (Populus trichocarpa). PLoS ONE 2015, 10, e0134753. [Google Scholar] [CrossRef]
- Hu, X.; Zhu, L.; Zhang, Y.; Xu, L.; Li, N.; Zhang, X.; Pan, Y. Genome-wide identification of C2H2 zinc-finger genes and their expression patterns under heat stress in tomato (Solanum lycopersicum L.). PeerJ 2019, 7, e7929. [Google Scholar] [CrossRef]
- Xie, M.; Sun, J.; Gong, D.; Kong, Y. The roles of Arabidopsis C1-2i subclass of C2H2-type zinc-finger transcription factors. Genes 2019, 10, 653. [Google Scholar] [CrossRef]
- Englbrecht, C.C.; Schoof, H.; Böhm, S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genom. 2004, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Takatsuji, H.; Mori, M.; Benfey, P.N.; Ren, L.; Chua, N.-H. Characterization of a zinc finger DNA-binding protein expressed specifically in Petunia petals and seedlings. EMBO J. 1992, 11, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Kubo, K.-I.; Sakamoto, A.; Kobayashi, A.; Rybka, Z.; Kanno, Y.; Nakagawa, H.; Nishino, T.; Takatsuji, H. Cys2/His2 zinc-finger protein family of petunia: Evolution and general mechanism of target-sequence recognition. Nucleic Acids Res. 1998, 26, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Dathan, N.; Zaccaro, L.; Esposito, S.; Isernia, C.; Omichinski, J.G.; Riccio, A.; Pedone, C.; Blasio, B.D.; Fattorusso, R.; Pedone, P.V. The Arabidopsis SUPERMAN protein is able to specifically bind DNA through its single Cys2-His2 zinc finger motif. Nucleic Acids Res. 2002, 30, 4945–4951. [Google Scholar] [CrossRef] [PubMed]
- Kiełbowicz-Matuk, A. Involvement of plant C2H2-type zinc finger transcription factors in stress responses. Plant Sci. 2012, 185–186, 78–85. [Google Scholar] [CrossRef]
- Liu, Y.; Khan, A.R.; Gan, Y. C2H2 zinc finger proteins response to abiotic stress in plants. Int. J. Mol. Sci. 2022, 23, 2730. [Google Scholar] [CrossRef]
- Sakamoto, H.; Maruyama, K.; Sakuma, Y.; Meshi, T.; Iwabuchi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 2004, 136, 2734–2746. [Google Scholar] [CrossRef]
- Rizhsky, L.; Davletova, S.; Liang, H.; Mittler, R. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 2004, 279, 11736–11743. [Google Scholar] [CrossRef]
- Ciftci-Yilmaz, S.; Morsy, M.R.; Song, L.; Coutu, A.; Krizek, B.A.; Lewis, M.W.; Warren, D.; Cushman, J.; Connolly, E.L.; Mittler, R. The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J. Biol. Chem. 2007, 282, 9260–9268. [Google Scholar] [CrossRef]
- Yin, M.; Wang, Y.; Zhang, L.; Li, J.; Quan, W.; Yang, L.; Wang, Q.; Chan, Z. The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J. Exp. Bot. 2017, 68, 2991–3005. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, Z.; Yang, C.; Li, G.; Zeng, H.; Li, Z.; Zhang, Y.; Yang, X. Pseudomonas syringae activates ZAT18 to inhibit salicylic acid accumulation by repressing EDS1 transcription for bacterial infection. New Phytol. 2022, 233, 1274–1288. [Google Scholar] [CrossRef]
- Kam, J.; Gresshoff, P.M.; Shorter, R.; Xue, G.-P. The Q-type C2H2 zinc finger subfamily of transcription factors in Triticum aestivum is predominantly expressed in roots and enriched with members containing an EAR repressor motif and responsive to drought stress. Plant Mol. Biol. 2008, 67, 305–322. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.-J.; Guo, S.-Q.; Yang, X.; Bao, Y.-M.; Tang, H.-J.; Sun, H.; Huang, J.; Zhang, H.-S. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J. Exp. Bot. 2010, 61, 2807–2818. [Google Scholar] [CrossRef]
- Lawrence, S.D.; Novak, N.G. Comparative analysis of the genetic variability within the Q-type C2H2 zinc-finger transcription factors in the economically important cabbage, canola and Chinese cabbage genomes. Hereditas 2018, 155, 29. [Google Scholar] [CrossRef] [PubMed]
- Pu, J.; Li, M.; Mao, P.; Zhou, Q.; Liu, E.; Liu, Z. Genome-wide identification of the Q-type C2H2 transcription factor family in alfalfa (Medicago sativa) and expression analysis under different abiotic stresses. Genes 2021, 12, 1906. [Google Scholar] [CrossRef]
- Dohm, J.C.; Minoche, A.E.; Holtgräwe, D.; Capella-Gutiérrez, S.; Zakrzewski, F.; Tafer, H.; Rupp, O.; Sörensen, T.R.; Stracke, R.; Reinhardt, R. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 2014, 505, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Alavilli, H.; Yolcu, S.; Skorupa, M.; Aciksoz, S.B.; Asif, M. Salt and drought stress-mitigating approaches in sugar beet (Beta vulgaris L.) to improve its performance and yield. Planta 2023, 258, 30. [Google Scholar] [CrossRef]
- Rašovský, M.; Pačuta, V.; Ducsay, L.; Lenická, D. Quantity and quality changes in sugar beet (Beta vulgaris Provar. Altissima Doel) induced by different sources of biostimulants. Plants 2022, 11, 2222. [Google Scholar] [CrossRef]
- Toscano, S.; Romano, D.; Ferrante, F. Molecular responses of vegetable, ornamental crops, and model plants to salinity stress. Int. J. Mol. Sci. 2023, 24, 3190. [Google Scholar] [CrossRef]
- Yolcu, S.; Alavilli, H.; Ganesh, P.; Panigrahy, M.; Song, K. Salt and drought stress responses in cultivated beets (Beta vulgaris L.) and wild beet (Beta maritima L.). Plants 2021, 10, 1834. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, V.; Willlems, G.; Darracq, A.; Galein, Y.; Liebe, S.; Varrelmann, M. The Beta vulgaris-derived resistance gene Rz2 confers broad-spectrum resistance against soilborne sugar beet-infecting viruses from different families by recognizing triple gene block protein 1. Mol. Plant Pathol. 2021, 22, 829–842. [Google Scholar] [CrossRef]
- EFSA Panel on Plant Health (PLH); Dehnen-Schmutz, K.; Serio, F.D.; Gonthier, P.; Jacques, M.-A.; Miret, J.A.J.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; Milonas, P. Pest categorisation of beet necrotic yellow vein virus. EFSA J. 2020, 18, e06360. [Google Scholar] [PubMed]
- Gong, Y.; Liu, X.; Chen, S.; Li, H.; Duanmu, H. Genome-wide identification and salt stress response analysis of the bZIP transcription factor family in sugar beet. Int. J. Mol. Sci. 2022, 23, 11573. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.Q.; Li, Z.-Q.; Cao, H.; Wang, J.-L. Genome-wide identification and expression analysis of the WRKY genes in sugar beet (Beta vulgaris L.) under alkaline stress. PeerJ 2019, 7, e7817. [Google Scholar] [CrossRef]
- McGrath, J.M.; Funk, A.; Galewski, P.; Ou, S.; Townsend, B.; Davenport, K.; Daligault, H.; Johnson, S.; Lee, J.; Hastie, A.; et al. A contiguous de novo genome assembly of sugar beet EL10 (Beta vulgaris L.). DNA Res. 2023, 30, dsac033. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Ayaz, A.; Saqib, S.; Huang, H.; Zaman, W.; Lü, S.; Zhao, H. Genome-wide comparative analysis of long-chain acyl-CoA synthetases (LACSs) gene family: A focus on identification, evolution and expression profiling related to lipid synthesis. Plant Physiol. Biochem. 2021, 161, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wascher, F.L.; Nancy Stralis-Pavese, N.-S.; McGrath, J.M.; Schulz, B.; Himmelbauer, H.; Dohm, J.C. Genomic distances reveal relationships of wild and cultivated beets. Nat. Commun. 2022, 13, 2021. [Google Scholar] [CrossRef] [PubMed]
- Roth, C.; Liberles, D.A. A systematic search for positive selection in higher plants (Embryophytes). BMC Plant Biol. 2006, 6, 12. [Google Scholar] [CrossRef][Green Version]
- Cui, J.; Li, J.; Dai, C.; Li, L. Transcriptome and metabolome analyses revealed the response mechanism of sugar beet to salt stress of different durations. Int. J. Mol. Sci. 2022, 23, 9599. [Google Scholar] [CrossRef]
- Gil, J.F.; Wibberg, D.; Eini, O.; Savenkov, E.I.; Varrelmann, M.; Liebe, S. Comparative transcriptome analysis provides molec ular insights into the interaction of beet necrotic yellow vein virus and beet soil-borne mosaic virus with their host sugar beet. Viruses 2020, 12, 76. [Google Scholar]
- Sharma, R.; Mahanty, B.; Mishra, R.; Joshi, R.K. Genome wide identification and expression analysis of pepper C2H2 zinc finger transcription factors in response to anthracnose pathogen Colletotrichum truncatum. 3 Biotech. 2021, 11, 118. [Google Scholar] [CrossRef]
- Li, J.; Zhang, L.; Yuan, Y.; Wang, Q.; Elbaiomy, R.G.; Zhou, W.; Wu, H.; Soaud, S.A.; Abbas, M.; Chen, B. In silico functional prediction and expression analysis of C2H2 zinc-finger family transcription factor revealed regulatory role of ZmZFP126 in maize growth. Front. Genet. 2021, 12, 770427. [Google Scholar] [CrossRef] [PubMed]
- Shuai, Y.; Feng, G.; Yang, Z.; Liu, Q.; Han, J.; Xu, X.; Nie, G.; Huang, L.; Zhang, X. Genome-wide identification of C2H2-type zinc finger gene family members and their expression during abiotic stress responses in orchardgrass (Dactylis glomerata). Genome 2022, 65, 189–203. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Bonthala, V.S.; Mishra, A.K.; Khandelwal, R.; Khan, Y.; Roy, R.; Prasad, M. C2H2 type of zinc finger transcription factors in foxtail millet define response to abiotic stresses. Funct. Integr. Genom. 2014, 14, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Faraji, S.; Rasouli, S.H.; Kazemitabar, S.K. Genome-wide exploration of C2H2 zinc finger family in durum wheat (Triticum turgidum ssp. Durum): Insights into the roles in biological processes especially stress response. Biometals 2018, 31, 1019–1042. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.L.; Chao, J.; Wang, D.; Hu, J.; Wu, H.; Gong, D.; Liu, G. Genome-wide identification and expression profiling of the C2H2-type zinc finger protein transcription factor family in tobacco. Hereditas 2016, 38, 337–349. [Google Scholar]
- Arrey-Salas, O.; Caris-Maldonado, J.C.; Hernández-Rojas, B.; Gonzalez, E. Comprehensive genome-wide exploration of C2H2 zinc finger family in grapevine (Vitis vinifera L.): Insights into the roles in the pollen development regulation. Genes 2021, 12, 302. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Coulter, J.A.; Li, Y.; Zhang, X.; Meng, J.; Zhang, J.; Liu, Y. Genome-wide identification and analysis of the Q-type C2H2 gene family in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 2020, 153, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. Corrigendum: C2H2 zinc finger proteins: Master regulators of abiotic stress responses in plants. Front. Plant Sci. 2020, 11, 298. [Google Scholar] [CrossRef]
- Yang, X.; Wu, G.; Wei, M.; Wang, B. Genome-wide identification of BvHAK gene family in sugar beet (Beta vulgaris) and their expression analysis under salt treatments. Sheng Wu Gong Cheng Xue Bao 2022, 38, 3773–3789. [Google Scholar]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Lorenz, P.; Kreutzer, M.; Li, Y.; Thiesen, H.-J. SysZNF: The C2H2 zinc finger gene database. Nucleic Acids Res. 2009, 37, 236–273. [Google Scholar] [CrossRef] [PubMed]
- Alam, I.; Batool, K.; Cui, D.-L.; Yang, Y.-Q.; Lu, Y.-H. Comprehensive genomic survey, structural classification and expression analysis of C2H2 zinc finger protein gene family in Brassica rapa L. PLoS ONE 2019, 14, e0216071. [Google Scholar] [CrossRef]
- Gourcilleau, D.; Lenne, C.; Armenise, C.; Moulia, B.; Julien, J.-L.; Bronner, G.; Leblanc-Fournier, N. Phylogenetic study of plant Q-type C2H2 zinc finger proteins and expression analysis of poplar genes in response to osmotic, cold and mechanical stresses. DNA Res. 2011, 18, 77–92. [Google Scholar] [CrossRef]
- Ohta, M.; Matsui, K.; Hiratsu, K.; Shinshi, H.; Ohme-Takag, M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 2001, 13, 1959–1968. [Google Scholar] [CrossRef]
- Lyu, T.; Cao, J. Cys2/His2 zinc-finger proteins in transcriptional regulation of flower development. Int. J. Mol. Sci. 2018, 19, 2589. [Google Scholar] [CrossRef]
- Hamel, L.-P.; Benchabane, M.; Nicole, M.-C.; Major, I.T.; Morency, M.-J.; Pelletier, G.; Beaudoin, N.; Sheen, J.; Séguin, A. Stress-responsive mitogen-activated protein kinases interact with the EAR motif of a poplar zinc finger protein and mediate its degradation through the 26S proteasome. Plant Physiol. 2011, 157, 1379–1393. [Google Scholar] [CrossRef]
- Weigel, R.R.; Pfitzner, U.M.; Gatz, C. Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 2005, 17, 1279–1291. [Google Scholar] [CrossRef]
- Han, G.; Wei, X.; Dong, X.; Wang, C.; Sui, N.; Guo, J.; Yuan, F.; Gong, Z.; Li, X.; Zhang, Y.; et al. Arabidopsis ZINC FINGER PROTEIN1 acts downstream of GL2 to repress root hair initiation and elongation by directly suppressing bHLH genes. Plant Cell 2020, 32, 206–225. [Google Scholar] [CrossRef]
- Krogan, N.T.; Hogan, K.; Long, J.A. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 2012, 139, 4180–4190. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, J.; Zhong, G.; Wang, B. Genome-wide identification and expression patterns of the C2H2-zinc finger gene family related to stress responses and catechins accumulation in Camellia sinensis [L.] O. Kuntze. Int. J. Mol. Sci. 2021, 22, 4197. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yue, M.; Jiang, L.; Liu, Y.; Zhang, N.; Liu, X.; Ye, Y.; Lin, X.; Zhang, Y.; Lin, Y.; et al. Genome-wide identification of strawberry C2H2-ZFP C1-2i subclass and the potential function of FaZAT10 in abiotic stress. Int. J. Mol. Sci. 2022, 23, 13079. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.R.; Yang, K.; Wang, X.; Lin, X.-L.; Rui, L.; Liu, H.-F.; Liu, D.-D.; You, C.-X. Overexpression of MdZAT5, an C2H2-type zinc finger protein, regulates anthocyanin accumulation and salt stress response in Apple Calli and Arabidopsis. Int. J. Mol. Sci. 2022, 23, 1897. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.-D.; Zhang, Y.; Liu, J.; Xie, C.-H. Novel potato C2H2-type zinc finger protein gene, StZFP1, which responds to biotic and abiotic stress, plays a role in salt tolerance. Plant Biol. 2010, 12, 689–697. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Ye, T.; Chen, F.; Deng, J.; Yang, P.; Zhang, Y.; Chan, Z. The Cysteine2/Histidine2-type transcription factor zinc finger of Arabidopsis thaliana 6 modulates biotic and abiotic stress responses by activating salicylic acid-related genes and C-REPEAT-BINDING FACTOR genes in Arabidopsis. Plant Physiol. 2014, 165, 1367–1379. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Gene ID | Amino Acid/aa | ORF/bp | Molecular Weight | pI | Instability Index | GRAVY | Location | Subcellular Localization | Group | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BvZFP1 | LOC104905560 | 541 | 1623 | 58,633.75 | 6.86 | 57.91 | −0.786 | 10,695,460 | 10,697,687 | Nucleus | 3i |
| BvZFP2 | LOC125495709 | 367 | 1101 | 41,279.25 | 8.02 | 55.83 | −0.862 | 10,706,247 | 10,708,429 | Nucleus | 3i |
| BvZFP3 | LOC104898669 | 456 | 1368 | 50,063.48 | 6.01 | 60.88 | −1.002 | 36,219,959 | 36,222,487 | Nucleus | 2i |
| BvZFP4 | LOC104887623 | 283 | 849 | 31,507.39 | 8.21 | 41.25 | −0.667 | 4,757,460 | 4,758,311 | Nucleus | 1i |
| BvZFP5 | LOC104906182 | 543 | 1629 | 60,544.44 | 7.17 | 59.45 | −0.701 | 14,363,882 | 14,366,296 | Nucleus | 3i |
| BvZFP6 | LOC104886969 | 186 | 558 | 20,667.45 | 8.86 | 57.6 | −0.506 | 29,939,770 | 29,940,614 | Nucleus | 2i |
| BvZFP7 | LOC104883124 | 278 | 834 | 30,386.53 | 6.13 | 59.97 | −0.644 | 45,479,878 | 45,480,934 | Nucleus | 1i |
| BvZFP8 | LOC104883624 | 179 | 537 | 20,615.07 | 8.3 | 54.73 | −0.976 | 52,215,194 | 52,216,003 | Nucleus | 1i |
| BvZFP9 | LOC104906611 | 170 | 510 | 18,959.88 | 6.65 | 65.03 | −0.922 | 11,875,979 | 11,876,976 | Nucleus | 1i |
| BvZFP10 | LOC104906657 | 255 | 765 | 28,493.69 | 9.22 | 57.02 | −0.867 | 11,959,217 | 11,959,984 | Nucleus | 1i |
| BvZFP11 | LOC104888927 | 232 | 696 | 26,701.97 | 6.89 | 67.24 | −0.791 | 12,886,752 | 12,888,260 | Nucleus | 1i |
| BvZFP12 | LOC104906753 | 304 | 912 | 33,684.31 | 8.85 | 43.09 | −1.037 | 43,115,842 | 43,118,169 | Nucleus | 1i |
| BvZFP13 | LOC104889553 | 323 | 969 | 35,834.41 | 8.56 | 58.9 | −0.805 | 51,214,957 | 51,215,928 | Nucleus | 1i |
| BvZFP14 | LOC104907233 | 277 | 831 | 31,391.42 | 6.2 | 58.48 | −0.972 | 8,025,899 | 8,027,395 | Nucleus | 1i |
| BvZFP15 | LOC104890281 | 572 | 1716 | 62,896.02 | 5.69 | 58.76 | −1.132 | 57,253,629 | 57,255,347 | Nucleus | 3i |
| BvZFP16 | LOC104894695 | 162 | 486 | 18,233.83 | 6.1 | 58.33 | −1.089 | 405,412 | 405,900 | Nucleus | 1i |
| BvZFP17 | LOC104893969 | 496 | 1488 | 54,927.45 | 8.48 | 66.19 | −0.718 | 10,823,448 | 10,825,115 | Nucleus | 3i |
| BvZFP18 | LOC104892924 | 177 | 531 | 20,328.6 | 5.97 | 61.13 | −0.759 | 43,735,292 | 43,736,429 | Nucleus | 1i |
| BvZFP19 | LOC104897252 | 226 | 678 | 25,693.54 | 9.1 | 57.43 | −0.892 | 10,200,725 | 10,201,830 | Nucleus | 1i |
| BvZFP20 | LOC104896450 | 247 | 741 | 25,939.68 | 8.64 | 46.76 | −0.604 | 34,971,928 | 34,972,995 | Nucleus | 1i |
| BvZFP21 | LOC104896549 | 220 | 660 | 24,314.02 | 7.24 | 54.26 | −0.921 | 36,683,224 | 36,687,039 | Nucleus | 1i |
| BvZFP22 | LOC104895813 | 613 | 1839 | 68,769.06 | 6.91 | 48.17 | −1.27 | 53,568,797 | 53,571,334 | Nucleus | 4i |
| BvZFP23 | LOC104895001 | 179 | 537 | 19,911.57 | 8.99 | 75.14 | −0.496 | 63,541,505 | 63,542,044 | Nucleus | 1i |
| BvZFP24 | LOC104885426 | 177 | 531 | 19,942.29 | 6.15 | 49.73 | −0.751 | 64,317,253 | 64,317,786 | Nucleus | 1i |
| BvZFP25 | LOC104885142 | 165 | 495 | 18,957.43 | 7.75 | 37.2 | −0.788 | 64,323,220 | 64,323,717 | Nucleus | 1i |
| BvZFP26 | LOC104884472 | 181 | 543 | 20,434.76 | 8.32 | 43.2 | −0.842 | 64,406,993 | 64,407,538 | Nucleus | 1i |
| BvZFP27 | LOC104900166 | 245 | 735 | 26,613.84 | 6.01 | 65.67 | −0.521 | 2,250,266 | 2,251,688 | Nucleus | 2i |
| BvZFP28 | LOC104908846 | 340 | 1020 | 37,135.81 | 7.75 | 62.32 | −0.839 | 13,404,684 | 13,406,826 | Nucleus | 2i |
| BvZFP29 | LOC104899451 | 204 | 612 | 22,977.76 | 8.79 | 47.89 | −0.777 | 47,440,915 | 47,443,048 | Nucleus | 1i |
| BvZFP30 | LOC104901462 | 242 | 726 | 25,661.67 | 8.09 | 62.93 | −0.545 | 21,006,400 | 21,007,514 | Nucleus | 2i |
| BvZFP31 | LOC104904162 | 237 | 711 | 25,331.76 | 8.55 | 54.27 | −0.358 | 19,929,210 | 19,930,057 | Nucleus | 3i |
| BvZFP32 | LOC104904895 | 299 | 897 | 33,574.34 | 8.24 | 50.67 | −0.682 | 33,331,932 | 33,333,327 | Nucleus | 1i |
| BvZFP33 | LOC104904924 | 185 | 555 | 21,039.05 | 8.76 | 59.64 | −0.511 | 33,657,441 | 33,658,524 | Nucleus | 1i |
| BvZFP34 | LOC104902980 | 342 | 1026 | 37,289.83 | 6.21 | 58.69 | −0.823 | 44,803,778 | 44,805,047 | Nucleus | 2i |
| BvZFP35 | LOC104893645 | 237 | 711 | 26,905.18 | 8.52 | 67.93 | −0.792 | 1,955,234 | 1,956,376 | Nucleus | 1i |
| Species | The Number of C2H2 ZFPs | The Number of Q-Type | The Number of EAR Motif | Reference |
|---|---|---|---|---|
| G. max | 321 | 130 | -- | [8] |
| B. rapa | 301 | 110 | -- | [9] |
| M. sativa | -- | 58 | -- | [34] |
| M. truncatula | 218 | 71 | 106 | [10] |
| Z. mays | 326 | 147 | -- | [52] |
| T. aestivum | 204 | 159 | 68 | [11] |
| O. sativa | 189 | 72 | 43 | [12] |
| A. thaliana | 176 | 64 | -- | [20] |
| S. bicolor | 145 | -- | -- | [15] |
| C. sativus | 129 | 52 | -- | [16] |
| Dactylis glomerata | 125 | 50 | -- | [53] |
| Setaria italica | 124 | 97 | -- | [54] |
| Triticum turgidum | 122 | 96 | 11 | [55] |
| Nicotiana tabacum | 118 | 118 | -- | [56] |
| P. trichocarpa | 109 | 62 | 16 | [17] |
| S. lycopersicum | 104 | 54 | -- | [18] |
| V. vinifera | 98 | 65 | 49 | [57] |
| Capsicum annuum | 79 | 37 | 41 | [51] |
| S. tuberosum | -- | 79 | 54 | [58] |
| B. oleracea | -- | 37 | -- | [33] |
| B. vulgaris | 104 | 35 | 32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Dong, X.; Long, G.; Zhang, Z.; Han, C.; Wang, Y. Genome-Wide Analysis of Q-Type C2H2 ZFP Genes in Response to Biotic and Abiotic Stresses in Sugar Beet. Biology 2023, 12, 1309. https://doi.org/10.3390/biology12101309
Li M, Dong X, Long G, Zhang Z, Han C, Wang Y. Genome-Wide Analysis of Q-Type C2H2 ZFP Genes in Response to Biotic and Abiotic Stresses in Sugar Beet. Biology. 2023; 12(10):1309. https://doi.org/10.3390/biology12101309
Chicago/Turabian StyleLi, Menglin, Xuanyu Dong, Guozhang Long, Zongying Zhang, Chenggui Han, and Ying Wang. 2023. "Genome-Wide Analysis of Q-Type C2H2 ZFP Genes in Response to Biotic and Abiotic Stresses in Sugar Beet" Biology 12, no. 10: 1309. https://doi.org/10.3390/biology12101309
APA StyleLi, M., Dong, X., Long, G., Zhang, Z., Han, C., & Wang, Y. (2023). Genome-Wide Analysis of Q-Type C2H2 ZFP Genes in Response to Biotic and Abiotic Stresses in Sugar Beet. Biology, 12(10), 1309. https://doi.org/10.3390/biology12101309






