Cancer Stem Cells and Glioblastoma: Time for Innovative Biomarkers of Radio-Resistance?
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Cancer Stem Cells
1.2. Radiobiology of CSCs
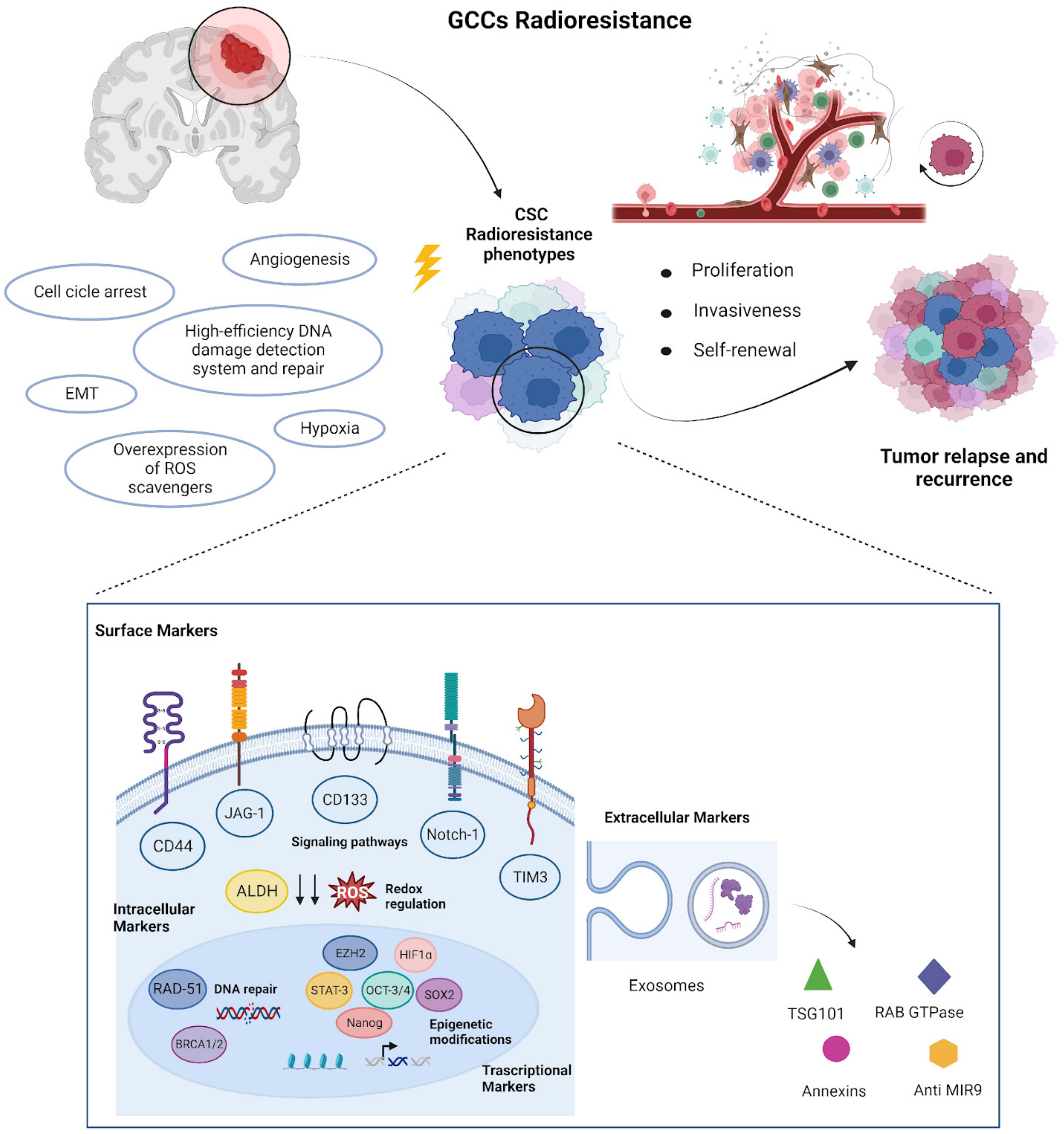
1.3. Biomarkers Related to CSCs Radioresistance
1.3.1. Surface Markers
- CD 133, also known as prominin-1, is a glycoprotein with five transmembrane regions first discovered as a hemopoietic cell biomarker. Chemoresistance has also been associated with the presence of CD133+ CSCs in oral cancer, lung cancer, and glioma (GB) (Table 1). Angelastro et al. propose that CD133 might contribute to the observed apoptosis resistance of CD133+ cancer progenitor cells. They demonstrate that ectopic overexpression of CD133 in rat C6 glioma cells results in significant resistance to camptothecin- and doxorubicin-induced apoptosis. Despite the fact that p53 was upregulated in CD133-overexpressing glioma cells treated with DNA-damaging agents, apoptosis appeared to be independent of p53. Tamura et al. obtained tumor samples from both the primary and secondary surgery of glioma 31 patients treated with postoperative RTCT [66]. The mean percentage of CD133-positive glioma cells in sections obtained during recurrence was 12.2% ± 10.3%, which was considerably higher than the percentage obtained during the initial surgery (1.08% ± 1.75%). The findings of the authors indicate that CD133-positive glioma stem cells can survive radiotherapy and chemotherapy, acquiring a proliferative cancer stem cell phenotype. This causes recurrence in de novo glioblastoma cases. Park et al. (2021) demonstrated that CD133 induces the PI3K/AKT-dependent activation of nuclear factor erythroid 2-related factor 2 (NRF2), an important transcription factor that protects the cell from ROS [92]. High NRF2 levels in spheroid-cultured HCT116 cells and the CD133-high subpopulation contributed to aggressive CSC phenotypes, such as anticancer radioresistance, sphere formation, anchorage-independent colony formation, and migration capability. Therefore, the NRF2 axis may be a promising target for inhibiting therapeutic radioresistance and enhancing survival capacity under stressful conditions in CD133-high CSCs.
- The cell surface glycoprotein CD44 is involved in cell adhesion, migration, and interaction [94]. CD44 plays a major role in both tumor neoangiogenesis and progression because of its affinity for messengers, such as growth hormones present in the tumor microenvironment as well as extracellular matrix elements including hyaluronan (HA) and osteopontin (OPN) [95]. According to a growing body of research, HA-CD44 interaction in the extracellular domain activates a number of signaling pathways, such as receptor tyrosine kinases (ErbB2 and EGFR) and transforming growth factor-receptor type 1 (TGF-R1) [96,97]. The study by Si et al. involving 62 patients with GB indicates that high CD44 expression is an indicator of a poor prognosis for GB patients. The median survival periods for those with high and low CD44 expressions were 3.5 and 18.5 months, respectively [67]. Liu et al. discovered, by analyzing primary cell lines obtained from GB patients, that CD44 is more abundantly present in radioresistant cells and acts a crucial role in stemness, cell proliferation, and angiogenesis. In fact, CD44+ cells expressed higher levels of ATM, Rad51, and CHK2 kinase phosphorilation compared to CD44-. These proteins are part of DNA damage response signaling pathway that is activated in response to cellular radiation damage [98].
- TIM-3 (T-cell immunoglobulin mucin-3) is a type 1 cell-surface glycoprotein known to be expressed on the surface of leukemic stem cells and in more than 70% of patients with GB [68]. Through interaction with its ligand Galectin-9, TIM-3 causes aberrant catenin accumulation and constitutive activation of the canonical Wnt pathway. This phenomenon permits the maintenance and enhancement of cancer stemness [47]. Zhang et al. examined Tim-3 expression and MGMT promoter methylation in 84 GBs [68]. Therein, 62 patients out of 84 (73.81%) demonstrated mesenchymal Tim-3 expression in GB tissues, which was classified as low 15.48% (13/84), moderate 7.14% (6/84), or strong 51.14% (4/84) expression. The tumors of 48 patients tested positive for MGMT promoter methylation, while the tumors of 36 individuals tested negative. Tim-3 expression and MGMT promoter methylation status were found to be an independent risk factor for survival in GB patients. Strong expression of Tim-3 in conjunction with an unmethylated MGMT promoter was substantially linked with shorter OS in each of the four categories (p 0.05). Patients with no or low Tim-3 expression experienced a median survival of 16.9 and 16.4 months, respectively, but those with high Tim-3 expression and MGMT promoter nonmethylation had a median survival of 7.6 months. The average survival time for patients with low Tim-3 expression and methylation of the MGMT promoter was 21.8 months.
1.3.2. Intracellular Markers
- The Notch pathway controls cell division, differentiation, proliferation, and death and plays a fundamental role in central nervous system development. Protein convertases at site 1 (S1) cleave Notch receptors after they are synthesized, controlling their trafficking and signaling function. The intensity and timing of Notch activity are regulated by posttranslational changes of the receptors and ligands to generate context-specific signals [69]. The expression Pattern of Notch Signaling in Glioblastoma has been widely studied in the recent years. Wang et al. demonstrated that the synthesis of SOX2 induced NOTCH1 expression with consequent increasing of the GSCs’ invasiveness, making it extremely difficult for radiotherapists to cover effectively the radiotherapy target [102]. On the other hand, inhibition of Notch signaling reduced GSCs’ affinity towards white matter tropism. Han et al. analyzed Notch1 using immunohistochemistry in 69 glioma tissue specimens and 8 normal brain tissue specimens [103]. Multivariate analysis revealed that Notch1 expression was an independent adverse prognostic factor for survival. The effect of NOTCH1 down regulation, investigated on two glioma cell lines (U87MG and U251), was correlated with the reduction of GB proliferation. Moreover, Notch1 downregulation affected clonogenicity of GB cells and increased the number of γH2AX foci at 30 min and 24 h after irradiation at the dose of 8 Gy enhancing radiosensitivity. Notch1 inhibition also reduced angiogenesis, VEGF, and the hypoxic response RT.
- There is evidence that JAG-1 is implicated in radioresistance. KIM et al. irradiated JAG1-depleted LN18 cells with 3 and 5 Gy. The results showed colony survival was significantly reduced compared to LN18 cells that normally expressed JAG1 [104]. Another interesting protein in the NOTCH pathway is Jagged1 (JAG1) that is able to promote glioma-initiating cells (GICs) in HGG. Several studies demonstrated that JAG1 staining was strongly expressed only in glioma tissue. Meanwhile, no evidence in nearby normal brain tissue were found [105,106]. Hai et al. observed that high expression of Jagged1 was correlated with poor prognosis [6]. It was also evidenced in vitro that Jagged1 improved the invasiveness of glioma cells through the stimulation of the Nf-kb pathway. This study also demonstrated in vivo that the suppression of Jagged 1 inhibited the tumorogenis of Glioma cells improving the OS of mice compared to the control group. According Jubb et al., high JAG1 expression is also associated with type I microvascular pattern (MVP) that is notoriously associated with poor PFS and OS [107,108]. This finding was later confirmed by X.X Qui et al., who reported the Jagged1 expression in tumor and endothelial cells (EC) was correlated at the multivariate analysis with shortened time to progression (p < 0.001 for TC, p < 0.001 for TC) and OS (p < 0.001 for TC, p < 0.001 for TC). Lastly, there is evidence that JAG1 is also implicated in radioresistance. Kim et al. irradiated JAG1-depleted LN18 cells with 3 and 5 Gy. The results showed colony survival was significantly reduced compared to LN18 cells that normally expressed JAG1 [104].
- Aldehyde dehydrogenase (ALDH) is a class of NAD(P)+-dependent enzymes that play an important role in the detoxification process catalyzing the oxidation of aldehyde substrates [109]. ALDH is implicated in several cellular pathways which contribute to cancer cells’ radio- and chemoresistance; for example, it is involved in retinoid as well as β-Catenin/Tcf signaling pathways, which have been related to the stemness of CSCs [110]. Furthermore, high ALDH activity is correlated with low ROS cellular concentration, suggesting a strong antioxidant activity [109]. In HGG glioblastoma CSCs (GCC), ALDH expression is correlated with expression of mesenchymal phenotype (MES) and radio-chemoresistance. Conversely, ALDH- GCC was correlated with a pro-neural phenotype (PN) and a better prognosis. Mao et al. analyzed the 40 specimens collected from high-grade glioma patients. Transcriptome array analyses showed that ALDH genes were the most significantly expressed with an higher glycolytic activity in Mes GSCs (p = 0.000315), compared with PN GSCs (p < 0.01) [85]. ALDH expression was also more expressed in HGG specimens compared to low-grade glioma or normal brain tissue. Moreover, this study showed that radiotherapy induces transition of PN GSCs into a Mes-like GSC phenotype (PMT) that is highly resistant to radiation treatment. At the same time, the inhibition of ALDH attenuates the transformation in the radiation-resistant phenotype of Mes GSCs. Another study analyzed 30 surgical specimens (n = 30) collected from adult patients with histopathologically confirmed diagnosis of GB. High ALDH mRNA expression was associated with the poorer OS (p < 0.01, HR = 3.170, 95% CI: 1.328–7.566) and higher grade of peritumoral edema compared to the low expression group. In addition, Wang et al. demonstrated that ALDH3B1 and ALDH16A1 affect proliferation and migration of HGG cells by inducing cell-cycle arrest and the epithelial–mesenchymal transition [86].
- Research into DNA repair pathways, particularly involving RAD51 and BRCA1/2, has shed light on their critical roles in glioblastoma. These pathways are central to maintaining genome stability and influencing the behavior of CSCs. A pivotal investigation conducted by Balbous et al. assessed the expression of RAD51 within glioblastoma stem-like cells and its consequential association with resistance to radiation [87]. This underscored the pronounced role of RAD51 in the realm of treatment resistance. Furthermore, RAD51 holds the potential for involvement in the perpetuation of glioblastoma stem-like cells, entities speculated to underlie tumor growth, recurrence, and resistance to therapeutic regimens. The prospective targeting of RAD51 could potentially disrupt the processes of self-renewal and survival within these stem-like cells.
- STAT3 (Signal transducer and activator of transcription 3) has been reported to be permanently activated in a variety of tumors, including GB, resulting in raised radio-resistance [82]. Masliantsev et al. demonstrated in 2018 that inhibiting STAT3 prior to cell irradiation reduced the surviving fraction of CSCs, implying that this technique could amplify radiation effects [83]. In addition, they used clinical specimens to evaluate STAT3 activation status in 61 GB patients, discovering a preferential phosphorylation of STAT3 on Serine727 (pS727). Furthermore, the investigators discovered that pS727 was linked to a significantly worse overall patient survival and was free of progression time. Taken together, these findings imply that pS727-STAT3 could be a prognosis marker as well as a therapeutic target for sensitizing highly radioresistant GSCs. Sherry et al. discovered in 2009 that treating GB-SC with two chemically separate small molecule STAT3 DNA-binding inhibitors reduces cell growth and the generation of new neurospheres from single cells [84]. STAT3 governs the proliferation and regeneration of GB CSCs, suggesting that it could be a viable target.
1.3.3. Transcription Factor as Biomarker
- Transcriptional regulator Sox2 (SRY-Box Transcription Factor 2) is responsible for the maintenance of an undifferentiated cellular phenotype [111]. SOX 2 is also linked to resistance to anti-tumor therapy through SOX2-mediated activation of ABC transporters, which can efflux drugs across the cell membrane [112]. According to an in vitro study conducted by Wang et al., Sox2 induced the dedifferentiation of differentiated glioma cells cultured in 1% of O2 [73]. Other studies also showed that SOx2 is implicated in white matter GSC tropism and also in the development of temozolomide resistance [113].
- The HIF-1 (Hypoxia-Inducible Factor 1-alpha) pathway plays an important and nuanced function in glioblastoma, particularly in response to the neoplasm’s hypoxic condition [114]. Under hypoxia, HIF-1 can be persistently expressed and works as an essential molecule in regulating the production of CSCs; however, the exact method is still unknown [114]. HIF-1 has been linked to the development of CSC markers such as OCT4, SOX2, NANOG, and KrÃ1⁄4ppel-like factor 4 (KLF4) [70,71,115]. Furthermore, suppressing HIF-1 can slow tumor progression by reducing the production of CSC biomarkers. For instance, HIF-1 has been found to attach directly to the CD47 promoter, facilitating gene transcription, assisting in the avoidance of macrophage phagocytosis, and maintaining the stem phenotype of breast CSCs [71]. Activation of HIF-1 in response to a hypoxic condition activates several genes that may increase the radioresistance of irradiated tumors (reducing the response to treatment) and is a negative prognostic factor [72].
- EZH2 (Enhancer of zeste 2 polycomb repressive complex 2 subunit) is a part of a multimeric proteic complex called PRC2 consisting of three other subunits termed EED, SUZ12, and RbAp46/4. EZH2 has also been implicated in radio-resistance. Kim et al. assessed the impact of ionizing radiation on three glioma sphere samples (GB83, GB1123, and GB528) and detected a significant increase in both mRNA expression and protein levels of the EZH2/MELK–FOXM1 axis [76]. The effect of radiation on mice xenografted with spheres of GB with EZH2/MELK–FOXM1 axis genes silenced. The mean OS was longer in these mice compared to that of the control animals with activated EZH2/MELK–FOXM1 axis genes. These data demonstrated that EZH2/MELK–FOXM1 axis protein upregulation could promote tumorigenesis in vivo models (162). Wang et al. also evidenced the role of EZH2 in radioresistance; the expression of NEK2 expression, a protein that protectsprotect EZH2 from ubiquitin degradation in GCSc, can induce radioresistence in animal models [116]. EZH2 catalyzes the trimethylation of H3K27 which is associated with transcriptional repression and heterochromatin formation. This enzyme has been associated also with poor prognosis in HGG. A preclinical study demonstrated that EZH2 expression is significantly upregulated in the U87 and U251 glioma cells compared to HA-1800 human astrocytes. In glioma tissues EZH2 expression is grade dependent. In fact, higher expression levels have been detected in GB cells. Finally, the analysis of the Chinese Glioma Genome Atlas (CGGA) data set revealed that patients in the high-EZH2 group had a worse prognosis compared to those in the low-EZH2 group [117]. A possible role of E2F7−EZH2 axis on AKT/mTOR activation through PTEN suppression emerged from in vitro and in vivo experimental models. Another study demonstrated that EZH2 promotes M2 macrophage polarization in HGG resulting in macrophage-dependent disease development [118]. EZH2 also enhances the surface NKG2D ligands suppression on NK cells thus preventing immune response against GCSc [74]. Furthermore, EZH2 enhances chemoresistance to TMZ through stabilization of PARP1 protein [119].
1.3.4. Extracellular Biomarkers
2. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grochans, S.; Cybulska, A.M.; Simińska, D.; Korbecki, J.; Kojder, K.; Chlubek, D.; Baranowska-Bosiacka, I. Epidemiology of Glioblastoma Multiforme-Literature Review. Cancers 2022, 14, 2412. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; van den Bent, M.; Preusser, M.; Le Rhun, E.; Tonn, J.C.; Minniti, G.; Bendszus, M.; Balana, C.; Chinot, O.; Dirven, L.; et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat. Rev. Clin. Oncol. 2021, 18, 170–186. [Google Scholar] [CrossRef] [PubMed]
- Vanderbeek, A.M.; Rahman, R.; Fell, G.; Ventz, S.; Chen, T.; Redd, R.; Parmigiani, G.; Cloughesy, T.F.; Wen, P.Y.; Trippa, L.; et al. The clinical trials landscape for glioblastoma: Is it adequate to develop new treatments? Neuro Oncol. 2018, 20, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Le Rhun, E.; Preusser, M.; Roth, P.; Reardon, D.A.; van den Bent, M.; Wen, P.; Reifenberger, G.; Weller, M. Molecular targeted therapy of glioblastoma. Cancer Treat. Rev. 2019, 80, 101896. [Google Scholar] [CrossRef] [PubMed]
- Piccirillo, S.G.M.; Alonso, M.M.; Pasqualetti, F. Basic and Translational Advances in Glioblastoma. Biomed Res. Int. 2018, 2018, 1820345. [Google Scholar] [CrossRef] [PubMed]
- Pasqualetti, F.; Barberis, A.; Zanotti, S.; Montemurro, N.; De Salvo, G.L.; Soffietti, R.; Mazzanti, C.M.; Ius, T.; Caffo, M.; Paiar, F.; et al. The Impact of Survivorship Bias in Glioblastoma Research. Crit. Rev. Oncol. Hematol. 2023, 104065. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.L.V.; Gomes, I.N.F.; Carloni, A.C.; Rosa, M.N.; da Silva, L.S.; Evangelista, A.F.; Reis, R.M.; Silva, V.A.O. Role of glioblastoma stem cells in cancer therapeutic resistance: A perspective on antineoplastic agents from natural sources and chemical derivatives. Stem Cell Res. Ther. 2021, 12, 206. [Google Scholar] [CrossRef]
- Agosti, E.; Panciani, P.P.; Zeppieri, M.; De Maria, L.; Pasqualetti, F.; Tel, A.; Zanin, L.; Fontanella, M.M.; Ius, T. Tumor Microenvironment and Glioblastoma Cell Interplay as Promoters of Therapeutic Resistance. Biology 2023, 12, 736. [Google Scholar] [CrossRef]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Kemper, K.; Prasetyanti, P.R.; De Lau, W.; Rodermond, H.; Clevers, H.; Medema, J.P. Monoclonal antibodies against Lgr5 identify human colorectal cancer stem cells. Stem Cells 2012, 30, 2378–2386. [Google Scholar] [CrossRef]
- Lee, T.K.; Guan, X.Y.; Ma, S. Cancer stem cells in hepatocellular carcinoma-from origin to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 26–44. [Google Scholar] [CrossRef] [PubMed]
- Eramo, A.; Lotti, F.; Sette, G.; Pilozzi, E.; Biffoni, M.; Di Virgilio, A.; Conticello, C.; Ruco, L.; Peschle, C.; De Maria, R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008, 15, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Bruschini, S.; Ciliberto, G.; Mancini, R. The emerging role of cancer cell plasticity and cell-cycle quiescence in immune escape. Cell Death Dis. 2020, 11, 471. [Google Scholar] [CrossRef] [PubMed]
- Nayak, A.; Warrier, N.M.; Kumar, P. Cancer Stem Cells and the Tumor Microenvironment: Targeting the Critical Crosstalk through Nanocarrier Systems. Stem Cell Rev. Rep. 2022, 18, 2209–2233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Lang, F.; Yang, C. IDH mutation and cancer stem cell. Essays Biochem. 2022, 66, 413–422. [Google Scholar] [CrossRef]
- Yao, Q.; Cai, G.; Yu, Q.; Shen, J.; Gu, Z.; Chen, J.; Shi, W.; Shi, J. IDH1 mutation diminishes aggressive phenotype in glioma stem cells. Int. J. Oncol. 2018, 52, 270–278. [Google Scholar] [CrossRef]
- Harland, A.; Liu, X.; Ghirardello, M.; Galan, M.C.; Perks, C.M.; Kurian, K.M. Glioma Stem-Like Cells and Metabolism: Potential for Novel Therapeutic Strategies. Front. Oncol. 2021, 11, 743814. [Google Scholar] [CrossRef]
- Ma, C.; Nguyen, H.P.T.; Jones, J.J.; Stylli, S.S.; Whitehead, C.A.; Paradiso, L.; Luwor, R.B.; Areeb, Z.; Hanssen, E.; Cho, E.; et al. Extracellular Vesicles Secreted by Glioma Stem Cells Are Involved in Radiation Resistance and Glioma Progression. Int. J. Mol. Sci. 2022, 23, 2770. [Google Scholar] [CrossRef] [PubMed]
- Prager, B.C.; Xie, Q.; Bao, S.; Rich, J.N. Cancer Stem Cells: The Architects of the Tumor Ecosystem. Cell Stem Cell 2019, 24, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Wang, Z.; Liu, H.; Pang, Z. Cytokine-mediated crosstalk between cancer stem cells and their inflammatory niche from the colorectal precancerous adenoma stage to the cancerous stage: Mechanisms and clinical implications. Front. Immunol. 2022, 13, 1057181. [Google Scholar] [CrossRef] [PubMed]
- Wei, H. Interleukin 6 signaling maintains the stem-like properties of bladder cancer stem cells. Transl. Cancer Res. 2019, 8, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Radharani, N.N.V.; Yadav, A.S.; Nimma, R.; Kumar, T.V.S.; Bulbule, A.; Chanukuppa, V.; Kumar, D.; Patnaik, S.; Rapole, S.; Kundu, G.C. Tumor-associated macrophage derived IL-6 enriches cancer stem cell population and promotes breast tumor progression via Stat-3 pathway. Cancer Cell Int. 2022, 22, 122. [Google Scholar] [CrossRef] [PubMed]
- Futakuchi, M.; Lami, K.; Tachibana, Y.; Yamamoto, Y.; Furukawa, M.; Fukuoka, J. The Effects of TGF-β Signaling on Cancer Cells and Cancer Stem Cells in the Bone Microenvironment. Int. J. Mol. Sci. 2019, 20, 5117. [Google Scholar] [CrossRef] [PubMed]
- López de Andrés, J.; Griñán-Lisón, C.; Jiménez, G.; Marchal, J.A. Cancer stem cell secretome in the tumor microenvironment: A key point for an effective personalized cancer treatment. J. Hematol. Oncol. 2020, 13, 136. [Google Scholar] [CrossRef]
- Bao, B.; Ahmad, A.; Azmi, A.S.; Ali, S.; Sarkar, F.H. Overview of cancer stem cells (CSCs) and mechanisms of their regulation: Implications for cancer therapy. Curr. Protoc. Pharmacol. 2013, 61, 14. [Google Scholar] [CrossRef]
- Schulz, A.; Meyer, F.; Dubrovska, A.; Borgmann, K. Cancer Stem Cells and Radioresistance: DNA Repair and Beyond. Cancers 2019, 11, 862. [Google Scholar] [CrossRef]
- Aker, M.; Ganeshan, B.; Afaq, A.; Wan, S.; Groves, A.M.; Arulampalam, T. Magnetic Resonance Texture Analysis in Identifying Complete Pathological Response to Neoadjuvant Treatment in Locally Advanced Rectal Cancer. Dis. Colon Rectum 2019, 62, 163–170. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat. Rev. 2018, 69, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, E.N.; Scaffidi, P. Epigenetics and Cancer Stem Cells: Unleashing, Hijacking, and Restricting Cellular Plasticity. Trends Cancer 2017, 3, 372–386. [Google Scholar] [CrossRef] [PubMed]
- Marusyk, A.; Janiszewska, M.; Polyak, K. Intratumor Heterogeneity: The Rosetta Stone of Therapy Resistance. Cancer Cell 2020, 37, 471–484. [Google Scholar] [CrossRef]
- Pasqualetti, F.; Rizzo, M.; Franceschi, S.; Lessi, F.; Paiar, F.; Buffa, F.M. New perspectives in liquid biopsy for glioma patients. Curr. Opin. Oncol. 2022, 34, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Holley, A.K.; Miao, L.; St Clair, D.K.; St Clair, W.H. Redox-modulated phenomena and radiation therapy: The central role of superoxide dismutases. Antioxid. Redox Signal. 2014, 20, 1567–1589. [Google Scholar] [CrossRef] [PubMed]
- Mladenov, E.; Magin, S.; Soni, A.; Iliakis, G. DNA double-strand break repair as determinant of cellular radiosensitivity to killing and target in radiation therapy. Front. Oncol. 2013, 3, 113. [Google Scholar] [CrossRef]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef]
- Begg, A.C.; Stewart, F.A.; Vens, C. Strategies to improve radiotherapy with targeted drugs. Nat. Rev. Cancer 2011, 11, 239–253. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef]
- Phillips, T.M.; McBride, W.H.; Pajonk, F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J. Natl. Cancer Inst. 2006, 98, 1777–1785. [Google Scholar] [CrossRef] [PubMed]
- Woodward, W.A.; Chen, M.S.; Behbod, F.; Alfaro, M.P.; Buchholz, T.A.; Rosen, J.M. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc. Natl. Acad. Sci. USA 2007, 104, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Daguenet, E.; Khalifa, J.; Tolédano, A.; Borchiellini, D.; Pointreau, Y.; Rodriguez-Lafrasse, C.; Chargari, C.; Magné, N. To exploit the 5 ‘R’ of radiobiology and unleash the 3 ‘E’ of immunoediting: ‘RE’-inventing the radiotherapy-immunotherapy combination. Ther. Adv. Med. Oncol. 2020, 12, 1758835920913445. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J.; Ganesh, K. Metastasis-Initiating Cells and Ecosystems. Cancer Discov. 2021, 11, 971–994. [Google Scholar] [CrossRef]
- Lu, W.; Kang, Y. Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev. Cell 2019, 49, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Malik, B.; Nie, D. Cancer stem cells and resistance to chemo and radio therapy. Front. Biosci. 2012, 4, 2142–2149. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, H.; Shiota, G. Immune evasion by cancer stem cells. Regen. Ther. 2021, 17, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Al-Assar, O.; Mantoni, T.; Lunardi, S.; Kingham, G.; Helleday, T.; Brunner, T.B. Breast cancer stem-like cells show dominant homologous recombination due to a larger S-G2 fraction. Cancer Biol. Ther. 2011, 11, 1028–1035. [Google Scholar] [CrossRef][Green Version]
- Abdullah, L.N.; Chow, E.K. Mechanisms of chemoresistance in cancer stem cells. Clin. Transl. Med. 2013, 2, 3. [Google Scholar] [CrossRef]
- Yang, Z.X.; Sun, Y.H.; He, J.G.; Cao, H.; Jiang, G.Q. Increased activity of CHK enhances the radioresistance of MCF-7 breast cancer stem cells. Oncol. Lett. 2015, 10, 3443–3449. [Google Scholar] [CrossRef]
- Yin, H.; Glass, J. The phenotypic radiation resistance of CD44+/CD24(-or low) breast cancer cells is mediated through the enhanced activation of ATM signaling. PLoS ONE 2011, 6, e24080. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Wu, S.P.; Liu, J.B.; Shi, Y.S.; Huang, X.; Zhang, Q.B.; Yao, K.T. MYC regulation of CHK1 and CHK2 promotes radioresistance in a stem cell-like population of nasopharyngeal carcinoma cells. Cancer Res. 2013, 73, 1219–1231. [Google Scholar] [CrossRef] [PubMed]
- Carruthers, R.; Ahmed, S.U.; Strathdee, K.; Gomez-Roman, N.; Amoah-Buahin, E.; Watts, C.; Chalmers, A.J. Abrogation of radioresistance in glioblastoma stem-like cells by inhibition of ATM kinase. Mol. Oncol. 2015, 9, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ma, Z.; Xiao, Z.; Liu, H.; Dou, Z.; Feng, X.; Shi, H. Chk1 knockdown confers radiosensitization in prostate cancer stem cells. Oncol. Rep. 2012, 28, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Rycaj, K.; Tang, D.G. Cancer stem cells and radioresistance. Int. J. Radiat. Biol. 2014, 90, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair. 2008, 7, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Moore, N.; Lyle, S. Quiescent, slow-cycling stem cell populations in cancer: A review of the evidence and discussion of significance. J. Oncol. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Giancotti, F.G. Mechanisms governing metastatic dormancy and reactivation. Cell 2013, 155, 750–764. [Google Scholar] [CrossRef]
- Diehn, M.; Cho, R.W.; Clarke, M.F. Therapeutic implications of the cancer stem cell hypothesis. Semin. Radiat. Oncol. 2009, 19, 78–86. [Google Scholar] [CrossRef]
- Lagadec, C.; Vlashi, E.; Della Donna, L.; Meng, Y.; Dekmezian, C.; Kim, K.; Pajonk, F. Survival and self-renewing capacity of breast cancer initiating cells during fractionated radiation treatment. Breast Cancer Res. 2010, 12, R13. [Google Scholar] [CrossRef]
- Konge, J.; Leteurtre, F.; Goislard, M.; Biard, D.; Morel-Altmeyer, S.; Vaurijoux, A.; Gruel, G.; Chevillard, S.; Lebeau, J. Breast cancer stem cell-like cells generated during TGFβ-induced EMT are radioresistant. Oncotarget 2018, 9, 23519–23531. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Blum, W.; Zhu, C.Q.; Yun, Z.; Pecze, L.; Kohno, M.; Chan, M.L.; Zhao, Y.; Felley-Bosco, E.; Schwaller, B.; et al. Putative cancer stem cells may be the key target to inhibit cancer cell repopulation between the intervals of chemoradiation in murine mesothelioma. BMC Cancer 2018, 18, 471. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Graham, P.; Hao, J.; Ni, J.; Deng, J.; Bucci, J.; Malouf, D.; Gillatt, D.; Li, Y. Cancer stem cells and signaling pathways in radioresistance. Oncotarget 2016, 7, 11002–11017. [Google Scholar] [CrossRef] [PubMed]
- Najafi, M.; Mortezaee, K.; Majidpoor, J. Cancer stem cell (CSC) resistance drivers. Life Sci. 2019, 234, 116781. [Google Scholar] [CrossRef] [PubMed]
- Diehn, M.; Cho, R.W.; Lobo, N.A.; Kalisky, T.; Dorie, M.J.; Kulp, A.N.; Qian, D.; Lam, J.S.; Ailles, L.E.; Wong, M.; et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 2009, 458, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Aoyagi, M.; Ando, N.; Ogishima, T.; Wakimoto, H.; Yamamoto, M.; Ohno, K. Expansion of CD133-positive glioma cells in recurrent de novo glioblastomas after radiotherapy and chemotherapy. J. Neurosurg. 2013, 119, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
- Si, D.; Yin, F.; Peng, J.; Zhang, G. High Expression of CD44 Predicts a Poor Prognosis in Glioblastomas. Cancer Manag. Res. 2020, 12, 769–775. [Google Scholar] [CrossRef]
- Zhang, J.; Sai, K.; Wang, X.L.; Ye, S.Q.; Liang, L.J.; Zhou, Y.; Chen, Z.J.; Hu, W.M.; Liu, J.M. Tim-3 Expression and MGMT Methylation Status Association With Survival in Glioblastoma. Front. Pharmacol. 2020, 11, 584652. [Google Scholar] [CrossRef]
- Kopan, R. Notch signaling. Cold Spring Harb. Perspect. Biol. 2012, 4, a011213. [Google Scholar] [CrossRef]
- Lin, Y.T.; Wu, K.J. Epigenetic regulation of epithelial-mesenchymal transition: Focusing on hypoxia and TGF-β signaling. J. Biomed. Sci. 2020, 27, 39. [Google Scholar] [CrossRef]
- Zhang, Q.; Han, Z.; Zhu, Y.; Chen, J.; Li, W. Role of hypoxia inducible factor-1 in cancer stem cells (Review). Mol. Med. Rep. 2021, 23, 1. [Google Scholar] [CrossRef] [PubMed]
- Kabakov, A.E.; Yakimova, A.O. Hypoxia-Induced Cancer Cell Responses Driving Radioresistance of Hypoxic Tumors: Approaches to Targeting and Radiosensitizing. Cancers 2021, 13, 1102. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Gong, S.; Liao, B.; Pan, J.; Wang, J.; Zou, D.; Zhao, L.; Xiong, S.; Deng, Y.; Yan, Q.; et al. HIF1α/HIF2α induces glioma cell dedifferentiation into cancer stem cells through Sox2 under hypoxic conditions. J. Cancer 2022, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Yang, X.; Chen, J.; He, K.; Gao, X.; Wu, X.; Zhang, M.; Zhou, H.; Xiao, F.; An, L.; et al. Circular EZH2-encoded EZH2-92aa mediates immune evasion in glioblastoma via inhibition of surface NKG2D ligands. Nat. Commun. 2022, 13, 4795. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Takahashi, H.; Inoue, A.; Harada, H.; Toshimori, S.; Kobayashi, Y.; Goto, K.; Sugimoto, K.; Yano, H.; Ohnishi, T.; et al. Oct-3/4 promotes migration and invasion of glioblastoma cells. J. Cell. Biochem. 2012, 113, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Joshi, K.; Ezhilarasan, R.; Myers, T.R.; Siu, J.; Gu, C.; Nakano-Okuno, M.; Taylor, D.; Minata, M.; Sulman, E.P.; et al. EZH2 protects glioma stem cells from radiation-induced cell death in a MELK/FOXM1-dependent manner. Stem Cell Rep. 2015, 4, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Tuncay Cagatay, S.; Mayah, A.; Mancuso, M.; Giardullo, P.; Pazzaglia, S.; Saran, A.; Daniel, A.; Traynor, D.; Meade, A.D.; Lyng, F.; et al. Phenotypic and Functional Characteristics of Exosomes Derived from Irradiated Mouse Organs and Their Role in the Mechanisms Driving Non-Targeted Effects. Int. J. Mol. Sci. 2020, 21, 8389. [Google Scholar] [CrossRef]
- Kaur, B.; Khwaja, F.W.; Severson, E.A.; Matheny, S.L.; Brat, D.J.; Van Meir, E.G. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro Oncol. 2005, 7, 134–153. [Google Scholar] [CrossRef]
- Courtnay, R.; Ngo, D.C.; Malik, N.; Ververis, K.; Tortorella, S.M.; Karagiannis, T.C. Cancer metabolism and the Warburg effect: The role of HIF-1 and PI3K. Mol. Biol. Rep. 2015, 42, 841–851. [Google Scholar] [CrossRef]
- Méndez, O.; Zavadil, J.; Esencay, M.; Lukyanov, Y.; Santovasi, D.; Wang, S.C.; Newcomb, E.W.; Zagzag, D. Knock down of HIF-1alpha in glioma cells reduces migration in vitro and invasion in vivo and impairs their ability to form tumor spheres. Mol. Cancer 2010, 9, 133. [Google Scholar] [CrossRef]
- Gabriely, G.; Wheeler, M.A.; Takenaka, M.C.; Quintana, F.J. Role of AHR and HIF-1α in Glioblastoma Metabolism. Trends Endocrinol. Metab. 2017, 28, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Hou, X.; Dong, L.; Hou, W. Roles of STAT3 in the pathogenesis and treatment of glioblastoma. Front. Cell Dev. Biol. 2023, 11, 1098482. [Google Scholar] [CrossRef] [PubMed]
- Masliantsev, K.; Pinel, B.; Balbous, A.; Guichet, P.O.; Tachon, G.; Milin, S.; Godet, J.; Duchesne, M.; Berger, A.; Petropoulos, C.; et al. Impact of STAT3 phosphorylation in glioblastoma stem cells radiosensitization and patient outcome. Oncotarget 2018, 9, 3968–3979. [Google Scholar] [CrossRef] [PubMed]
- Sherry, M.M.; Reeves, A.; Wu, J.K.; Cochran, B.H. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells 2009, 27, 2383–2392. [Google Scholar] [CrossRef] [PubMed]
- Mao, P.; Joshi, K.; Li, J.; Kim, S.H.; Li, P.; Santana-Santos, L.; Luthra, S.; Chandran, U.R.; Benos, P.V.; Smith, L.; et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc. Natl. Acad. Sci. USA 2013, 110, 8644–8649. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Mo, Y.; Tan, Y.; Wen, Z.; Dai, Z.; Zhang, H.; Zhang, X.; Feng, S.; Liang, X.; Song, T.; et al. The ALDH Family Contributes to Immunocyte Infiltration, Proliferation and Epithelial-Mesenchymal Transformation in Glioma. Front. Immunol. 2021, 12, 756606. [Google Scholar] [CrossRef] [PubMed]
- Balbous, A.; Cortes, U.; Guilloteau, K.; Rivet, P.; Pinel, B.; Duchesne, M.; Godet, J.; Boissonnade, O.; Wager, M.; Bensadoun, R.J.; et al. A radiosensitizing effect of RAD51 inhibition in glioblastoma stem-like cells. BMC Cancer 2016, 16, 604. [Google Scholar] [CrossRef] [PubMed]
- Bernal, A.; Arranz, L. Nestin-expressing progenitor cells: Function, identity and therapeutic implications. Cell. Mol. Life Sci. 2018, 75, 2177–2195. [Google Scholar] [CrossRef]
- Ishiwata, T.; Teduka, K.; Yamamoto, T.; Kawahara, K.; Matsuda, Y.; Naito, Z. Neuroepithelial stem cell marker nestin regulates the migration, invasion and growth of human gliomas. Oncol. Rep. 2011, 26, 91–99. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Yu, T.S.; McKay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012, 488, 522–526. [Google Scholar] [CrossRef]
- Guadagno, E.; Borrelli, G.; Califano, M.; Calì, G.; Solari, D.; Del Basso De Caro, M. Immunohistochemical expression of stem cell markers CD44 and nestin in glioblastomas: Evaluation of their prognostic significance. Pathol. Res. Pract. 2016, 212, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, S.K.; Hallis, S.P.; Choi, B.H.; Kwak, M.K. Role of CD133/NRF2 Axis in the Development of Colon Cancer Stem Cell-Like Properties. Front. Oncol. 2021, 11, 808300. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.G.; Xue, X.Y.; Wang, L.; Zhang, X.; Yan, M.; Tu, Y.Y.; Lin, W.; Jiang, X.F.; Ren, H.G.; Zhang, W.; et al. CDH5 is specifically activated in glioblastoma stemlike cells and contributes to vasculogenic mimicry induced by hypoxia. Neuro Oncol. 2013, 15, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Thapa, R.; Wilson, G.D. The Importance of CD44 as a Stem Cell Biomarker and Therapeutic Target in Cancer. Stem Cells Int. 2016, 2016, 2087204. [Google Scholar] [CrossRef] [PubMed]
- Morath, I.; Hartmann, T.N.; Orian-Rousseau, V. CD44: More than a mere stem cell marker. Int. J. Biochem. Cell Biol. 2016, 81, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.H.; Lin, J.C.; Chou, Y.C.; Li, M.H.; Tsai, J.T. CD44-associated radioresistance of glioblastoma in irradiated brain areas with optimal tumor coverage. Cancer Med. 2020, 9, 350–360. [Google Scholar] [CrossRef]
- Zhang, M.; Song, T.; Yang, L.; Chen, R.; Wu, L.; Yang, Z.; Fang, J. Nestin and CD133: Valuable stem cell-specific markers for determining clinical outcome of glioma patients. J. Exp. Clin. Cancer Res. 2008, 27, 85. [Google Scholar] [CrossRef]
- Calabrese, C.; Poppleton, H.; Kocak, M.; Hogg, T.L.; Fuller, C.; Hamner, B.; Oh, E.Y.; Gaber, M.W.; Finklestein, D.; Allen, M.; et al. A perivascular niche for brain tumor stem cells. Cancer Cell 2007, 11, 69–82. [Google Scholar] [CrossRef]
- Maderna, E.; Salmaggi, A.; Calatozzolo, C.; Limido, L.; Pollo, B. Nestin, PDGFRbeta, CXCL12 and VEGF in glioma patients: Different profiles of (pro-angiogenic) molecule expression are related with tumor grade and may provide prognostic information. Cancer Biol. Ther. 2007, 6, 1018–1024. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, S.L.; Duan, J.J.; Yi, L.; Guo, Y.F.; Shi, Y.; Li, L.; Yang, Z.Y.; Liao, X.M.; Cai, J.; et al. Invasion of white matter tracts by glioma stem cells is regulated by a NOTCH1-SOX2 positive-feedback loop. Nat. Neurosci. 2019, 22, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Hu, G.; Shi, L.; Long, G.; Yang, L.; Xi, Q.; Guo, Q.; Wang, J.; Dong, Z.; Zhang, M. Notch1 ablation radiosensitizes glioblastoma cells. Oncotarget 2017, 8, 88059–88068. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Kim, S.O.; Jin, X.; Ham, S.W.; Kim, J.; Park, J.B.; Lee, J.Y.; Kim, S.C.; Kim, H. Epidermal growth factor receptor variant III renders glioma cancer cells less differentiated by JAGGED1. Tumour Biol. 2015, 36, 2921–2928. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.X.; Chen, L.; Wang, C.H.; Lin, Z.X.; Chen, B.J.; You, N.; Chen, Y.; Wang, X.F. The Vascular Notch Ligands Delta-Like Ligand 4 (DLL4) and Jagged1 (JAG1) Have Opposing Correlations with Microvascularization but a Uniform Prognostic Effect in Primary Glioblastoma: A Preliminary Study. World Neurosurg. 2016, 88, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.X.; Wang, C.H.; You, N.; Chen, B.J.; Wang, X.F.; Chen, Y.P.; Lin, Z.X. High Jagged1 expression is associated with poor outcome in primary glioblastoma. Med. Oncol. 2015, 32, 341. [Google Scholar] [CrossRef] [PubMed]
- Jubb, A.M.; Browning, L.; Campo, L.; Turley, H.; Steers, G.; Thurston, G.; Harris, A.L.; Ansorge, O. Expression of vascular Notch ligands Delta-like 4 and Jagged-1 in glioblastoma. Histopathology 2012, 60, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lin, Z.X.; Lin, G.S.; Zhou, C.F.; Chen, Y.P.; Wang, X.F.; Zheng, Z.Q. Classification of microvascular patterns via cluster analysis reveals their prognostic significance in glioblastoma. Hum. Pathol. 2015, 46, 120–128. [Google Scholar] [CrossRef]
- Singh, S.; Brocker, C.; Koppaka, V.; Chen, Y.; Jackson, B.C.; Matsumoto, A.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic. Biol. Med. 2013, 56, 89–101. [Google Scholar] [CrossRef]
- Zanoni, M.; Bravaccini, S.; Fabbri, F.; Arienti, C. Emerging Roles of Aldehyde Dehydrogenase Isoforms in Anti-cancer Therapy Resistance. Front. Med. 2022, 9, 795762. [Google Scholar] [CrossRef]
- Liu, K.; Lin, B.; Zhao, M.; Yang, X.; Chen, M.; Gao, A.; Liu, F.; Que, J.; Lan, X. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell. Signal. 2013, 25, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Huang, S.; Chen, S.; Chen, J.; Wang, Z.; Wang, Y.; Zheng, H. SOX2 promotes chemoresistance, cancer stem cells properties, and epithelial-mesenchymal transition by β-catenin and Beclin1/autophagy signaling in colorectal cancer. Cell Death Dis. 2021, 12, 449. [Google Scholar] [CrossRef] [PubMed]
- Garros-Regulez, L.; Aldaz, P.; Arrizabalaga, O.; Moncho-Amor, V.; Carrasco-Garcia, E.; Manterola, L.; Moreno-Cugnon, L.; Barrena, C.; Villanua, J.; Ruiz, I.; et al. mTOR inhibition decreases SOX2-SOX9 mediated glioma stem cell activity and temozolomide resistance. Expert Opin. Ther. Targets 2016, 20, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Lee, H.K. Current Understanding of Hypoxia in Glioblastoma Multiforme and Its Response to Immunotherapy. Cancers 2022, 14, 1176. [Google Scholar] [CrossRef] [PubMed]
- Rankin, E.B.; Nam, J.M.; Giaccia, A.J. Hypoxia: Signaling the Metastatic Cascade. Trends Cancer 2016, 2, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cheng, P.; Pavlyukov, M.S.; Yu, H.; Zhang, Z.; Kim, S.H.; Minata, M.; Mohyeldin, A.; Xie, W.; Chen, D.; et al. Targeting NEK2 attenuates glioblastoma growth and radioresistance by destabilizing histone methyltransferase EZH2. J. Clin. Investig. 2017, 127, 3075–3089. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.N.; Hou, S.Q.; Jiang, R.; Sun, J.L.; Cheng, C.D.; Qian, Z.R. EZH2 is a potential prognostic predictor of glioma. J. Cell. Mol. Med. 2021, 25, 925–936. [Google Scholar] [CrossRef]
- Qi, B.; Yang, C.; Zhu, Z.; Chen, H. EZH2-Inhibited MicroRNA-454-3p Promotes M2 Macrophage Polarization in Glioma. Front. Cell Dev. Biol. 2020, 8, 574940. [Google Scholar] [CrossRef]
- Han, B.; Meng, X.; Wu, P.; Li, Z.; Li, S.; Zhang, Y.; Zha, C.; Ye, Q.; Jiang, C.; Cai, J.; et al. ATRX/EZH2 complex epigenetically regulates FADD/PARP1 axis, contributing to TMZ resistance in glioma. Theranostics 2020, 10, 3351–3365. [Google Scholar] [CrossRef]
- Yang, Z.; Zhong, W.; Yang, L.; Wen, P.; Luo, Y.; Wu, C. The emerging role of exosomes in radiotherapy. Cell Commun. Signal. 2022, 20, 171. [Google Scholar] [CrossRef]
- Jenjaroenpun, P.; Kremenska, Y.; Nair, V.M.; Kremenskoy, M.; Joseph, B.; Kurochkin, I.V. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ 2013, 1, e201. [Google Scholar] [CrossRef] [PubMed]
- Duijvesz, D.; Luider, T.; Bangma, C.H.; Jenster, G. Exosomes as biomarker treasure chests for prostate cancer. Eur. Urol. 2011, 59, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Silva, J.; Garcia, V.; Rodriguez, M.; Compte, M.; Cisneros, E.; Veguillas, P.; Garcia, J.M.; Dominguez, G.; Campos-Martin, Y.; Cuevas, J.; et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer 2012, 51, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Bucci, J.; Malouf, D.; Knox, M.; Graham, P.; Li, Y. Exosomes in Cancer Radioresistance. Front. Oncol. 2019, 9, 869. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Du, S.; Liu, L.; Gan, F.; Jiang, X.; Wangrao, K.; Lyu, P.; Gong, P.; Yao, Y. Radiation-Induced Bystander Effect can be Transmitted Through Exosomes Using miRNAs as Effector Molecules. Radiat. Res. 2020, 194, 89–100. [Google Scholar] [CrossRef]
- Kumar, D.; Gupta, D.; Shankar, S.; Srivastava, R.K. Biomolecular characterization of exosomes released from cancer stem cells: Possible implications for biomarker and treatment of cancer. Oncotarget 2015, 6, 3280–3291. [Google Scholar] [CrossRef] [PubMed]
- Munoz, J.L.; Bliss, S.A.; Greco, S.J.; Ramkissoon, S.H.; Ligon, K.L.; Rameshwar, P. Delivery of Functional Anti-miR-9 by Mesenchymal Stem Cell-derived Exosomes to Glioblastoma Multiforme Cells Conferred Chemosensitivity. Mol. Ther. Nucleic Acids 2013, 2, e126. [Google Scholar] [CrossRef]
- Dai, B.; Xiao, Z.; Mao, B.; Zhu, G.; Huang, H.; Guan, F.; Su, H.; Lin, Z.; Peng, W.; Hu, Z. lncRNA AWPPH promotes the migration and invasion of glioma cells by activating the TGF-β pathway. Oncol. Lett. 2019, 18, 5923–5929. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, L.; Zhou, Y.; Dong, L.; Ma, W.; Lv, L.; Zhang, J.; Wang, X. Glioblastoma Stem Cell-Derived Exosomes Enhance Stemness and Tumorigenicity of Glioma Cells by Transferring Notch1 Protein. Cell. Mol. Neurobiol. 2020, 40, 767–784. [Google Scholar] [CrossRef]
| Surface Biomarker | Biological Action | References |
|---|---|---|
| CD133 | Antioxidant scavenger system CD133-induced hypoxia | [66] |
| CD44 | CD44 interaction with extracellular domain activates a number of signaling pathways implicated in tumor angiogenesis, proliferation and stemness | [67] |
| TIM 3 | Galectin-TIM-3 interaction causes canonical Wnt pathway and permits the maintenance and enhancement of cancer stemness | [68] |
| Notch-1 and Jagged-1 | Promote CSC’s invasiveness and white matter tropism, proliferation, angiogenesisand glioma-initiating cells (GICs) | [69] |
| Transcription Biomarker | ||
| NANOG | Induces suppression of differentiation and cellular stamness | [70,71,72] |
| SOX2 | Increases white matter GSC tropisim, drug resistance, epithelial-mesenchymal transition andangiogenesis | [73] |
| OCT3/4 | Drug efflux pump Increases invasiveness, migration and cell proliferation Induces tumor angiogenesis regulating the homologous recombination factors | [74,75] |
| EZH2 | AKT/mTOR activation Epithelial–mesenchymal transition (EMT) Silences transcription through trimethylation of histone H3 lysine 27 Stabilizes DDB2 and promotes nucleotide excision repair | [74,76,77] |
| HIF1- alfa | Angiogenesis Metabolic reprogramming (Warburg effect) Increases the invasion of cancer cells Immune system suppression | [78,79,80,81] |
| STAT-3 | Proliferation Immune evasion Therapy resistance through upregulation of DNA repair proteins | [82,83,84] |
| Intracellular Biomarker | ||
| ALDH | Activates retinoid as well as β-Catenin/Tcf signaling pathways related to the stemness of CSCs Antioxidant activity Expression of mesenchymal phenotype (MES) Affect proliferation and migration of cells by inducing cell-cycle arrest and the epithelial-mesenchymal transition | [85,86] |
| RAD51 and BRCA 1-2 | DNA repair through homologous recombination Perpetuation of CSCs Sensitivity to chemotherapy | [87] |
| Nestin | Enhances invasiveness Generates transient populations of intensely proliferative cells Angiogenesis | [88,89,90,91] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pasqualetti, F.; Miniati, M.; Gonnelli, A.; Gadducci, G.; Giannini, N.; Palagini, L.; Mancino, M.; Fuentes, T.; Paiar, F. Cancer Stem Cells and Glioblastoma: Time for Innovative Biomarkers of Radio-Resistance? Biology 2023, 12, 1295. https://doi.org/10.3390/biology12101295
Pasqualetti F, Miniati M, Gonnelli A, Gadducci G, Giannini N, Palagini L, Mancino M, Fuentes T, Paiar F. Cancer Stem Cells and Glioblastoma: Time for Innovative Biomarkers of Radio-Resistance? Biology. 2023; 12(10):1295. https://doi.org/10.3390/biology12101295
Chicago/Turabian StylePasqualetti, Francesco, Mario Miniati, Alessandra Gonnelli, Giovanni Gadducci, Noemi Giannini, Laura Palagini, Maricia Mancino, Taiusha Fuentes, and Fabiola Paiar. 2023. "Cancer Stem Cells and Glioblastoma: Time for Innovative Biomarkers of Radio-Resistance?" Biology 12, no. 10: 1295. https://doi.org/10.3390/biology12101295
APA StylePasqualetti, F., Miniati, M., Gonnelli, A., Gadducci, G., Giannini, N., Palagini, L., Mancino, M., Fuentes, T., & Paiar, F. (2023). Cancer Stem Cells and Glioblastoma: Time for Innovative Biomarkers of Radio-Resistance? Biology, 12(10), 1295. https://doi.org/10.3390/biology12101295







