Profiling the Spatial Expression Pattern and ceRNA Network of lncRNA, miRNA, and mRNA Associated with the Development of Intermuscular Bones in Zebrafish
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
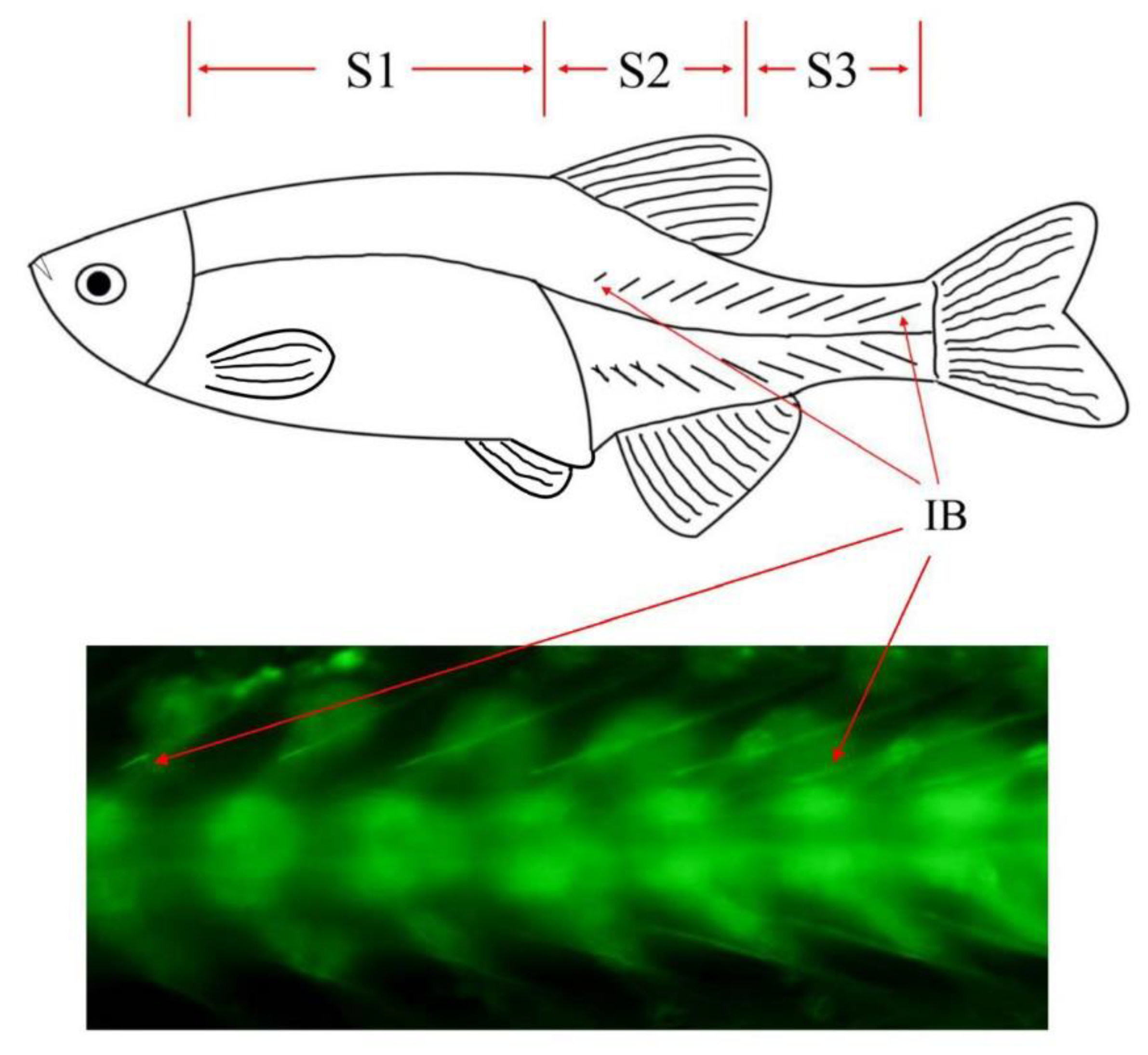
2.1. Experimental Fish
2.2. Sample Collection and RNA Extraction
2.3. Library Construction and Sequencing
2.4. Assembly and Annotation of Transcriptomes
2.5. Differential Expression, Gene Expression Pattern, and Function Enrichment Analysis
2.6. Identification of Genes Associated with the Development of IBs and Construction of ceRNA Networks
2.7. Verification of Gene Expression Using RT-qPCR
3. Results
3.1. Transcriptome Assembly and Statistics
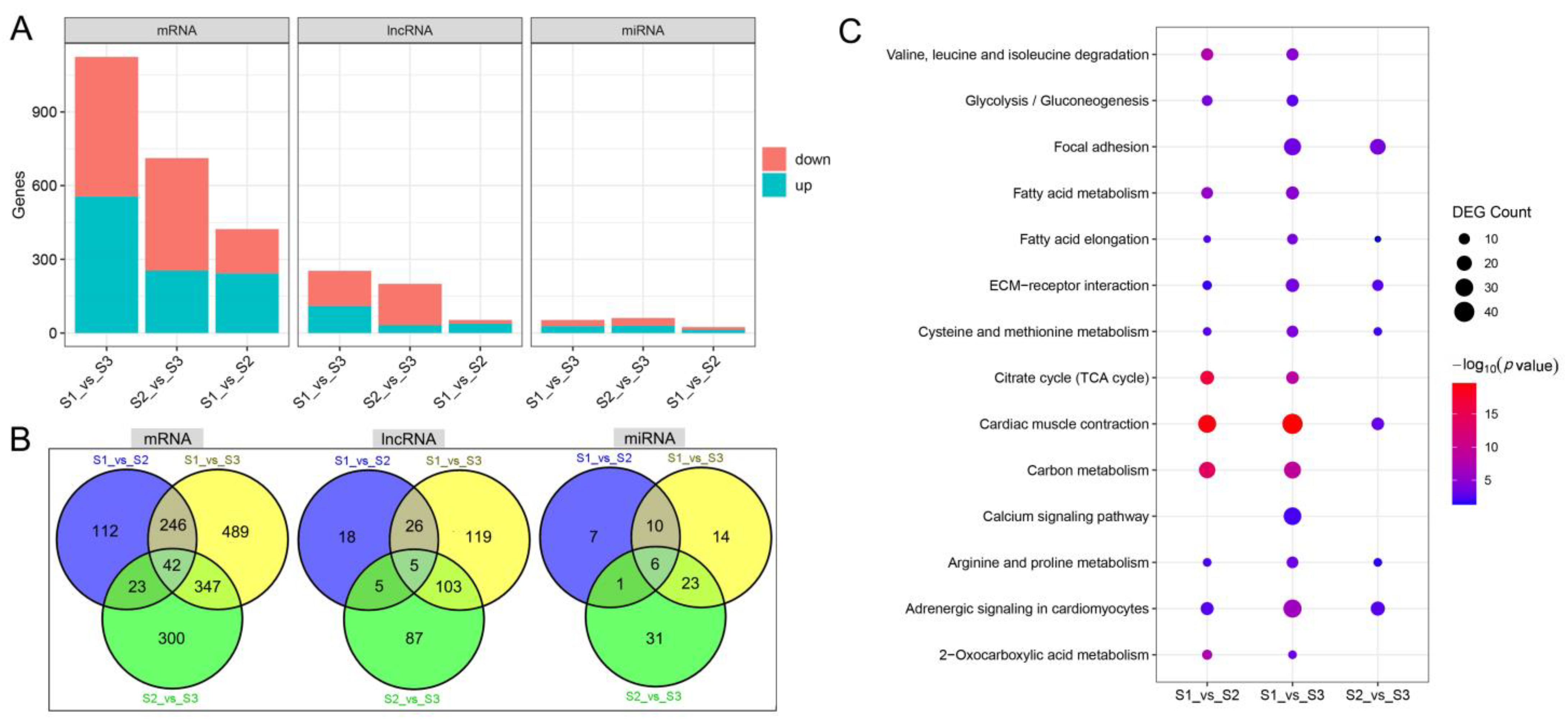
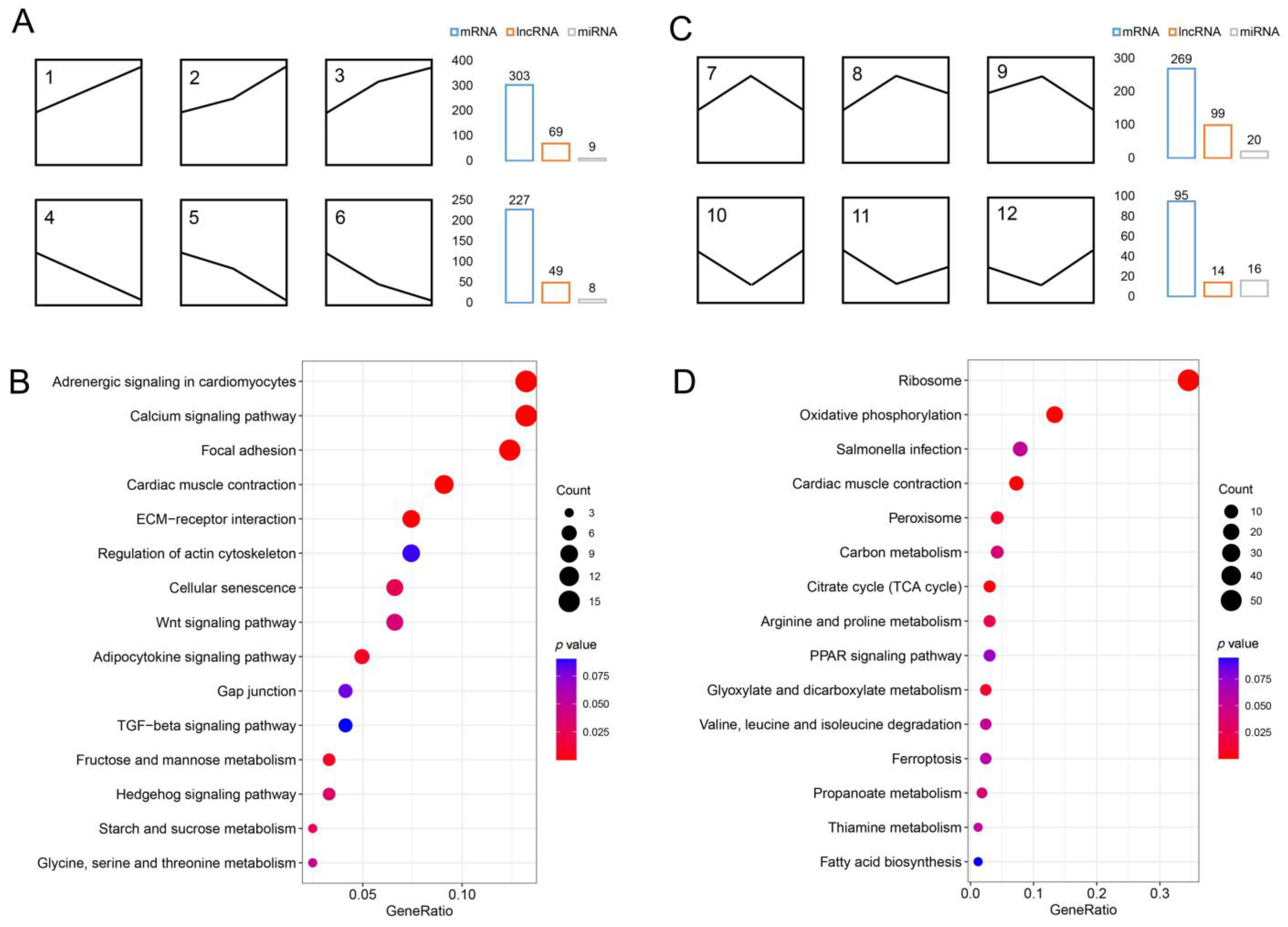
3.2. Identification of DEGs and Functional Enrichment Analysis
3.3. Identification of mRNA, lncRNA, and miRNA Associated with IB Development
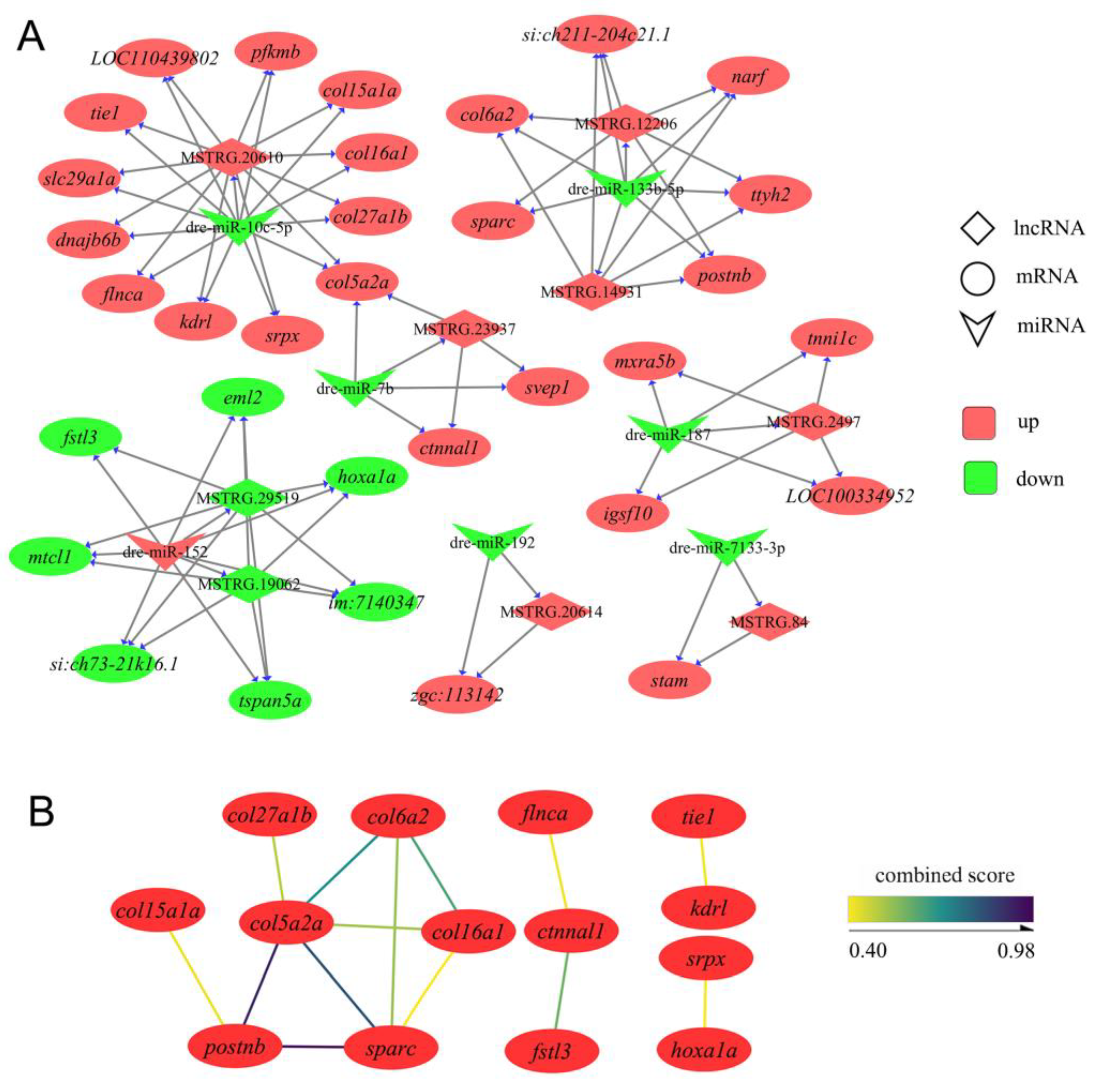
3.4. Construction of ceRNA Networks Associated with IB Growth
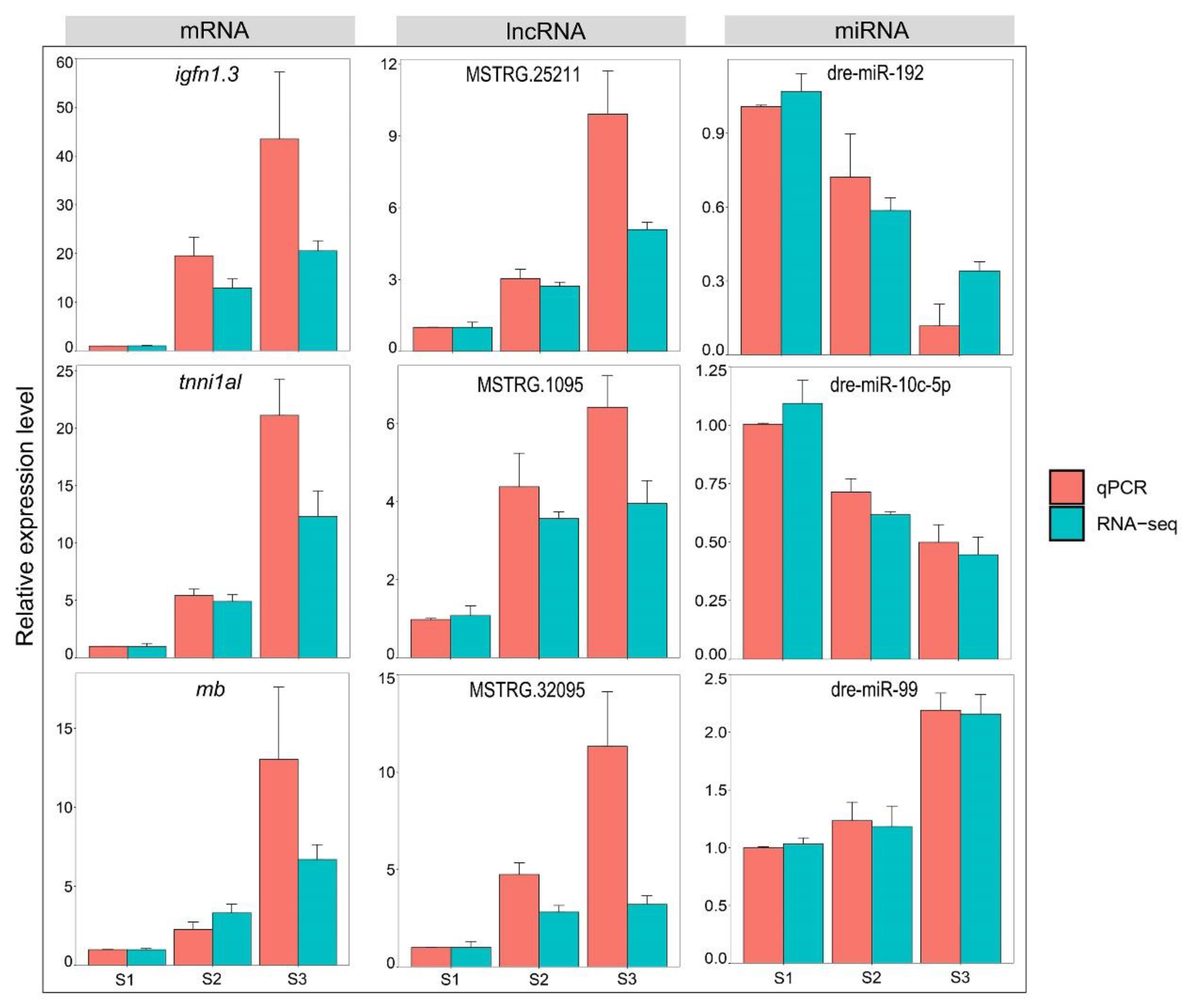
3.5. Validation of RNA-Seq Data via RT-qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patterson, C.; Johnson, G.D. The Intermuscular Bones and Ligaments of Teleostean Fishes; Smithsonian Institution Press: Washington, DC, USA, 1995. [Google Scholar]
- Nie, C.H.; Hilsdorf, A.W.; Wan, S.M.; Gao, Z.X. Understanding the development of intermuscular bones in teleost: Status and future directions for aquaculture. Rev. Aquac. 2020, 12, 759–772. [Google Scholar] [CrossRef]
- Xu, X.; Zheng, J.; Qian, Y.; Luo, C. Normally grown and developed intermuscular bone-deficient mutant in grass carp, Ctenopharyngodon idellus. Chin. Sci. Bull. 2015, 60, 52–57. [Google Scholar]
- Perazza, C.A.; de Menezes, J.T.B.; Ferraz, J.B.S.; Pinaffi, F.L.V.; Silva, L.A.; Hilsdorf, A.W.S. Lack of intermuscular bones in specimens of Colossoma macropomum: An unusual phenotype to be incorporated into genetic improvement programs. Aquaculture 2017, 472, 57–60. [Google Scholar] [CrossRef]
- Yang, J.; Tong, G.; Sun, Z.; Zheng, X.; Lv, W.; Cao, D.; Sun, X.; Kuang, Y. Comparative analysis of muscle development in zebrafish with different intermuscular-bones patterns. Pak. J. Zool. 2021, 53, 313. [Google Scholar]
- Nie, C.; Wan, S.; Chen, Y.; Zhu, D.; Wang, X.; Dong, X.; Gao, Z.-X. Loss of scleraxis leads to distinct reduction of mineralized intermuscular bone in zebrafish. Aquac. Fish. 2021, 6, 169–177. [Google Scholar] [CrossRef]
- Zhong, Z.; Niu, P.; Wang, M.; Huang, G.; Xu, S.; Sun, Y.; Xu, X.; Hou, Y.; Sun, X.; Yan, Y. Targeted disruption of sp7 and myostatin with CRISPR-Cas9 results in severe bone defects and more muscular cells in common carp. Sci. Rep. 2016, 6, 22953. [Google Scholar] [CrossRef]
- Nie, C.-H.; Wan, S.-M.; Chen, Y.-L.; Huysseune, A.; Wu, Y.-M.; Zhou, J.-J.; Hilsdorf, A.W.S.; Wang, W.-m.; Witten, P.E.; Lin, Q. Single-cell transcriptomes and runx2b−/− mutants reveal the genetic signatures of intermuscular bone formation in zebrafish. Natl. Sci. Rev. 2022, 9, nwac152. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X.; Huang, X.; Huang, G.; Gao, Z.; Wang, W.; Liu, H. Genome-wide analysis of intermuscular bone development reveals changes of key genes expression and signaling pathways in blunt snout bream (Megalobrama amblycephala). Genomics 2021, 113, 654–663. [Google Scholar] [CrossRef]
- Huynh, N.P.; Anderson, B.A.; Guilak, F.; McAlinden, A. Emerging roles for long noncoding RNAs in skeletal biology and disease. Connect. Tissue Res. 2017, 58, 116–141. [Google Scholar] [CrossRef]
- Fang, S.; Deng, Y.; Gu, P.; Fan, X. MicroRNAs regulate bone development and regeneration. Int. J. Mol. Sci. 2015, 16, 8227–8253. [Google Scholar] [CrossRef]
- Chen, Y.; Wan, S.; Li, Q.; Dong, X.; Diao, J.; Liao, Q.; Wang, G.-Y.; Gao, Z.-X. Genome-Wide Integrated Analysis Revealed Functions of lncRNA–miRNA–mRNA Interaction in Growth of Intermuscular Bones in Megalobrama amblycephala. Front. Cell. Dev. Biol. 2021, 8, 603815. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.-M.; Yi, S.-K.; Zhong, J.; Nie, C.-H.; Guan, N.-N.; Zhang, W.-Z.; Gao, Z.-X. Dynamic mRNA and miRNA expression analysis in response to intermuscular bone development of blunt snout bream (Megalobrama amblycephala). Sci. Rep. 2016, 6, 31050. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zeng, D.; He, P.; Wei, P.; Hui, W.; Wu, T.; Zhuo, X.; Lin, Y. mRNA and microRNA transcriptomics analyses in intermuscular bones of two carp species, rice flower carp (Cyprinus carpio var. Quanzhounensis) and Jian carp (Cyprinus carpio var. Jian). Comp. Biochem. Physiol. D-Genom. Proteom. 2019, 30, 71–80. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Duan, Y.; Zhang, W.; Cheng, Y.; Shi, M.; Xia, X.-Q. Comprehensive analysis of hub mRNA, lncRNA and miRNA, and associated ceRNA networks implicated in grass carp (Ctenopharyngodon idella) growth traits. Genomics 2021, 113, 4004–4014. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Z.; Su, H.; Chen, F.; Ma, M.; Yu, W.; Ye, G.; Cen, S.; Mi, R.; Wu, X. Effects of long-term culture on the biological characteristics and RNA profiles of human bone-marrow-derived mesenchymal stem cells. Mol. Ther.-Nucl. Acids 2021, 26, 557–574. [Google Scholar] [CrossRef]
- Wang, X.; Hu, K.B.; Zhang, Y.Q.; Yang, C.J.; Yao, H.H. Comprehensive analysis of aberrantly expressed profiles of lncRNAs, miRNAs and mRNAs with associated ceRNA network in cholangiocarcinoma. Cancer Biomark. 2018, 23, 549–559. [Google Scholar] [CrossRef]
- Nie, C.; Chen, Z.; Dai, C.; Wan, S.; Gao, Z. Ossification patterns of intermuscular bones in different fish species. Acta. Hydrob. Sin. 2018, 42, 131–137. [Google Scholar]
- Du, S.J.; Frenkel, V.; Kindschi, G.; Zohar, Y. Visualizing normal and defective bone development in zebrafish embryos using the fluorescent chromophore calcein. Dev. Biol. 2001, 238, 239–246. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, Y.; Zhang, Y.; Ning, Z.; Li, Y.; Zhao, Q.; Lu, H.; Huang, R.; Xia, X.; Feng, Q. The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat. Genet. 2015, 47, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.-P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-J.; Yang, D.-C.; Kong, L.; Hou, M.; Meng, Y.-Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Duan, Y.; Zhang, W.; Cheng, Y.; Shi, M.; Xia, X.-Q. A systematic evaluation of bioinformatics tools for identification of long noncoding RNAs. RNA 2021, 27, 80–98. [Google Scholar] [CrossRef]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinf. 2010, 32, 11.17.1–11.17.14. [Google Scholar] [CrossRef]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018, 46, D335–D342. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Saini, H.K.; Van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2007, 36, D154–D158. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.-S.; Tam, W.-L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef]
- Cline, M.S.; Smoot, M.; Cerami, E.; Kuchinsky, A.; Landys, N.; Workman, C.; Christmas, R.; Avila-Campilo, I.; Creech, M.; Gross, B. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007, 2, 2366. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016, 45, D362–D368. [Google Scholar] [CrossRef]
- Salhotra, A.; Shah, H.N.; Levi, B.; Longaker, M.T. Mechanisms of bone development and repair. Nat. Rev. Mol. Cell Biol. 2020, 21, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, B.; Jonsson, N. Early environment influences later performance in fishes. J. Fish Biol. 2014, 85, 151–188. [Google Scholar] [CrossRef]
- Nishiwaki, T.; Yamaguchi, T.; Zhao, C.; Amano, H.; Hankenson, K.D.; Bornstein, P.; Toyama, Y.; Matsuo, K. Reduced expression of thrombospondins and craniofacial dysmorphism in mice overexpressing Fra1. J. Bone Miner. Res. 2006, 21, 596–604. [Google Scholar] [CrossRef]
- Eijken, M.; Swagemakers, S.; Koedam, M.; Steenbergen, C.; Derkx, P.; Uitterlinden, A.G.; van der Spek, P.J.; Visser, J.A.; de Jong, F.H.; Pols, H.A. The activin A-follistatin system: Potent regulator of human extracellular matrix mineralization. FASEB J. 2007, 21, 2949–2960. [Google Scholar] [CrossRef]
- Shi, C.; Iura, A.; Terajima, M.; Liu, F.; Lyons, K.; Pan, H.; Zhang, H.; Yamauchi, M.; Mishina, Y.; Sun, H. Deletion of BMP receptor type IB decreased bone mass in association with compromised osteoblastic differentiation of bone marrow mesenchymal progenitors. Sci. Rep. 2016, 6, 24256. [Google Scholar] [CrossRef] [PubMed]
- Yasui, T.; Kadono, Y.; Nakamura, M.; Oshima, Y.; Matsumoto, T.; Masuda, H.; Hirose, J.; Omata, Y.; Yasuda, H.; Imamura, T. Regulation of RANKL-induced osteoclastogenesis by TGF-β through molecular interaction between Smad3 and Traf6. J. Bone Miner. Res. 2011, 26, 1447–1456. [Google Scholar] [CrossRef] [PubMed]
- Shieh, A.; Han, W.; Ishii, S.; Greendale, G.A.; Crandall, C.J.; Karlamangla, A.S. Quantifying the balance between total bone formation and total bone resorption: An index of net bone formation. J. Clin. Endocrinol. Metab. 2016, 101, 2802–2809. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, I.; Zeveleva, S.; Duarte, A.; Zhao, X.; Depalle, B.; Cardoso, L.; Jin, S.; Berteau, J. Microstructure, mineral and mechanical properties of teleost intermuscular bones. J. Biomech. 2019, 94, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Lv, Y.; Gong, X.; Wu, J.; Bao, B. Different ossification patterns of intermuscular bones in fish with different swimming modes. Biol. Open 2015, 4, 1727–1732. [Google Scholar] [CrossRef]
- Zhou, J.-J.; Chang, Y.-J.; Chen, Y.-L.; Wang, X.-D.; Liao, Q.; Shi, R.-H.; Gao, Z.-X. Comparison of Myosepta Development and Transcriptome Profiling between Blunt Snout Bream with and Tilapia without Intermuscular Bones. Biology 2021, 10, 1311. [Google Scholar] [CrossRef]
- Gupton, S.L.; Waterman-Storer, C.M. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell 2006, 125, 1361–1374. [Google Scholar] [CrossRef]
- Su, P.; Tian, Y.; Yang, C.; Ma, X.; Wang, X.; Pei, J.; Qian, A. Mesenchymal stem cell migration during bone formation and bone diseases therapy. Int. J. Mol. Sci. 2018, 19, 2343. [Google Scholar] [CrossRef]
- Luo, H.; Gao, H.; Liu, F.; Qiu, B. Regulation of Runx2 by microRNA-9 and microRNA-10 modulates the osteogenic differentiation of mesenchymal stem cells. Int. J. Mol. Med. 2017, 39, 1046–1052. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, L.; Zhang, Z.; Ren, F.; Chen, L.; Lan, Z. miR-10a-5p inhibits osteogenic differentiation of bone marrow-derived mesenchymal stem cells. Mol. Med. Rep. 2020, 22, 135–144. [Google Scholar] [CrossRef]
- Li, X.; Zheng, Y.; Zheng, Y.; Huang, Y.; Zhang, Y.; Jia, L.; Li, W. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res. Ther. 2018, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Ren, W.; Zheng, Z.; Huang, Z.; Liang, T.; Li, F.; Shi, Z.; Jiang, Q.; Yang, X.; Guo, L. Mmu_circ_003795 regulates osteoblast differentiation and mineralization in MC3T3-E1 and MDPC23 by targeting COL15A1. Mol. Med. Rep. 2020, 22, 1737–1746. [Google Scholar] [CrossRef] [PubMed]
- Lisignoli, G.; Codeluppi, K.; Todoerti, K.; Manferdini, C.; Piacentini, A.; Zini, N.; Grassi, F.; Cattini, L.; Piva, R.; Rizzoli, V. Gene array profile identifies collagen type XV as a novel human osteoblast-secreted matrix protein. J. Cell. Physiol. 2009, 220, 401–409. [Google Scholar] [CrossRef]
- Michalickova, K.; Susic, M.; Willing, M.C.; Wenstrup, R.J.; Cole, W.G. Mutations of the α2 (V) chain of type V collagen impair matrix assembly and produce Ehlers-Danlos syndrome type I. Hum. Mol. Genet. 1998, 7, 249–255. [Google Scholar] [CrossRef]
- Yamazaki, M.; Majeska, R.J.; Yoshioka, H.; Moriya, H.; Einhorn, T.A. Spatial and temporal expression of fibril-forming minor collagen genes (types V and XI) during fracture healing. J. Orthop. Res. 1997, 15, 757–764. [Google Scholar] [CrossRef]
- Gonzaga-Jauregui, C.; Gamble, C.N.; Yuan, B.; Penney, S.; Jhangiani, S.; Muzny, D.M.; Gibbs, R.A.; Lupski, J.R.; Hecht, J.T. Mutations in COL27A1 cause Steel syndrome and suggest a founder mutation effect in the Puerto Rican population. Eur. J. Hum. Genet. 2015, 23, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Rosset, E.M.; Bradshaw, A.D. SPARC/osteonectin in mineralized tissue. Matrix Biol. 2016, 52, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, N.; Conway, S.J.; Ferrari, S.L. Regulation of beta catenin signaling and parathyroid hormone anabolic effects in bone by the matricellular protein periostin. Proc. Natl. Acad. Sci. USA 2012, 109, 15048–15053. [Google Scholar] [CrossRef]
- Gerbaix, M.; Vico, L.; Ferrari, S.L.; Bonnet, N. Periostin expression contributes to cortical bone loss during unloading. Bone 2015, 71, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Iwai, K.; Ishii, M.; Ohshima, S.; Miyatake, K.; Saeki, Y. Expression and function of transmembrane-4 superfamily (tetraspanin) proteins in osteoclasts: Reciprocal roles of Tspan-5 and NET-6 during osteoclastogenesis. Allergol. Int. 2007, 56, 457–463. [Google Scholar] [CrossRef]
- Zhou, J.; Fujiwara, T.; Ye, S.; Li, X.; Zhao, H. Downregulation of Notch modulators, tetraspanin 5 and 10, inhibits osteoclastogenesis in vitro. Calcif. Tissue Int. 2014, 95, 209–217. [Google Scholar] [CrossRef] [PubMed]
| Pathway | Upregulated Genes | Downregulated Genes |
|---|---|---|
| ECM–receptor interactions | col6a1, col6a2, itga11b, thbs1a, thbs3a, thbs4a, itga9 | col6a4a, lama5 |
| Calcium signaling pathway | adrb2a, pde1ca, cacna1sa, pdgfbb, plce1, kdrl, pdgfra, ryr1a, atp2a2a, slc25a5, si:dkey-163m14.2, tnnc1b, zgc:56235 | slc8a4b, calm3a, ppp3r1b |
| Hedgehog signaling pathway | cdon, hhatlb | lrp2a, csnk1da |
| Wnt signaling pathway | wnt10b, rspo3, sfrp2 | ctnnbip1, sfrp5, mycb, ppp3r1b, smad3b |
| TGF-β signaling pathway | inhbaa, thbs1a, bmpr1bb | mycb, smad3b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, W.; Shi, M.; Ren, K.; Liu, Y.; Duan, Y.; Cheng, Y.; Zhang, W.; Xia, X.-Q. Profiling the Spatial Expression Pattern and ceRNA Network of lncRNA, miRNA, and mRNA Associated with the Development of Intermuscular Bones in Zebrafish. Biology 2023, 12, 75. https://doi.org/10.3390/biology12010075
Ye W, Shi M, Ren K, Liu Y, Duan Y, Cheng Y, Zhang W, Xia X-Q. Profiling the Spatial Expression Pattern and ceRNA Network of lncRNA, miRNA, and mRNA Associated with the Development of Intermuscular Bones in Zebrafish. Biology. 2023; 12(1):75. https://doi.org/10.3390/biology12010075
Chicago/Turabian StyleYe, Weidong, Mijuan Shi, Keyi Ren, Yuhang Liu, You Duan, Yingyin Cheng, Wanting Zhang, and Xiao-Qin Xia. 2023. "Profiling the Spatial Expression Pattern and ceRNA Network of lncRNA, miRNA, and mRNA Associated with the Development of Intermuscular Bones in Zebrafish" Biology 12, no. 1: 75. https://doi.org/10.3390/biology12010075
APA StyleYe, W., Shi, M., Ren, K., Liu, Y., Duan, Y., Cheng, Y., Zhang, W., & Xia, X.-Q. (2023). Profiling the Spatial Expression Pattern and ceRNA Network of lncRNA, miRNA, and mRNA Associated with the Development of Intermuscular Bones in Zebrafish. Biology, 12(1), 75. https://doi.org/10.3390/biology12010075





