Simple Summary
A highly virulent, multidrug-resistant Enterococcus faecalis IRMC827A strain was found in a Saudi Arabian hospital. The strain carries antimicrobial resistance genes and mobile genetic elements, making it resistant to various antibiotics. It also carries virulence factors associated with adherence, biofilm formation, and spreading multidrug resistance. The study highlights the importance of monitoring multidrug-resistant E. faecalis colonization and infection in hospitalized patients, as it is a serious pathogen.
Abstract
We report on a highly virulent, multidrug-resistant strain of Enterococcus faecalis IRMC827A that was found colonizing a long-term male patient at a tertiary hospital in Khobar, Saudi Arabia. The E. faecalis IRMC827A strain carries several antimicrobial drug resistance genes and harbours mobile genetic elements such as Tn6009, which is an integrative conjugative element that can transfer resistance genes between bacteria and ISS1N via an insertion sequence. Whole-genome-sequencing-based antimicrobial susceptibility testing on strains from faecal samples revealed that the isolate E. faecalis IRMC827A is highly resistant to a variety of antibiotics, including tetracycline, doxycycline, minocycline, dalfopristin, virginiamycin, pristinamycin, chloramphenicol, streptomycin, clindamycin, lincomycin, trimethoprim, nalidixic acid and ciprofloxacin. The isolate IRMC827A carries several virulence factors that are significantly associated with adherence, biofilm formation, sortase-assembled pili, manganese uptake, antiphagocytosis, and spreading factor of multidrug resistance. The isolate also encompasses two mutations (G2576T and G2505A) in the 23S rRNA gene associated with linezolid resistance and three more mutations (gyrA p.S83Y, gyrA p.D759N and parC p.S80I) of the antimicrobial resistance phenotype. The findings through next-generation sequencing on the resistome, mobilome and virulome of the isolate in the study highlight the significance of monitoring multidrug-resistant E. faecalis colonization and infection in hospitalized patients. As multidrug-resistant E. faecalis is a serious pathogen, it is particularly difficult to treat and can cause fatal infections. It is important to have quick and accurate diagnostic tests for multidrug-resistant E. faecalis, to track the spread of multidrug-resistant E. faecalis in healthcare settings, and to improve targeted interventions to stop its spread. Further research is necessary to develop novel antibiotics and treatment strategies for multidrug-resistant E. faecalis infections.
1. Introduction
Enterococcus faecalis is a non-sporulating, facultatively anaerobic, and Gram-positive bacteria []. It is a causative agent of several pathological conditions, including surgical wound infections, infective endocarditis and central line-associated bloodstream infections [,]. Enterococci was first discovered by Thiercelin in 1899. However, until 1984 they were considered part of Streptococcus [,,]. Contemporarily, more than 50 morphologically and biochemically diverse species belong to Enterococcus [,].
E. faecalis was first isolated from a patient with infective endocarditis in 1906 []. Moreover, E. faecalis has been isolated from plants, water, soil, sewage, fermented cheese, and dairy food []. In the last century, E. faecalis was one of the leading causes of hospital-acquired infections because of its multidrug resistance nature, with few therapeutic options. The dramatic increase in prevalence is due to the bacteria’s versatility in accommodating nutrition-poor environments and diverse ecological niches, such as pH, hypertonic and hypotonic conditions and temperature, as well as its ability to defeat the infection control interventions employed in hospitals [,,,,]. Enterococcus is a known human intestinal flora inhabitant. Its opportunistic infections in immune-compromised individuals, along with patients receiving broad-spectrum antibiotic treatment or requiring extended hospitalization, are well studied [,]. Antimicrobial drugs have been the cornerstone of medical treatment during the last few decades. Since then, the infection survival rate has declined. Yet, the widespread utilization of antimicrobial drugs has been the selective pressure leading to antimicrobial resistance [,,]. Despite the various applied infectious control interventions across the hospital settings, as well as the wild environments concerning antimicrobial drug use, antimicrobial resistance still continues to rise [,]. A comprehensive study in European countries has demonstrated that Enterococci species accounted for approximately 6.1% to 17.5% of the isolated pathogens and were associated with the highest rate of mortality []. Likewise, a global meta-analysis study has highlighted the acceleration of bloodstream infections associated with antimicrobial resistance in Southeast Asian and Eastern Mediterranean countries compared to the world []. It is specifically noted that there has been a rapid growth in the vancomycin-resistant Enterococci (VRE) rate since it was first reported in 1988 []. VRE is a serious dilemma as its infections are not easily treated, owing to the fact that vancomycin is a drug of last resort []. India has experienced an upsurge in the prevalence of VRE, estimated to have risen up to 10% since 2000 []. Furthermore, the latest evidence has revealed the presence of VRE in food, animals, and wild environments which holds the risk of interstrain transmission of resistance genes and might require more complicated interventions and multisectoral collaboration []. The expansion of antimicrobial resistance has encompassed other antimicrobial drugs, such as tigecycline, linezolid, and daptomycin, yet they are still considered reasonable options against enterococcal infections [].
The translocation process of E. faecalis remains controversial. Some studies have proposed that it occurs when E. faecalis crosses the lymphatic system after failing to be neutralized through phagocytosis by intestinal epithelial cells, dendritic cells, or other tissue-resident leukocytes. Others have claimed it happens when a slight quantity of bacteria diffuses across the intestinal barrier into the bloodstream. [,,]. However, this minor leakage is not a threat in immunocompetent individuals; the innate system is sufficient to conquer such invaders [,,]. Over-colonization of bacteria is a significant risk factor for developing intestinal infection, and it is certainly associated with E. faecalis emergences; it competes with other commensals to colonize human intestines [].
The spread of multidrug-resistant microorganisms is not uncommon in Saudi Arabia. In recent years, several hospitals across the kingdom have been reporting resistant bacteria. The vancomycin-resistant genes in Enterococcus species were first identified in Riyadh in 1993, and ever since, the provenance of these genes has steadily increased []. The following genes—van A, van B, and van C—were identified in the central region and are estimated to occur in approximately 3.5% to 4% of the detected isolates [,], while in the eastern region, the prevalence rate of vancomycin-resistant genes is higher compared to other regions of the country at 6.1% []. However, a genomic analysis study of E. faecalis in the western region has identified over 34 resistance-associated genes linked to various types of commonly used antibiotics [].
Aggregation substance is the foremost virulence factor for pathogenesis. It permits the bacteria to adhere to and colonize the host’s epithelial tissues []. E. faecalis secretes substances that present bactericidal and cytocidal activities. In addition, virulent E. faecalis strains express a pore-forming exotoxin named cytolysin []. For instance, Cytolysin coded by cylLL and cylLS genes hemolyze the host’s cells [,,,]. Gelatinase encoded by the gelE gene hydrolyzes gelatin [,,]. Lastly, serine protease encoded by the sprE gene disintegrates casein [,]. Conjointly, these virulence factors play a role in forming biofilm communities. Biofilm communities are cells encased in an exopolymer matrix and can adhere to biotic and abiotic surfaces, exchange genetic material, and facilitate the spread to extra-intestinal sites. Most importantly, it gives them the beneficial feature of being resistant to antibiotics and immunological responses [,,].
Various studies including a recent systematic review were conducted to give prominence to the prevalence and emergence of multidrug-resistant bacteria and fungi from the Arabian Peninsula, including Saudi Arabia [,,,]. Around 80 species of bacteria and fungi were reported in a recent systematic review from the Arabian region []. Unfortunately, Saudi Arabia reported the highest number of multidrug-resistant bacteria among the other Arabian countries and demonstrated the highest mortality rate. E. faecalis accounts for 256 out of 533 cases caused by Enterococcus species from the Arabian Peninsula []. Considering the great genomic plasticity of E. faecalis, which allows the bacteria to disseminate the resistant genes [,], an increase in the incidence of E. faecalis after drug administration for treating COVID-19 patients was reported from the study region—the Eastern Province of Saudi []. A thorough genomic analysis of E. faecalis in the eastern region of Saudi Arabia has not been previously investigated in the literature. Hence, the objective of this study is to sequence the whole genome of E. faecalis IRMC827A and to perform analysis using various bioinformatics platforms. The isolate was evaluated for the presence of genes associated with multidrug-resistant virulent factors as well as phenotypic mutations.
2. Materials and Methods
2.1. Ethical Approval
Imam Abdulrahman Bin Faisal University’s ethical committee reviewed and approved this project (IRB-2022-01-398). The 1964 Helsinki Declaration and its following revisions, as well as comparable ethical principles, were followed during every procedure.
2.2. Isolation of Bacteria and DNA Extraction
Using cycloserine, cefoxitin, and fructose agar media and a faecal sample from a male patient who was clinically suspected of having a gastrointestinal infection and had recently experienced antibiotic exposure and diarrhoea, a pathogenic strain was isolated. Six predisposing antimicrobial agents (ciprfloxacin, gentamicin, flagyl, meropenem, vancomycin, and tazocin) have been associated with infection. The pathogenic strain was isolated using cycloserine, cefoxitin, and fructose agar media. A positive strain was cultivated on acyclo-serine cefoxitin fructose agar selective medium (CCFA) (MOLEQULE-ON, Auckland, New Zealand). The Gentra Puregene Yeast/Bact. Kit (Qiagen, Hilden, Germany) was used to extract the whole DNA. Thermo Scientific’s Nanodrop 2000 (Waltham, MA, USA) was used to evaluate the purity, quality, and amount of genomic DNA in accordance with the manufacturer’s recommendations. The isolate was PCR amplified, and the 16S rRNA gene was sequenced (GenBank Accession No: OR533998) and analysed as we described earlier to confirm the strain as bacteria [,].
2.3. Genome Mining for Multidrug-Resistant Genes
The genome of the isolate was sequenced using an Illumina HiSeq system (Illumina, San Diego, CA, USA). Genomic DNA was sheared randomly to construct three-read libraries. The paired-end fragment libraries were sequenced according to the Illumina HiSeq system’s protocol. Raw reads of low quality from paired-end sequencing were discarded and other reads were assembled using SOAPdenovo v1.05 software. The paired readings were put together and annotated as previously described [,] using the RAST tool kit (RASTtk 1.3.0) [] and PATRIC (BV-BRC 3.28.5) []. We determined the taxonomy of the IRMC827A’s genome and estimated the average G + C content and contig count using the predictions for the proteins and their roles in gene ontology (GO) [], enzyme commission (EC) [], pathways [], subsystems of protein complexes [] and protein family types []. Specific source databases for known transporters [], antibiotic-resistant genes [], virulence factors [,], and drug targets [,], were used for identifying speciality genes in the IRMC827A genome. Plasmid multilocus sequence typing was used for detecting known plasmid types of IRMC827A []. Anti-microbial resistance (AMR) genes were detected using k-mer-based methods []. Phylogenetic analysis was completed using 100 genes from the NCBI reference for the IRMC827A’s genome in addition to representative genomes by Mash/MinHash (Mash v2.3) [] aligned with MUSCLE [] and a matrix analysis with fast bootstrapping [,]. Metagenomic read mapping was conducted through k-mer alignment against the selected template using the VFDB (2019) and CARD (2020) databases [].
Resistance phenotypes of IRMC827A were predicted using ResFinderFG (Version 2.0) using a functional metagenomic antibiotic resistance database. LRE-Finder (Version 1.0) was applied to detect the mutations in the 23S rRNA gene and genes encoding linezolid resistance (optrA, cfr, cfr(B) and poxtA) in Enterococci [,]. Pathogenic protein families and mobile genetic elements associated with antibiotic resistance in the IRMC827A were predicted using PathogenFinder (Version 1.1) [] and MGE [], respectively.
3. Results
The collected stool sample was subjected to isolating anaerobic bacteria and the isolated bacterial strain, IRMC827A, was initially identified as Enterococcus IRMC827A using 16S rRNA gene sequencing and analysis. In order to identify the genetic impact in the genome of IRMC827A, the whole genome of the strain was sequenced successfully, and annotated (Table 1).

Table 1.
Assembly details and annotated features of Enterococcus faecalis IRMC827A.
The IRMC827A genome (GenBank accession No: JAVLSN000000000; SubmissionID: SUB13827866; BioProject ID: PRJNA1014890) was assembled into 42 contigs with an average G+C content of 37.34% and a total length of 2,899,764 bp (Table 1). This IRMC827A genome belongs to the superkingdom Bacteria, and its taxonomy is cellular organisms > Bacteria > Terrabacteria group > Firmicutes > Bacilli > Lactobacillales > Enterococcaceae > Enterococcus > Enterococcus faecalis. There are 2 ribosomal RNA (rRNA) genes, 41 transfer RNA (tRNA) genes, and 2889 protein-coding sequences (CDS) in this E. faecalis IRMC827A genome (Table 1). Figure 1 displays a circular graphical representation of the distribution of E. faecalis IRMC827A genome annotations as well as a summary of the distinct biological process or structural complex for the genome. The phylogenetic tree (Figure 2), the number of speciality genes (Table 2 and Table 3), the antimicrobial resistance gene details (Table 2), the functional classification (Figure 1), and the genome comparison between Enterococcus faecalis IRMC827A and Enterococcus faecalis V583 (Figure 2) all show that the IRMC827A genome is similar to that of Enterococcus faecalis. In the examination of the proteins, 2250 proteins with known functions and 639 hypothetical proteins were found (Table 1). A total of 686 proteins had EC numbers and 564 had GO designations among the proteins with functional assignments. There are 2813 genus-specific protein families (PLFams) and 2848 cross-genus protein families (PGFams) in the genome of E. faecalis IRMC827A.
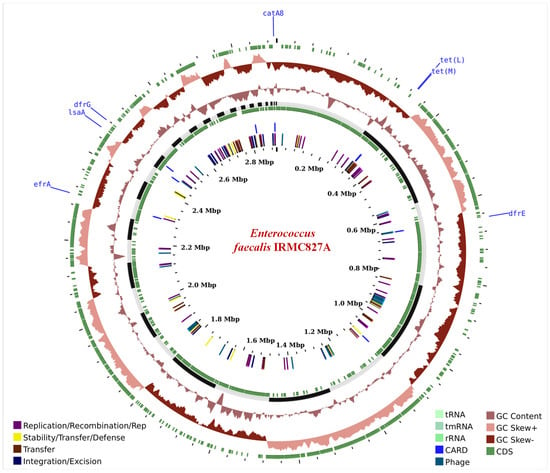
Figure 1.
A circular graphical display of the distribution of the genome annotations in Enterococcus faecalis IRMC827A. This includes the antimicrobial resistance genes projected on the outer ring with names coloured blue.
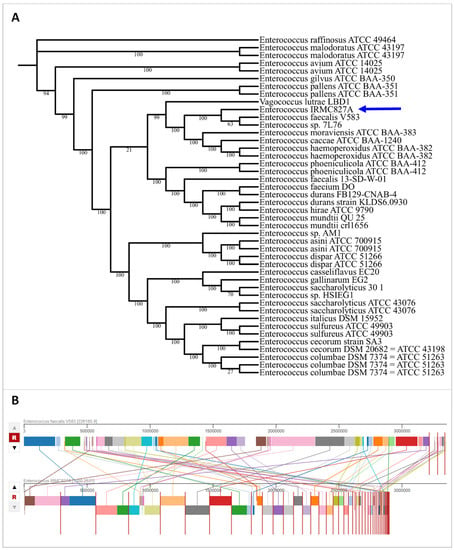
Figure 2.
Phylogenetic tree and genome comparison of IRMC827A genome. (A) Phylogenetic tree of IRMC827A genome. Blue arrow indicates the genome of IRMC827A. Statistics used for phylogenetic tree analysis: alignment program—mafft; branch support method—RAxML fast bootstrapping; requested genomes and number of genomes with data—44; single-copy genes requested, found and number of protein aligned—100; max allowed deletions and duplications—0; number of aligned amino acids—39,722; protein alignment time—346.3 s; number of aligned nucleotides—119,166; number CDS alignments—100; RAxML time—8526.6 s; RAxML likelihood -2,334,654.7903; (B) genome comparison between E. faecalis IRMC827A and E. faecalis V583 representing homologous regions.

Table 2.
Antimicrobial resistance genes and associated antimicrobial resistance mechanisms identified in the genome of E. faecalis IRMC827A.

Table 3.
List of virulence factors in the genome of E. faecalis IRMC827A.
WGS-Based Antimicrobial Susceptibility for Antimicrobial Resistance
The CARD (Comprehensive Antibiotic Resistance Database), PATRIC (Pathosystems Resource Integration Center), NDARO (National Database of Antibiotic Resistant Organisms) databases and ResFinder were used for identifying acquired antimicrobial resistance genes using the whole genome of E. faecalis IRMC827A, and revealed more than 30 antimicrobial resistance genes with various antimicrobial resistance mechanisms (Table 2 and Figure 3). The E. faecalis IRMC827A genome has the highest number (n = 19) of genes connected to antibiotic targets in susceptible species. Three genes (LiaF, LiaR, and LiaS) involved in regulator modulating expression of antibiotic resistance genes were also identified in the IRMC827A genome. Two genes were found in each category such as antibiotic inactivation enzyme, antibiotic target protection protein and efflux pump conferring antibiotic resistance in association with the mechanism of resistance in the genome of IRMC827A.
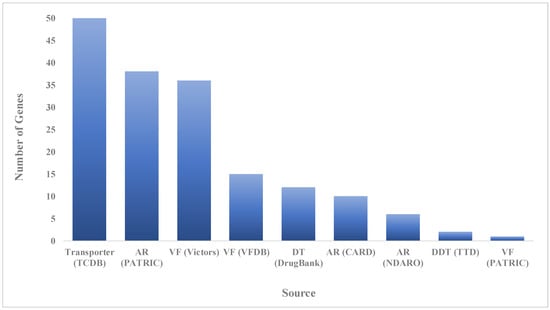
Figure 3.
Number of speciality genes identified in the genome mining of E. faecalis IRMC827A and the homology identified from the specific source database. AR: antibiotic resistance; VF: virulence factor; DT: drug target; CARD: Comprehensive Antibiotic Resistance Database; NDARO: National Database of Antibiotic Resistant Organisms; PATRIC: Pathosystems Resource Integration Center; TCDB: Transporter Classification Database; VFDB: Virulence Factor Database; NDARO: National Database of Antibiotic Resistant Organisms; TTD: Therapeutic Target Database.
Fifty-two virulence factors were identified in the genome of IRMC827A through ResFinder, VFDB, Victors and PATRIC (Table 3). Virulence factors are significantly associated with adherence, biofilm formation, sortase-assembled pili, manganese uptake, ABC transporter, antiphagocytosis, exoenzyme, and spreading factor of multidrug resistance in the genome of IRMC827A (Table 3 and Table S1). Metagenomic read mapping of the genome of E. faecalis IRMC827A against the template genomes (Staphylococcus aureus; Streptococcus pneumoniae; Enterococcus faecium; E. faecalis; Clostridium difficile; Geobacillus stearothermophilus and E. faecalis) using antibiotic resistance database exposed significant genes (p value = 1 × 10−26) (Table S2).
Plasmid multilocus sequence typing of IRMC827A revealed the presence of genes repA (GenBank: AB374546.1; Location: 83,003..84,010) and HMPREF0351_12738 (GenBank: CP003584.1; Location: 24,026..25,231) in E. faecalis plasmid pMG2200 and E. faecium DO plasmid 1, respectively, based on known plasmid types with 100% identity. ResFinderFG-based analysis for identifying resistance phenotypes of IRMC827A identified a resistance phenotype to chloramphenicol (Hit name: cat; 100% identity), tetracycline (Hit name: tet_efflux; 100% identity) and cotrimoxazole (Hit name: dfr; 98.18% identity) (Figure S1) based on a functional metagenomic database. Antimicrobial-resistant phenotype analysis exposed resistance phenotype to 13 antimicrobial agents (tetracycline, doxycycline, minocycline, dalfopristin, virginiamycin m, pristinamycin iia, chloramphenicol, streptomycin, clindamycin, lincomycin, trimethoprim, nalidixic acid and ciprofloxacin) in E. faecalis IRMC827A (Tables S3 and S4). Tetracycline, ciprofloxacin, and chloramphenicol-resistant phenotypes specific to E. faecalis were observed in the IRMC827A genome (Table S4). Two mutations G2576T and G2505A in the 23S rRNA gene were identified as associated with linezolid resistance; however, no mutations were detected in genes such as optrA, cfr, cfr(B) and poxtA in E. faecalis IRMC827A encoding linezolid resistance. There were three resistance-phenotype-associated mutations—gyrA p.S83Y, gyrA p.D759N and parC p.S80I—identified in the genome of E. faecalis IRMC827A (Table 4).

Table 4.
List of resistance-phenotype-associated mutations in the genome of E. faecalis IRMC827A.
Pathogenic protein families of the IRMC827A revealed the strain as a human pathogen (probability score 0.891) with a proteome coverage of 1.97% and 55 matched pathogenic protein families (Table S5). Alignment to reference-based prediction for mobile genetic elements (MGE) associated with antibiotic resistance of the IRMC827A revealed the presence of two MGEs—integrative conjugative element (name of the MGE: Tn6009; Accession: EU399632) and insertion sequence (name of the MGE: ISS1N; Accession: M37395) (Table S6). Tn6009 (position in contig: 31342-33230) showed alignment coverage of 100% containing 1889/1889 bp with sequence identity of 99.95% and one substitution. The ISS1N (position in contig: 16162-16969) showed alignment coverage of 100% containing 808/808 bp with a sequence identity of 98.51% and 12 substitutions.
4. Discussion
Recent studies have found various strains of E. faecalis from different resources that are highly resistant to antibiotics and carry several virulence factors [,,,,,,]. Due to its multidrug resistance, adaptability to nutrient-poor environments and a variety of ecological niches and limited therapy choices, E. faecalis was one of the main causes of hospital-acquired infections and saw dramatic increase in prevalence [,,,,]. The presence of genes and alleles involved in antimicrobial resistance observed in the isolate IRMC827A were reported earlier from various bacterial organisms, such as multidrug-resistant Moraxella catarrhalis [], a heavy-metal-resistant bacterium Cupriavidus campinensis S14E4C [], Bacillus cereus isolated from eye shadow cosmetic products [], Brucella abortus isolated from aborted fetal sheep [], Bacillus velezensis CMU008 [], multidrug-resistant Clostridium perfringens [], a high-lead-resistance bacterium Raoultella planticola [], a human pathogenic strain from Malaysia Chromobacterium violaceum [], and multidrug-resistant Salmonella enterica [], including E. faecalis [].
The analysis revealed the presence of two V domain mutations such as G2576T and G2505A in the 23S rRNA gene which are resistant to linezolid in Enterococci [,]. Recent studies reported an increase in the occurrence of linezolid resistance among Enterococci [], which is in line with the observations of the current study on the linezolid resistance-associated mutations in E. faecalis IRMC827A. Resistance-associated mutations in various genes and matched pathogenic protein families clearly support the multidrug-resistant phenotype of E. faecalis IRMC827A.
One ISS1N insertion sequence, first identified in Lactococcus lactis, was found in the study isolate IRMC827A. These sequences are thought to be crucial for the conjugal transfer of genes involved in lactose processing between different lactic acid bacteria species. However, it is not clear in any descriptions of ISS1N mediating the transfer of virulence or resistance genes. Weissella paramesenteroides were found to be substantially related to IS [,,]. Luna Colagrossi and colleagues reported that the insertion sequence ISS1N is an important factor for bacterial genome shaping and exogenous genetic content integration []. The transposase gene ISS1N was significantly abundant in microorganisms from urban wastewater compared to hospital wastewater []. The presence of insertion sequence ISS1N was reported in Bacteroidetes incertae sedis, Opitutae, and Nitrospira in the resistome analysis of microbial communities in river biofilms [] and Listeria monocytogenes isolated from ready-to-eat foods in Chile []. To our knowledge, this is the first report of E. faecalis in Saudi Arabia that is also carrying Tn6009, along with ISS1N and other resistant genes []. E. faecalis with a mobile genetic element, the integrative conjugative element Tn6009, was reported recently in South Africa’s characteristic vancomycin-resistant phenotype []. Tn6009 is known to be a mobile genetic element that can transfer resistance genes between bacteria []. According to a Norwegian study, E. faecalis, which is responsible for peripheral periodontitis in hospitalised patients, has Tn916 linked with the integrase genes []. Another study found that Tn6009 was linked to Tn916-like components in S. aureus that helped spread MDR determinants that could be acquired from a variety of bacteria, with Enterococcus spp. having the highest rate of transmissibility []. The presence of transposase genes does not necessarily mean that the transposases are active. However, it is important to note that transposons can play a role in the spread of antibiotic resistance genes. Our research suggests that the presence of Tn6009 in the microbiome of the strain in combination with various antimicrobial-resistant genes may facilitate the transfer of resistance and virulence factors and subsequently contribute to the fitness and pathogenicity of E. faecalis IRMC827A. This could result in outbreaks in the future caused by other bacteria in the microbiome in addition to E. faecalis. In order to reduce outbreak conditions, additional molecular research is required to track the genomic and pathogenicity trends of clinical and carriage isolates across geographical areas. Antibiotic use should be considered carefully in both clinical and community settings in order to prevent the emergence and spread of antimicrobial resistance.
A recent study identified common and novel mutations in the gyrA and parC genes in Pseudomonas spp. clinical isolates from Saudi Arabia and provided insight into the genetic background of quinolone resistance []. A Saudi Arabian study found that the DNA gyrase in clinical isolates of E. coli targets quinolones, and that a single amino acid change in gyrA can make E. coli resistant to nalidixic acid and less sensitive to ciprofloxacin []. Another study on the qnr-positive isolates found various mutations in gyrA, but high-level ciprofloxacin resistance was linked to double mutations in gyrA among Enterobacteriaceae from Saudi Arabia []. Fluoroquinolone resistance was found to be linked to gyrA and parC gene mutations in Salmonella enterica from Riyadh, Saudi Arabia []. E. faecalis isolated from the western region of Saudi Arabia possessed gyrA and parC genes and exhibited resistance to quinolone antibiotics like ciprofloxacin, levofloxacin, and moxifloxacin, commonly used for UTIs, enteric infections, and respiratory tract infections []. These studies clearly indicate the presence of the gyrA gene in various bacterial isolates from the study country, which supports the resistance to antimicrobial agents as observed in the present study.
The international spread of multidrug-resistant microorganisms is a challenge facing healthcare providers today. It is crucial to gain some knowledge regarding local microorganisms in the country. The findings of this study clarified and shed light on the understanding of the molecular characteristics of E. faecalis. Nevertheless, the implementation of an active surveillance system to refine hospital protocols and policies, as well as introducing more legislation on the accessibility and usability of antimicrobial drugs, might be feasible strategies for now. Thus, future research must focus on developing new techniques for these infections, such as drugs that could target the resistance sites or rather an advanced treatment that acts on the resistance gene itself. Further studies are needed by either RNA sequencing or RT-PCR analysis of the most important genes to confirm their state of expression to ensure phenotype. Genotypes are the genetic makeup of an organism. WGS can be used to determine the genotype of an organism, but it cannot be used to directly determine the phenotype. The lack of phenotypic identification through antibiotic susceptibility tests from the isolate is one of the limitations of the study. Genetic makeup can be used to develop new diagnostic tests and treatments for antibiotic-resistant infections. It can also be used to track the spread of antibiotic resistance and to develop strategies to prevent it.
5. Conclusions
Many of the antibiotics commonly used to treat infections brought on by this pathogen, including E. faecalis IRMC827A, are highly resistant to it. The strain carries several virulence factors, including those that promote adherence to host cells, biofilm formation, and resistance to phagocytosis. Various mutations and mobile genetic elements (Tn6009 and ISS1N) in IRMC827A are associated with antibiotic resistance, which may contribute to the high level of resistance to antibiotics due to their ability to cause serious and fatal infections and be difficult to treat. The current observations of this study suggest that multidrug-resistant E. faecalis IRMC827A is a serious concern for public health. Further research is needed to develop novel antibiotics and effective treatment strategies for this pathogen.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology12101296/s1, Table S1: List of virulence factors in the genome of E. faecalis IRMC827A. Table S2: Metagenomic read mapping of E. faecalis IRMC827A through various databases. Table S3: Antimicrobial-resistant phenotype results observed in E. faecalis IRMC827A. Table S4: Antimicrobial-resistant phenotype specific for Enterococcus faecalis results observed in E. faecalis IRMC827A. Table S5: List of pathogenic protein families of the IRMC827A. Table S6: List of mobile genetic elements associated with antibiotic resistance of the IRMC827A. Figure S1: Multiple sequence alignment of dfrG gene from Enterococcus faecalis IRMC827A.
Author Contributions
R.A. (Reem AlJindan), D.M.A., N.M. and J.F.B. conceived and designed the research; R.A. (Reem AlJindan), N.M., D.M.A., N.B.A., S.A. and J.F.B. designed the experiments; L.H.A., R.A. (Rahaf Alquwaie), R.A. (Razan Aldahhan), N.F.A., N.B.A., S.A. and J.F.B. performed the whole-genome analysis; R.A. (Reem AlJindan), D.M.A. and N.M. performed clinical analysis; L.H.A., R.A. (Rahaf Alquwaie), R.A. (Razan Aldahhan), N.F.A., N.B.A., S.A. and J.F.B. wrote the manuscript; R.A. (Reem AlJindan), D.M.A., N.M., N.B.A., S.A. and J.F.B. reviewed and approved the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Imam Abdulrahman Bin Faisal University (IRB-2022-01-398).
Informed Consent Statement
Not applicable.
Data Availability Statement
All data will be available on reasonable request from the corresponding author.
Acknowledgments
The authors thank The Dean, Institute for Research and Medical Consultations (IRMC), for her continuous support and encouragement and the administrative staff and facilities provided at IRMC, especially the technical assistance from Ranilo M. Tumbaga, Horace T. Pacifico, and Jee E. Aquino.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Murray, B.E. The Life and Times of the Enterococcus. Clin. Microbiol. Rev. 1990, 3, 46–65. [Google Scholar] [CrossRef] [PubMed]
- Kundra, S.; Lam, L.N.; Kajfasz, J.K.; Casella, L.G.; Andersen, M.J.; Abranches, J.; Flores-Mireles, A.L.; Lemos, J.A. C-Di-AMP Is Essential for the Virulence of Enterococcus faecalis. Infect. Immun. 2021, 89, e0036521. [Google Scholar] [CrossRef] [PubMed]
- Kwit, R.; Zając, M.; Śmiałowska-Węglińska, A.; Skarżyńska, M.; Bomba, A.; Lalak, A.; Skrzypiec, E.; Wojdat, D.; Koza, W.; Mikos-Wojewoda, E.; et al. Prevalence of Enterococcus spp. and the Whole-Genome Characteristics of Enterococcus faecium and Enterococcus faecalis Strains Isolated from Free-Living Birds in Poland. Pathogens 2023, 12, 836. [Google Scholar] [CrossRef]
- Byappanahalli, M.N.; Nevers, M.B.; Korajkic, A.; Staley, Z.R.; Harwood, V.J. Enterococci in the Environment. Microbiol. Mol. Biol. Rev. 2012, 76, 685–706. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, F.; Willems, R.J.L.; Gilmore, M.S. Enterococcus Diversity, Origins in Nature, and Gut Colonization. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- García-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to Its Environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Willems, R.J.L.; Friedrich, A.W.; Rossen, J.W.A.; Bathoorn, E. Enterococcus faecium: From Microbiological Insights to Practical Recommendations for Infection Control and Diagnostics. Antimicrob. Resist. Infect. Control 2020, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial Resistance in Enterococcus Spp. of Animal Origin. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Ruoff, K.L.; de la Maza, L.; Murtagh, M.J.; Spargo, J.D.; Ferraro, M.J. Species Identities of Enterococci Isolated from Clinical Specimens. J. Clin. Microbiol. 1990, 28, 435–437. [Google Scholar] [CrossRef] [PubMed]
- Huycke, M.M.; Sahm, D.F.; Gilmore, M.S. Multiple-Drug Resistant Enterococci: The Nature of the Problem and an Agenda for the Future. Emerg. Infect. Dis. 1998, 4, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Le Breton, Y.; Boël, G.; Benachour, A.; Prévost, H.; Auffray, Y.; Rincé, A. Molecular Characterization of Enterococcus faecalis Two-Component Signal Transduction Pathways Related to Environmental Stresses. Environ. Microbiol. 2003, 5, 329–337. [Google Scholar] [CrossRef]
- Hancock, L.E.; Perego, M. Systematic Inactivation and Phenotypic Characterization of Two-Component Signal Transduction Systems of Enterococcus faecalis V583. J. Bacteriol. 2004, 186, 7951–7958. [Google Scholar] [CrossRef] [PubMed]
- Fiore, E.; Van Tyne, D.; Gilmore, M.S. Pathogenicity of Enterococci. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, B.; Wityk, P.; Gałęcka, M.; Michalik, M. The Many Faces of Enterococcus spp.-Commensal, Probiotic and Opportunistic Pathogen. Microorganisms 2021, 9, 1900. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Teng, F.; Weinstock, G.M.; Murray, B.E. Translocation of Enterococcus faecalis Strains across a Monolayer of Polarized Human Enterocyte-like T84 Cells. J. Clin. Microbiol. 2004, 42, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R.; Matsoso, P.; Pant, S.; Brower, C.; Røttingen, J.-A.; Klugman, K.; Davies, S. Access to Effective Antimicrobials: A Worldwide Challenge. Lancet 2016, 387, 168–175. [Google Scholar] [CrossRef]
- Bondi, M.; Laukova, A.; de Niederhausern, S.; Messi, P.; Papadopoulou, C.; Economou, V. Controversial Aspects Displayed by Enterococci: Probiotics or Pathogens? Biomed. Res. Int. 2020, 2020, 9816185. [Google Scholar] [CrossRef]
- Hou, J.; Long, X.; Wang, X.; Li, L.; Mao, D.; Luo, Y.; Ren, H. Global Trend of Antimicrobial Resistance in Common Bacterial Pathogens in Response to Antibiotic Consumption. J. Hazard. Mater. 2023, 442, 130042. [Google Scholar] [CrossRef]
- Uchil, R.R.; Kohli, G.S.; Katekhaye, V.M.; Swami, O.C. Strategies to Combat Antimicrobial Resistance. J. Clin. Diagn. Res. 2014, 8, ME01–ME04. [Google Scholar] [CrossRef]
- Brinkwirth, S.; Ayobami, O.; Eckmanns, T.; Markwart, R. Hospital-Acquired Infections Caused by Enterococci: A Systematic Review and Meta-Analysis, WHO European Region, 1 January 2010 to 4 February 2020. Euro. Surveill. 2021, 26, 2001628. [Google Scholar] [CrossRef]
- Jabbari Shiadeh, S.M.; Pormohammad, A.; Hashemi, A.; Lak, P. Global Prevalence of Antibiotic Resistance in Blood-Isolated Enterococcus faecalis and Enterococcus faecium: A Systematic Review and Meta-Analysis. Infect. Drug. Resist. 2019, 12, 2713–2725. [Google Scholar] [CrossRef]
- Uttley, A.H.; Collins, C.H.; Naidoo, J.; George, R.C. Vancomycin-Resistant Enterococci. Lancet 1988, 1, 57–58. [Google Scholar] [CrossRef]
- Smout, E.; Palanisamy, N.; Valappil, S.P. Prevalence of Vancomycin-Resistant Enterococci in India between 2000 and 2022: A Systematic Review and Meta-Analysis. Antimicrob. Resist. Infect. Control 2023, 12, 79. [Google Scholar] [CrossRef]
- Dadashi, M.; Sharifian, P.; Bostanshirin, N.; Hajikhani, B.; Bostanghadiri, N.; Khosravi-Dehaghi, N.; van Belkum, A.; Darban-Sarokhalil, D. The Global Prevalence of Daptomycin, Tigecycline, and Linezolid-Resistant Enterococcus faecalis and Enterococcus faecium Strains from Human Clinical Samples: A Systematic Review and Meta-Analysis. Front. Med. 2021, 8, 720647. [Google Scholar] [CrossRef] [PubMed]
- Wells, C.L.; Maddaus, M.A.; Simmons, R.L. Proposed Mechanisms for the Translocation of Intestinal Bacteria. Rev. Infect. Dis. 1988, 10, 958–979. [Google Scholar] [CrossRef] [PubMed]
- Wells, C.L.; Jechorek, R.P.; Erlandsen, S.L. Evidence for the Translocation of Enterococcus faecalis across the Mouse Intestinal Tract. J. Infect. Dis. 1990, 162, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Kao, P.H.N.; Kline, K.A. Dr. Jekyll and Mr. Hide: How Enterococcus faecalis Subverts the Host Immune Response to Cause Infection. J. Mol. Biol. 2019, 431, 2932–2945. [Google Scholar] [CrossRef]
- Hashem, Y.A.; Abdelrahman, K.A.; Aziz, R.K. Phenotype-Genotype Correlations and Distribution of Key Virulence Factors in Enterococcus faecalis Isolated from Patients with Urinary Tract Infections. Infect. Drug Resist. 2021, 14, 1713–1723. [Google Scholar] [CrossRef]
- Qadri, S.H.; Qunibi, W.Y.; Al-Ballaa, S.R.; Kadhi, Y.; Burdette, J.M. Vancomycin Resistant Enterococcus: A Case Report and Review of the Literature. Ann. Saudi. Med. 1993, 13, 289–293. [Google Scholar] [CrossRef]
- Salem-Bekhit, M.M.; Moussa, I.M.I.; Muharram, M.M.; Alanazy, F.K.; Hefni, H.M. Prevalence and Antimicrobial Resistance Pattern of Multidrug-Resistant Enterococci Isolated from Clinical Specimens. Indian J. Med. Microbiol. 2012, 30, 44–51. [Google Scholar] [CrossRef]
- Somily, A.M.; Al-Mohizea, M.M.; Absar, M.M.; Fatani, A.J.; Ridha, A.M.; Al-Ahdal, M.N.; Senok, A.C.; Al-Qahtani, A.A. Molecular Epidemiology of Vancomycin Resistant Enterococci in a Tertiary Care Hospital in Saudi Arabia. Microb. Pathog. 2016, 97, 79–83. [Google Scholar] [CrossRef]
- Kankalil George, S.; Suseela, M.R.; El Safi, S.; Ali Elnagi, E.; Al-Naam, Y.A.; Adlan Mohammed Adam, A.; Mary Jacob, A.; Al-Maqati, T.; Kumar Ks, H. Molecular Determination of van Genes among Clinical Isolates of Enterococci at a Hospital Setting. Saudi J. Biol. Sci. 2021, 28, 2895–2899. [Google Scholar] [CrossRef] [PubMed]
- Farman, M.; Yasir, M.; Al-Hindi, R.R.; Farraj, S.A.; Jiman-Fatani, A.A.; Alawi, M.; Azhar, E.I. Genomic Analysis of Multidrug-Resistant Clinical Enterococcus faecalis Isolates for Antimicrobial Resistance Genes and Virulence Factors from the Western Region of Saudi Arabia. Antimicrob. Resist. Infect. Control 2019, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Kreft, B.; Marre, R.; Schramm, U.; Wirth, R. Aggregation Substance of Enterococcus faecalis Mediates Adhesion to Cultured Renal Tubular Cells. Infect. Immun. 1992, 60, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Van Tyne, D.; Martin, M.J.; Gilmore, M.S. Structure, Function, and Biology of the Enterococcus faecalis Cytolysin. Toxins 2013, 5, 895–911. [Google Scholar] [CrossRef]
- Chow, J.W.; Thal, L.A.; Perri, M.B.; Vazquez, J.A.; Donabedian, S.M.; Clewell, D.B.; Zervos, M.J. Plasmid-Associated Hemolysin and Aggregation Substance Production Contribute to Virulence in Experimental Enterococcal Endocarditis. Antimicrob. Agents Chemother. 1993, 37, 2474–2477. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; van der Donk, W.A. The Sequence of the Enterococcal cytolysin Imparts Unusual Lanthionine Stereochemistry. Nat. Chem. Biol. 2013, 9, 157–159. [Google Scholar] [CrossRef]
- Doss Susai Backiam, A.; Duraisamy, S.; Karuppaiya, P.; Balakrishnan, S.; Chandrasekaran, B.; Kumarasamy, A.; Raju, A. Antibiotic Susceptibility Patterns and Virulence-Associated Factors of Vancomycin-Resistant Enterococcal Isolates from Tertiary Care Hospitals. Antibiotics 2023, 12, 981. [Google Scholar] [CrossRef]
- Del Papa, M.F.; Hancock, L.E.; Thomas, V.C.; Perego, M. Full Activation of Enterococcus faecalis Gelatinase by a C-Terminal Proteolytic Cleavage. J. Bacteriol. 2007, 189, 8835–8843. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Guzmán-Soto, I.; McTiernan, C.; Gonzalez-Gomez, M.; Ross, A.; Gupta, K.; Suuronen, E.J.; Mah, T.-F.; Griffith, M.; Alarcon, E.I. Mimicking Biofilm Formation and Development: Recent Progress in in Vitro and in Vivo Biofilm Models. iScience 2021, 24, 102443. [Google Scholar] [CrossRef]
- Khalil, M.A.; Alorabi, J.A.; Al-Otaibi, L.M.; Ali, S.S.; Elsilk, S.E. Antibiotic Resistance and Biofilm Formation in Enterococcus spp. Isolated from Urinary Tract Infections. Pathogens 2022, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- AlJindan, R.; AlEraky, D.M.; Mahmoud, N.; Abdalhamid, B.; Almustafa, M.; AbdulAzeez, S.; Borgio, J.F. Drug Resistance-Associated Mutations in ERG11 of Multidrug-Resistant Candida auris in a Tertiary Care Hospital of Eastern Saudi Arabia. J. Fungi 2020, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- AlJindan, R.; AlEraky, D.M.; Borgio, J.F.; AbdulAzeez, S.; Abdalhamid, B.; Mahmoud, N.; Farhat, M. Diagnostic Deficiencies of C. difficile Infection among Patients in a Tertiary Hospital in Saudi Arabia: A Laboratory-Based Case Series. Saudi J. Biol. Sci. 2021, 28, 4472–4477. [Google Scholar] [CrossRef] [PubMed]
- AlJindan, R.; AlEraky, D.M.; Farhat, M.; Almandil, N.B.; AbdulAzeez, S.; Borgio, J.F. Genomic Insights into Virulence Factors and Multi-Drug Resistance in Clostridium perfringens IRMC2505A. Toxins 2023, 15, 359. [Google Scholar] [CrossRef]
- Borgio, J.F.; Rasdan, A.S.; Sonbol, B.; Alhamid, G.; Almandil, N.B.; AbdulAzeez, S. Emerging Status of Multidrug-Resistant Bacteria and Fungi in the Arabian Peninsula. Biology 2021, 10, 1144. [Google Scholar] [CrossRef]
- Zhong, Z.; Kwok, L.-Y.; Hou, Q.; Sun, Y.; Li, W.; Zhang, H.; Sun, Z. Comparative Genomic Analysis Revealed Great Plasticity and Environmental Adaptation of the Genomes of Enterococcus faecium. BMC Genom. 2019, 20, 602. [Google Scholar] [CrossRef]
- Al-Qaaneh, A.M.; Al-Ghamdi, F.H.; AbdulAzeez, S.; Borgio, J.F. Safety of Tocilizumab in COVID-19 Patients and Benefit of Single-Dose: The Largest Retrospective Observational Study. Pharmaceutics 2022, 14, 624. [Google Scholar] [CrossRef]
- AlEraky, D.M.; Madi, M.; El Tantawi, M.; AlHumaid, J.; Fita, S.; AbdulAzeez, S.; Borgio, J.F.; Al-Harbi, F.A.; Alagl, A.S. Predominance of Non-Streptococcus Mutans Bacteria in Dental Biofilm and Its Relation to Caries Progression. Saudi J. Biol. Sci. 2021, 28, 7390–7395. [Google Scholar] [CrossRef]
- Borgio, J.F.; Alhujaily, R.; Alquwaie, R.; Alabdullah, M.J.; AlHasani, E.; Alothman, W.; Alaqeel, R.K.; Alfaraj, A.S.; Kaabi, A.; Alhur, N.F.; et al. Mining the Nanotube-Forming Bacillus amyloliquefaciens MR14M3 Genome for Determining Anti-Candida auris and Anti-Candida albicans Potential by Pathogenicity and Comparative Genomics Analysis. Comput. Struct. Biotechnol. J. 2023, 21, 4261–4276. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A Modular and Extensible Implementation of the RAST Algorithm for Building Custom Annotation Pipelines and Annotating Batches of Genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the All-Bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Schomburg, I.; Chang, A.; Ebeling, C.; Gremse, M.; Heldt, C.; Huhn, G.; Schomburg, D. BRENDA, the Enzyme Database: Updates and Major New Developments. Nucleic Acids Res. 2004, 32, D431–D433. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Overbeek, R.; Begley, T.; Butler, R.M.; Choudhuri, J.V.; Chuang, H.-Y.; Cohoon, M.; de Crécy-Lagard, V.; Diaz, N.; Disz, T.; Edwards, R.; et al. The Subsystems Approach to Genome Annotation and Its Use in the Project to Annotate 1000 Genomes. Nucleic Acids Res. 2005, 33, 5691–5702. [Google Scholar] [CrossRef]
- Davis, J.J.; Gerdes, S.; Olsen, G.J.; Olson, R.; Pusch, G.D.; Shukla, M.; Vonstein, V.; Wattam, A.R.; Yoo, H. PATtyFams: Protein Families for the Microbial Genomes in the PATRIC Database. Front. Microbiol. 2016, 7, 118. [Google Scholar] [CrossRef]
- Saier, M.H.; Reddy, V.S.; Tsu, B.V.; Ahmed, M.S.; Li, C.; Moreno-Hagelsieb, G. The Transporter Classification Database (TCDB): Recent Advances. Nucleic Acids Res. 2016, 44, D372–D379. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and Refined Dataset for Big Data Analysis—10 Years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Mao, C.; Abraham, D.; Wattam, A.R.; Wilson, M.J.C.; Shukla, M.; Yoo, H.S.; Sobral, B.W. Curation, Integration and Visualization of Bacterial Virulence Factors in PATRIC. Bioinformatics 2015, 31, 252–258. [Google Scholar] [CrossRef]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding New Light on Drug Metabolism. Nucleic Acids Res. 2014, 42, D1091–D1097. [Google Scholar] [CrossRef]
- Zhu, F.; Han, B.; Kumar, P.; Liu, X.; Ma, X.; Wei, X.; Huang, L.; Guo, Y.; Han, L.; Zheng, C.; et al. Update of TTD: Therapeutic Target Database. Nucleic Acids Res. 2010, 38, D787–D791. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast Genome and Metagenome Distance Estimation Using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and Precise Alignment of Raw Reads against Redundant Databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef]
- Hasman, H.; Clausen, P.T.L.C.; Kaya, H.; Hansen, F.; Knudsen, J.D.; Wang, M.; Holzknecht, B.J.; Samulioniené, J.; Røder, B.L.; Frimodt-Møller, N.; et al. LRE-Finder, a Web Tool for Detection of the 23S rRNA Mutations and the optrA, Cfr, Cfr(B) and poxtA Genes Encoding Linezolid Resistance in Enterococci from Whole-Genome Sequences. J. Antimicrob. Chemother. 2019, 74, 1473–1476. [Google Scholar] [CrossRef]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder—Distinguishing Friend from Foe Using Bacterial Whole Genome Sequence Data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of Mobile Genetic Elements Associated with Antibiotic Resistance in Salmonella enterica Using a Newly Developed Web Tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef]
- Founou, R.C.; Founou, L.L.; Allam, M.; Ismail, A.; Essack, S.Y. Enterococcus faecalis ST21 Harbouring Tn6009 Isolated from a Carriage Sample in South Africa. S. Afr. Med. J. 2021, 111, 98–99. [Google Scholar] [CrossRef]
- Freitas, A.d.A.R.; Souza, S.D.S.R.; Faria, A.R.; Planet, P.J.; Merquior, V.L.C.; Teixeira, L.M. Draft Genome Sequences of Two Commensal Enterococcus faecalis Strains Isolated from American Black Vultures (Coragyps atratus) in Brazil. Microbiol. Resour. Announc. 2022, 11, e0005722. [Google Scholar] [CrossRef]
- Khan, A.; Miller, W.R.; Axell-House, D.; Munita, J.M.; Arias, C.A. Antimicrobial Susceptibility Testing for Enterococci. J. Clin. Microbiol. 2022, 60, e0084321. [Google Scholar] [CrossRef]
- Kuwabara, M.; Irimajiri, R.; Togo, S.; Fujino, Y.; Honsho, M.; Mawatari, S.; Fujino, T.; Doi, K. Complete Genome Sequence of the Thermophilic Enterococcus faecalis Strain K-4, Isolated from a Grass Silage in Thailand. Microbiol. Resour. Announc. 2023, 12, e0081422. [Google Scholar] [CrossRef]
- Segawa, T.; Hisatsune, J.; Ishida-Kuroki, K.; Sugawara, Y.; Masuda, K.; Tadera, K.; Kashiyama, S.; Yokozaki, M.; Le, M.N.-T.; Kawada-Matsuo, M.; et al. Complete Genome Sequence of optrA-Carrying Enterococcus faecalis Isolated from Open Pus in a Japanese Patient. J. Glob. Antimicrob. Resist. 2023, 33, 276–278. [Google Scholar] [CrossRef]
- Wardal, E.; Żabicka, D.; Hryniewicz, W.; Sadowy, E. VanA-Enterococcus faecalis in Poland: Hospital Population Clonal Structure and vanA Mobilome. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 1245–1261. [Google Scholar] [CrossRef]
- Bristy, S.A.; Hossain, M.A.; Hasan, M.I.; Mahmud, S.M.H.; Moni, M.A.; Rahman, M.H. An Integrated Complete-Genome Sequencing and Systems Biology Approach to Predict Antimicrobial Resistance Genes in the Virulent Bacterial Strains of Moraxella catarrhalis. Brief. Funct. Genom. 2023, 22, 375–391. [Google Scholar] [CrossRef]
- Abbaszade, G.; Szabó, A.; Vajna, B.; Farkas, R.; Szabó, C.; Tóth, E. Whole Genome Sequence Analysis of Cupriavidus campinensis S14E4C, a Heavy Metal Resistant Bacterium. Mol. Biol. Rep. 2020, 47, 3973–3985. [Google Scholar] [CrossRef]
- Yossa, N.; Bell, R.; Tallent, S.; Brown, E.; Binet, R.; Hammack, T. Genomic Characterization of Bacillus cereus Sensu Stricto 3A ES Isolated from Eye Shadow Cosmetic Products. BMC Microbiol. 2022, 22, 240. [Google Scholar] [CrossRef]
- Bolotin, V.; Kovalenko, G.; Marchenko, N.; Solodiankin, O.; Rudova, N.; Kutsenko, V.; Bortz, E.; Gerilovych, A.; Drown, D.M. Complete Genome Sequence of Brucella abortus 68, Isolated from Aborted Fetal Sheep in Ukraine. Microbiol. Resour. Announc. 2021, 10, e01436-20. [Google Scholar] [CrossRef] [PubMed]
- Rangseekaew, P.; Ua-Arak, N.; Pathom-Aree, W. Draft Genome Sequence Data of Plant Growth Promoting and Calcium Carbonate Precipitating Bacillus velezensis CMU008. Data Brief 2023, 47, 108965. [Google Scholar] [CrossRef] [PubMed]
- Elarabi, N.I.; Halema, A.A.; Abdelhadi, A.A.; Henawy, A.R.; Samir, O.; Abdelhaleem, H.A.R. Draft Genome of Raoultella planticola, a High Lead Resistance Bacterium from Industrial Wastewater. AMB Express 2023, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, V.K.; Mazlan, Z.; Mastor, N.N.; Hoque, M.Z. Whole Genome Sequence Data of Chromobacterium violaceum WCH4, a Human Pathogenic Strain from Sabah, Malaysia. Data Brief 2021, 39, 107533. [Google Scholar] [CrossRef] [PubMed]
- Mohakud, N.K.; Panda, R.K.; Patra, S.D.; Sahu, B.R.; Ghosh, M.; Kushwaha, G.S.; Misra, N.; Suar, M. Genome Analysis and Virulence Gene Expression Profile of a Multi Drug Resistant Salmonella enterica Serovar Typhimurium Ms202. Gut Pathog. 2022, 14, 28. [Google Scholar] [CrossRef]
- Akter, T.; Haque, M.N.; Ehsan, R.; Paul, S.I.; Foysal, M.J.; Tay, A.C.Y.; Islam, M.T.; Rahman, M.M. Virulence and Antibiotic-Resistance Genes in Enterococcus faecalis Associated with Streptococcosis Disease in Fish. Sci. Rep. 2023, 13, 1551. [Google Scholar] [CrossRef]
- Apostolakos, I.; Paramithiotis, S.; Mataragas, M. Functional and Safety Characterization of Weissella paramesenteroides Strains Isolated from Dairy Products through Whole-Genome Sequencing and Comparative Genomics. Dairy 2022, 3, 799–813. [Google Scholar] [CrossRef]
- Haandrikman, A.J.; van Leeuwen, C.; Kok, J.; Vos, P.; de Vos, W.M.; Venema, G. Insertion Elements on Lactococcal proteinase Plasmids. Appl. Environ. Microbiol. 1990, 56, 1890–1896. [Google Scholar] [CrossRef]
- Harmer, C.J.; Hall, R.M. An Analysis of the IS6/IS26 Family of Insertion Sequences: Is It a Single Family? Microb. Genom. 2019, 5, e000291. [Google Scholar] [CrossRef]
- Colagrossi, L.; Costabile, V.; Scutari, R.; Agosta, M.; Onori, M.; Mancinelli, L.; Lucignano, B.; Onetti Muda, A.; Del Baldo, G.; Mastronuzzi, A.; et al. Evidence of Pediatric Sepsis Caused by a Drug Resistant Lactococcus garvieae Contaminated Platelet Concentrate. Emerg. Microbes. Infect. 2022, 11, 1325–1334. [Google Scholar] [CrossRef]
- Buelow, E.; Rico, A.; Gaschet, M.; Lourenço, J.; Kennedy, S.P.; Wiest, L.; Ploy, M.-C.; Dagot, C. Hospital Discharges in Urban Sanitation Systems: Long-Term Monitoring of Wastewater Resistome and Microbiota in Relationship to Their Eco-Exposome. Water Res. X 2020, 7, 100045. [Google Scholar] [CrossRef] [PubMed]
- Matviichuk, O.; Mondamert, L.; Geffroy, C.; Gaschet, M.; Dagot, C.; Labanowski, J. River Biofilms Microbiome and Resistome Responses to Wastewater Treatment Plant Effluents Containing Antibiotics. Front. Microbiol. 2022, 13, 795206. [Google Scholar] [CrossRef] [PubMed]
- Parra-Flores, J.; Holý, O.; Bustamante, F.; Lepuschitz, S.; Pietzka, A.; Contreras-Fernández, A.; Castillo, C.; Ovalle, C.; Alarcón-Lavín, M.P.; Cruz-Córdova, A.; et al. Virulence and Antibiotic Resistance Genes in Listeria monocytogenes Strains Isolated from Ready-to-Eat Foods in Chile. Front. Microbiol. 2021, 12, 796040. [Google Scholar] [CrossRef] [PubMed]
- Soge, O.O.; Beck, N.K.; White, T.M.; No, D.B.; Roberts, M.C. A Novel Transposon, Tn6009, Composed of a Tn916 Element Linked with a Staphylococcus aureus Mer Operon. J. Antimicrob. Chemother. 2008, 62, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Sun, J.; Mikalsen, T.; Roberts, A.P.; Sundsfjord, A. Characterisation of the Plasmidome within Enterococcus faecalis Isolated from Marginal Periodontitis Patients in Norway. PLoS ONE 2013, 8, e62248. [Google Scholar] [CrossRef]
- El-Badawy, M.F.; Eed, E.M.; Sleem, A.S.; El-Sheikh, A.A.K.; Maghrabi, I.A.; Abdelwahab, S.F. The First Saudi Report of Novel and Common Mutations in the gyrA and parC Genes among Pseudomonas spp. Clinical Isolates Recovered from Taif Area. Infect Drug Resist 2022, 15, 3801–3814. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.M. Detection of Mutations in Quinolone-Resistant Determining Regions in Clinical Isolates of Escherichia coli from Saudi Arabia. Afr. J. Biotechnol. 2012, 11. [Google Scholar] [CrossRef]
- Shibl, A.M.; Al-Agamy, M.H.; Khubnani, H.; Senok, A.C.; Tawfik, A.F.; Livermore, D.M. High Prevalence of Acquired Quinolone-Resistance Genes among Enterobacteriaceae from Saudi Arabia with CTX-M-15 β-Lactamase. Diagn. Microbiol. Infect. Dis. 2012, 73, 350–353. [Google Scholar] [CrossRef]
- El-Tayeb, M.A.; Ibrahim, A.S.S.; Al-Salamah, A.A.; Almaary, K.S.; Elbadawi, Y.B. Prevalence, Serotyping and Antimicrobials Resistance Mechanism of Salmonella enterica Isolated from Clinical and Environmental Samples in Saudi Arabia. Braz. J. Microbiol. 2017, 48, 499–508. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).