KDM6B Variants May Contribute to the Pathophysiology of Human Cerebral Folate Deficiency
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Compliance
2.2. Human Subject
2.3. Sequencing Analysis
2.4. Plasmids
2.5. Cell Culture and Transfection
2.6. Immunofluorescence
2.7. Western Blotting Assay
2.8. Folate Receptor Alpha (FOLR1) Autoantibodies
2.9. Statistical Analysis
3. Results
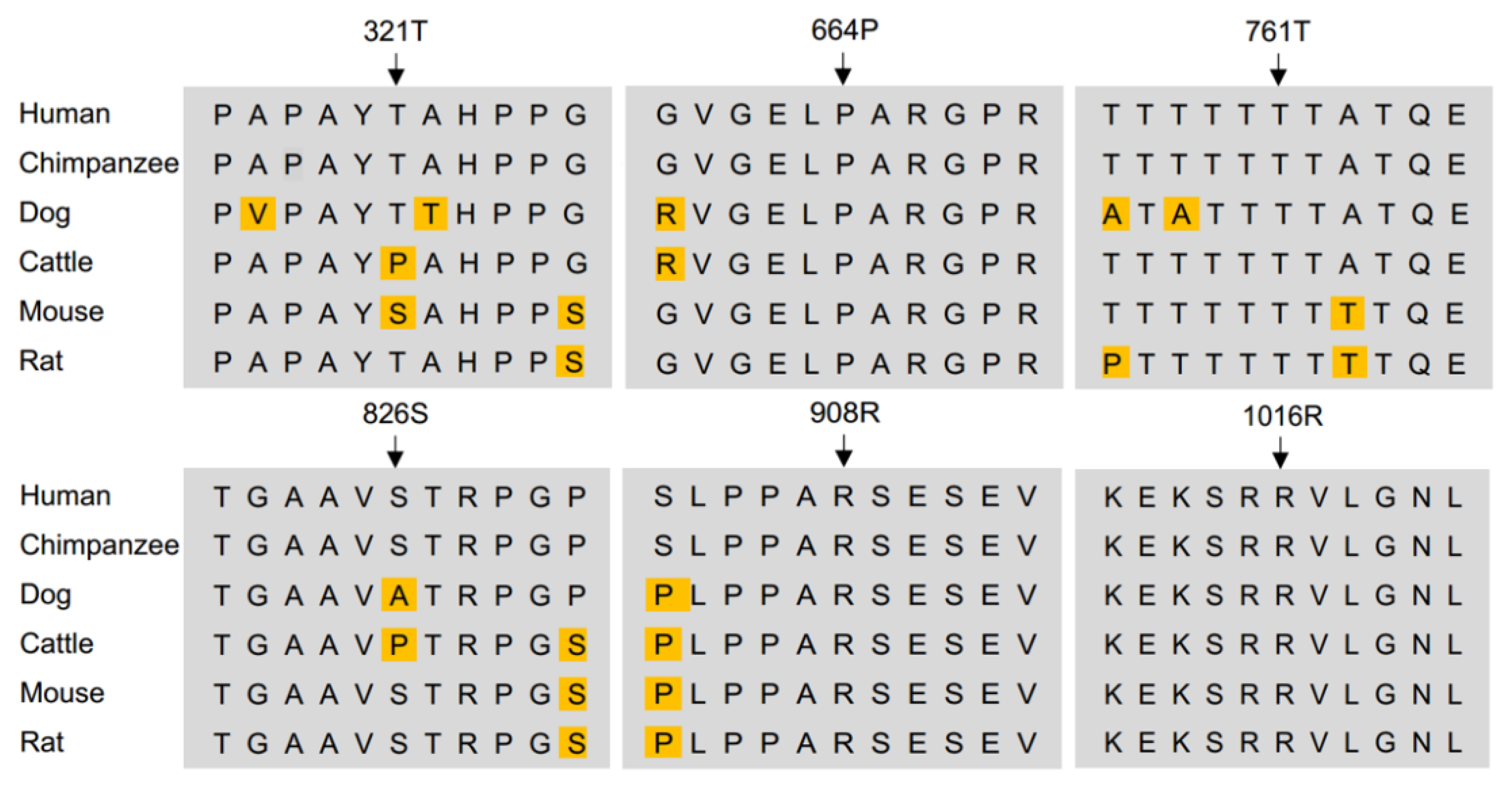
3.1. Missense Rare Variants in KDM6B Contribute to Human CFD
3.2. Clinical Features of CFD Patients with KDM6B Variants
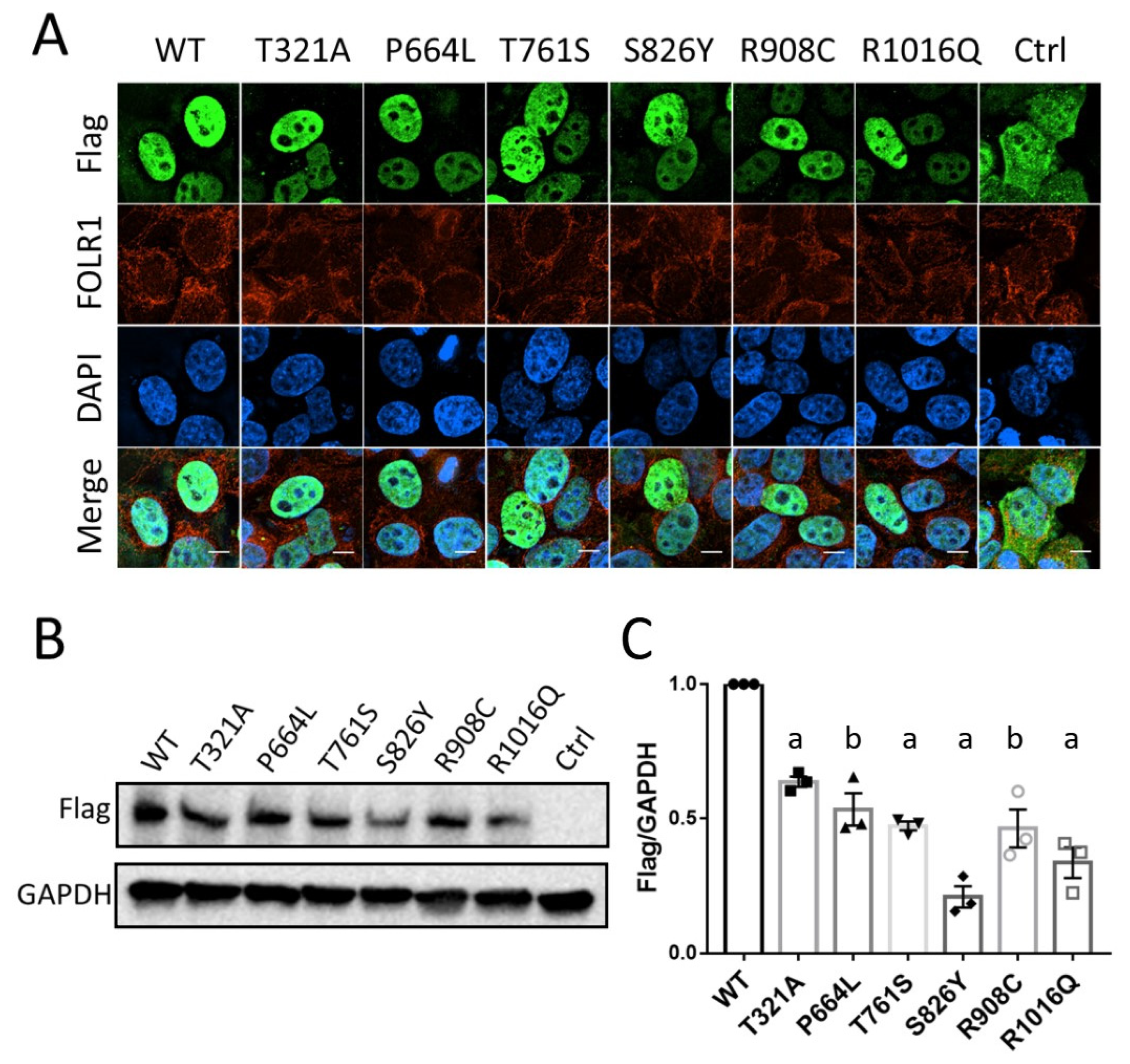
3.3. KDM6B Missense Variants Affect Protein Expression with No Effect on Localization
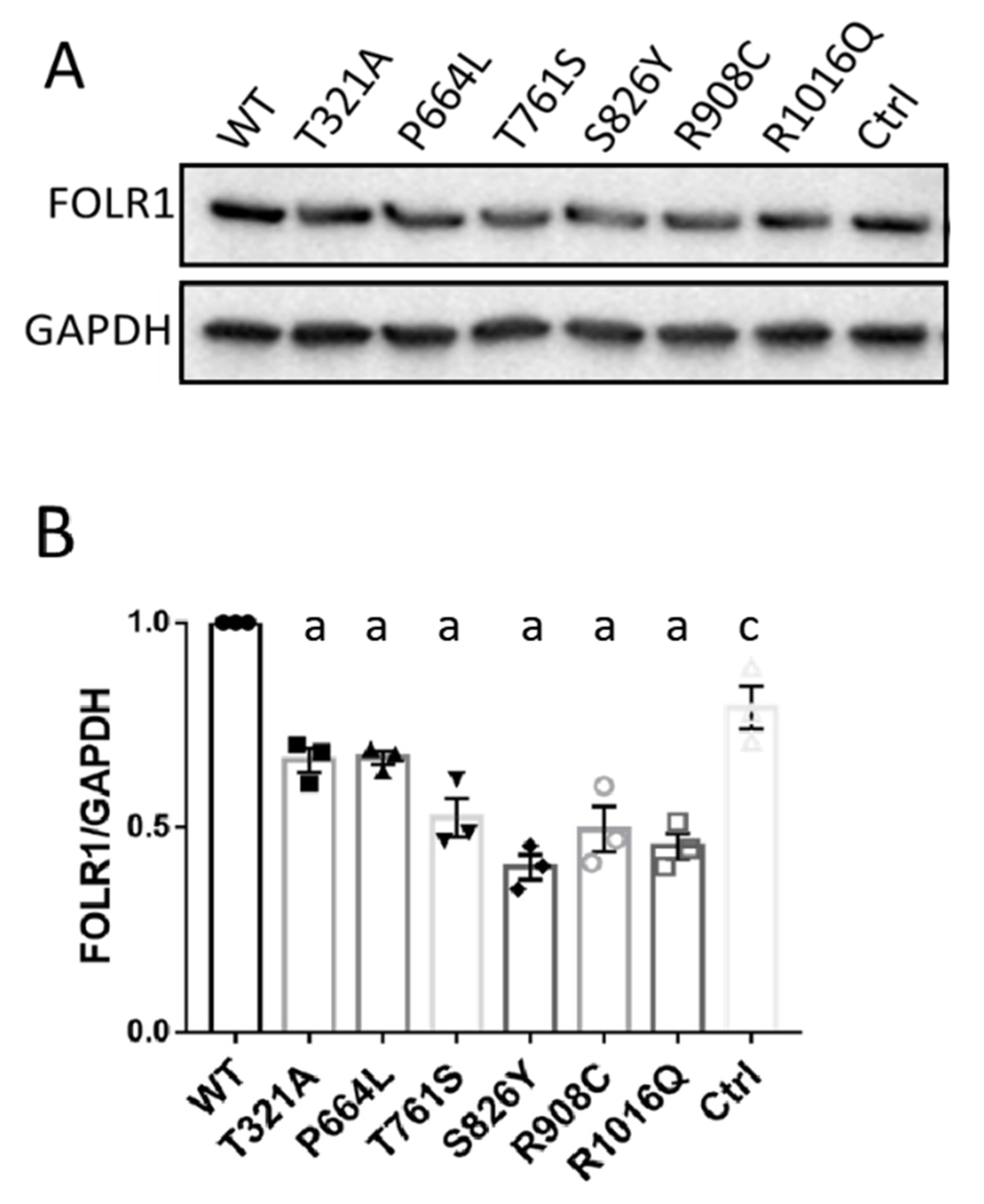
3.4. KDM6B Missense Variants Downregulate FOLR1 Protein Level
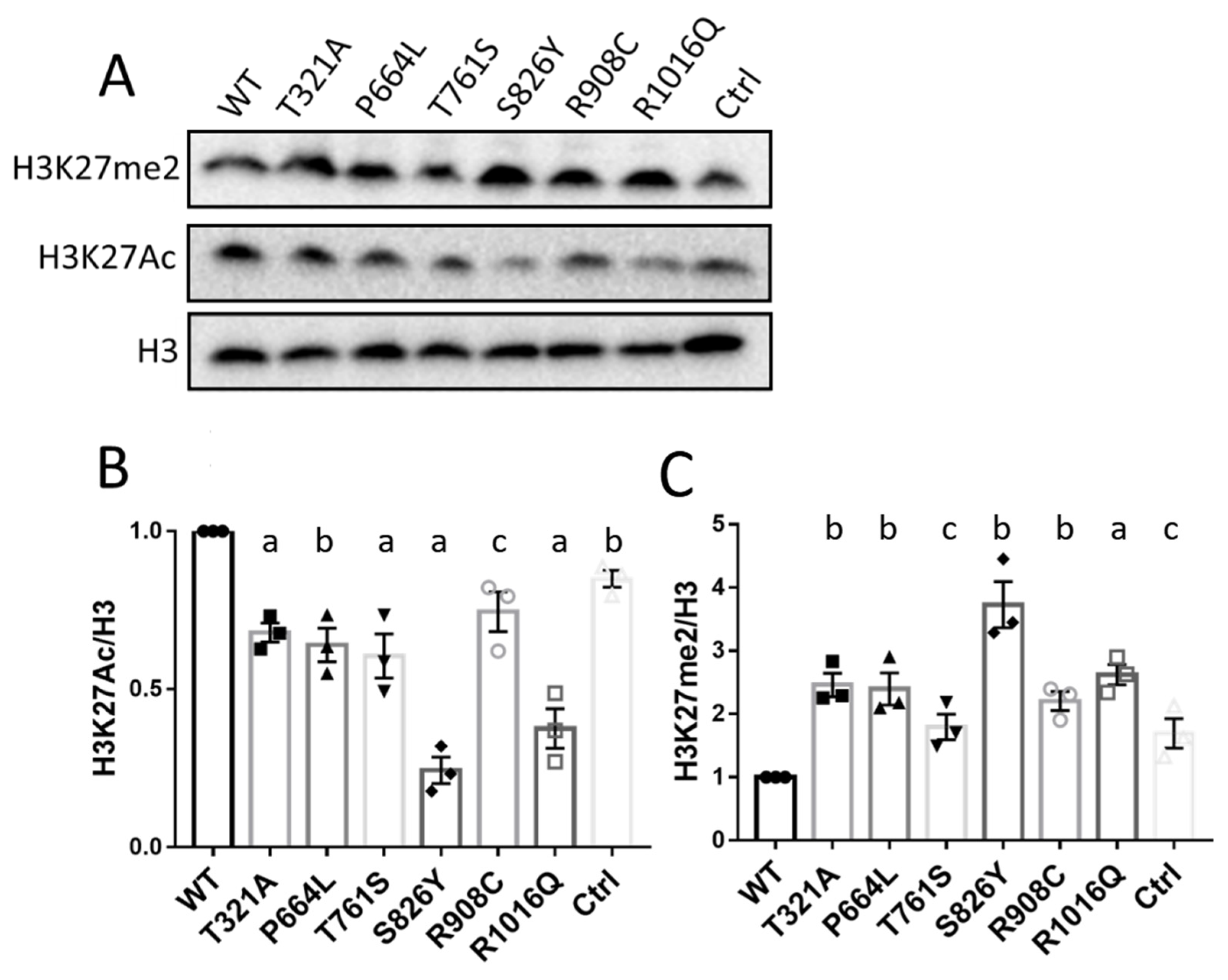
3.5. KDM6B Missense Variants Upregulated H3K27me2 and Downregulated H3K27Ac
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Molero-Luis, M.; Serrano, M.; O’Callaghan, M.M.; Sierra, C.; Pérez-Dueñas, B.; Garcia-Cazorla, A.; Artuch, R. Clinical, etiological and therapeutic aspects of cerebral folate deficiency. Expert Rev. Neurother. 2015, 15, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Ramaekers, V.T.; Blau, N. Cerebral folate deficiency. Dev. Med. Child Neurol. 2004, 46, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Hyland, K.; Shoffner, J.; Heales, S.J. Cerebral folate deficiency. J. Inherit. Metab. Dis. 2010, 33, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Spector, R.; Johanson, C.E. Vectorial ligand transport through mammalian choroid plexus. Pharm. Res. 2010, 27, 2054–2062. [Google Scholar] [CrossRef] [PubMed]
- Stover, P.J.; Durga, J.; Field, M.S. Folate nutrition and blood–Brain barrier dysfunction. Curr. Opin. Biotechnol. 2017, 44, 146–152. [Google Scholar] [CrossRef]
- Cao, X.; Wolf, A.; Kim, S.-E.; Cabrera, R.M.; Wlodarczyk, B.J.; Zhu, H.; Parker, M.; Lin, Y.; Steele, J.W.; Han, X.; et al. CIC de novo loss of function variants contribute to cerebral folate deficiency by downregulating FOLR1 expression. J. Med. Genet. 2020, 58, 484–494. [Google Scholar] [CrossRef]
- Mangold, S.; Blau, N.; Opladen, T.; Steinfeld, R.; Weßling, B.; Zerres, K.; Häusler, M. Cerebral folate deficiency: A neurometabolic syndrome? Mol. Genet. Metab. 2011, 104, 369–372. [Google Scholar] [CrossRef]
- Moretti, P.; Sahoo, T.; Hyland, K.; Bottiglieri, T.; Peters, S.; Del Gaudio, D.; Roa, B.; Curry, S.; Zhu, H.; Finnell, R.; et al. Cerebral folate deficiency with developmental delay, autism, and response to folinic acid. Neurology 2005, 64, 1088–1090. [Google Scholar] [CrossRef]
- Djukic, A. Folate-responsive neurologic diseases. Pediatr. Neurol. 2007, 37, 387–397. [Google Scholar] [CrossRef]
- Qiu, A.; Jansen, M.; Sakaris, A.; Min, S.H.; Chattopadhyay, S.; Tsai, E.; Sandoval, C.; Zhao, R.; Akabas, M.H.; Goldman, I.D. Identification of an intestinal folate transporter and the molecular basis for hereditary folate malabsorption. Cell 2006, 127, 917–928. [Google Scholar] [CrossRef]
- Grapp, M.; Just, I.A.; Linnankivi, T.; Wolf, P.; Lücke, T.; Häusler, M.; Gärtner, J.; Steinfeld, R. Molecular characterization of folate receptor 1 mutations delineates cerebral folate transport deficiency. Brain 2012, 135, 2022–2031. [Google Scholar] [CrossRef] [PubMed]
- Steinfeld, R.; Grapp, M.; Kraetzner, R.; Dreha-Kulaczewski, S.; Helms, G.; Dechent, P.; Wevers, R.; Grosso, S.; Gärtner, J. Folate receptor alpha defect causes cerebral folate transport deficiency: A treatable neurodegenerative disorder associated with disturbed myelin metabolism. Am. J. Hum. Genet. 2009, 85, 354–363. [Google Scholar] [CrossRef]
- Pérez-Duenas, B.; Ormazábal, A.; Toma, C.; Torrico, B.; Cormand, B.; Serrano, M.; Sierra, C.; De Grandis, E.; Marfa, M.P.; García-Cazorla, A.; et al. Cerebral folate deficiency syndromes in childhood: Clinical, analytical, and etiologic aspects. Arch. Neurol. 2011, 68, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Opladen, T.; Blau, N.; Ramaekers, V.T. Effect of antiepileptic drugs and reactive oxygen species on folate receptor 1 (FOLR1)-dependent 5-methyltetrahydrofolate transport. Mol. Genet. Metab. 2010, 101, 48–54. [Google Scholar] [CrossRef]
- Lan, F.; Bayliss, P.E.; Rinn, J.L.; Whetstine, J.R.; Wang, J.K.; Chen, S.; Iwase, S.; Alpatov, R.; Issaeva, I.; Canaani, E.; et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 2007, 449, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Wiles, E.T.; Selker, E.U. H3K27 methylation: A promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 2017, 43, 31–37. [Google Scholar] [CrossRef]
- De Santa, F.; Totaro, M.G.; Prosperini, E.; Notarbartolo, S.; Testa, G.; Natoli, G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007, 130, 1083–1094. [Google Scholar] [CrossRef]
- Stolerman, E.S.; Francisco, E.; Stallworth, J.L.; Jones, J.R.; Monaghan, K.G.; Keller-Ramey, J.; Person, R.; Wentzensen, I.M.; McWalter, K.; Keren, B.; et al. Genetic variants in the KDM6B gene are associated with neurodevelopmental delays and dysmorphic features. Am. J. Med. Genet. Part A 2019, 179, 1276–1286. [Google Scholar] [CrossRef]
- Cao, Z.; Shi, X.; Tian, F.; Fang, Y.; Wu, J.B.; Mrdenovic, S.; Nian, X.; Ji, J.; Xu, H.; Kong, C.; et al. KDM6B is an androgen regulated gene and plays oncogenic roles by demethylating H3K27me3 at cyclin D1 promoter in prostate cancer. Cell Death Dis. 2021, 12, 2. [Google Scholar] [CrossRef]
- Sequeira, J.M.; Ramaekers, V.T.; Quadros, E.V. The diagnostic utility of folate receptor autoantibodies in blood. Clin. Chem. Lab. Med. 2013, 51, 545–554. [Google Scholar] [CrossRef]
- Pasini, D.; Malatesta, M.; Jung, H.R.; Walfridsson, J.; Willer, A.; Olsson, L.; Skotte, J.; Wutz, A.; Porse, B.; Jensen, O.N.; et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010, 38, 4958–4969. [Google Scholar] [CrossRef] [PubMed]
- Ramaekers, V.T.; Rothenberg, S.P.; Sequeira, J.M.; Opladen, T.; Blau, N.; Quadros, E.V.; Selhub, J. Autoantibodies to folate receptors in the cerebral folate deficiency syndrome. N. Engl. J. Med. 2005, 352, 1985–1991. [Google Scholar] [CrossRef]
- Zhang, C.; Deng, X.; Wen, Y.; He, F.; Yin, F.; Peng, J. First case report of cerebral folate deficiency caused by a novel mutation of FOLR1 gene in a Chinese patient. BMC Med. Genet. 2020, 21, 2. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Dueñas, B.; Toma, C.; Ormazábal, A.; Muchart, J.; Sanmartí, F.; Bombau, G.; Serrano, M.; García-Cazorla, A.; Cormand, B.; Artuch, R. Progressive ataxia and myoclonic epilepsy in a patient with a homozygous mutation in the FOLR1 gene. J. Inherit. Metab. Dis. Off. J. Soc. Study Inborn Errors Metab. 2010, 33, 795–802. [Google Scholar] [CrossRef]
- Foda, B.M.; Singh, U. Dimethylated H3K27 is a repressive epigenetic histone mark in the protist entamoeba histolytica and is significantly enriched in genes silenced via the RNAi pathway. J. Biol. Chem. 2015, 290, 21114–21130. [Google Scholar] [CrossRef]
- Agger, K.; Cloos, P.A.; Rudkjær, L.; Williams, K.; Andersen, G.; Christensen, J.; Helin, K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A–ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009, 23, 1171–1176. [Google Scholar] [CrossRef]
- Juan, A.H.; Wang, S.; Ko, K.D.; Zare, H.; Tsai, P.-F.; Feng, X.; Vivanco, K.O.; Ascoli, A.M.; Gutierrez-Cruz, G.; Krebs, J.; et al. Roles of H3K27me2 and H3K27me3 examined during fate specification of embryonic stem cells. Cell Rep. 2016, 17, 1369–1382. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, Z.; Dong, Q.; Xiong, J.; Zhu, B. Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells. Genome Biol. 2020, 21, 45. [Google Scholar] [CrossRef]
- Marzi, S.; Leung, S.K.; Ribarska, T.; Hannon, E.; Smith, A.R.; Pishva, E.; Poschmann, J.; Moore, K.; Troakes, C.; Al-Sarraj, S.; et al. A histone acetylome-wide association study of Alzheimer’s disease identifies disease-associated H3K27ac differences in the entorhinal cortex. Nat. Neurosci. 2018, 21, 1618–1627. [Google Scholar] [CrossRef] [PubMed]
- Yoshitomi, R.; Nakayama, K.; Yamashita, S.; Kumazoe, M.; Lin, T.-A.; Mei, C.-Y.; Marugame, Y.; Fujimura, Y.; Maeda-Yamamoto, M.; Kuriyama, S.; et al. Plasma homocysteine concentration is associated with the expression level of folate receptor 3. Sci. Rep. 2020, 10, 10283. [Google Scholar] [CrossRef]
- Manna, S.; Kim, J.K.; Baugé, C.; Cam, M.; Zhao, Y.; Shetty, J.; Vacchio, M.S.; Castro, E.; Tran, B.; Tessarollo, L.; et al. Histone H3 Lysine 27 demethylases Jmjd3 and Utx are required for T-cell differentiation. Nat. Commun. 2015, 6, 8152. [Google Scholar] [CrossRef] [PubMed]
- Northrup, D.; Yagi, R.; Cui, K.; Proctor, W.R.; Wang, C.; Placek, K.; Pohl, L.R.; Wang, R.; Ge, K.; Zhu, J.; et al. Histone demethylases UTX and JMJD3 are required for NKT cell development in mice. Cell Biosci. 2017, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; Takeuchi, O.; Vandenbon, A.; Yasuda, K.; Tanaka, Y.; Kumagai, Y.; Miyake, T.; Matsushita, K.; Okazaki, T.; Saitoh, T.; et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 2010, 11, 936–944. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, H.; Hu, Y.; Liu, D.; Li, L.; Li, C.; Wang, Q.; Huo, J.; Liu, H.; Xie, N.; et al. Critical role of histone H3 lysine 27 demethylase Kdm6b in the homeostasis and function of medullary thymic epithelial cells. Cell Death Differ. 2020, 27, 2843–2855. [Google Scholar] [CrossRef] [PubMed]
| Features | Patient 1 * | Patient 2 | Patient 3 ** | Patient 4 *** | Patient 5 |
|---|---|---|---|---|---|
| Gender | F | M | F | F | F |
| Age (years at evaluation) | ¼ | 5 ½ | 4 | 11 ½ | 11 |
| KDM6B gene variant | c.C2282G p.T761S; c.G3047A p.R1016Q | c.C2722T p.R908C | c.C2722T p.R908C | c.C2477A p.S826Y | c.C1991T p.P664L |
| Pregnancy | Epilepsy, treated with VPA b + FA c | normal | normal | N/A a | Pre-eclampsia |
| Growth parameters Height Weight Head circumference | 75–90th 75th 50–70th | 3rd 3rd 3rd | 25th 25th <3rd | 3rd <3rd <3rd | N/A N/A 50th |
| Dysmorphic features | none | none | none | Coarse facial features | Facial; Pulmonary valve stenosis |
| Neurological features ° Unrest, insomnia ° Decelerating head growth ° Psychomotor delay and Regression ° Hypotonia and ataxia ° Pyramidal dysfunction ° Dyskinesias (chorea, athetosis) ° Epilepsy | + - + - + - - - | + + + + + + - + | - + - + - - + At 9 months ** Epileptic status and liver failure | + + + - - - - + | + N/A + + - - - + |
| Cognitive functions ° Language delays ° Intellectual disability | - - | + + | + + | + + | + + |
| Autism spectrum disorder | + | - | - | - | + |
| Neuro-imaging | Normal | LTBL d; partial recovery of white matter changes | Progressive cortical/cerebellar atrophy | Subcortical white matter lesions | N/A |
| Spinal fluid folate (nmol/L) | 14 | 24 | 38 | 30 | 34 |
| % of bottom reference CSF | 22.2% | 58.5% | 60.3% | 73.2% | 83% |
| Serum FRα antibodies | + | + | - | + | + |
| Start folinic acid treatment | 2 ½ months | 5 years | 3 ½ years | 11 ½ months | None |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, X.; Cao, X.; Cabrera, R.M.; Pimienta Ramirez, P.A.; Zhang, C.; Ramaekers, V.T.; Finnell, R.H.; Lei, Y. KDM6B Variants May Contribute to the Pathophysiology of Human Cerebral Folate Deficiency. Biology 2023, 12, 74. https://doi.org/10.3390/biology12010074
Han X, Cao X, Cabrera RM, Pimienta Ramirez PA, Zhang C, Ramaekers VT, Finnell RH, Lei Y. KDM6B Variants May Contribute to the Pathophysiology of Human Cerebral Folate Deficiency. Biology. 2023; 12(1):74. https://doi.org/10.3390/biology12010074
Chicago/Turabian StyleHan, Xiao, Xuanye Cao, Robert M. Cabrera, Paula Andrea Pimienta Ramirez, Cuilian Zhang, Vincent T. Ramaekers, Richard H. Finnell, and Yunping Lei. 2023. "KDM6B Variants May Contribute to the Pathophysiology of Human Cerebral Folate Deficiency" Biology 12, no. 1: 74. https://doi.org/10.3390/biology12010074
APA StyleHan, X., Cao, X., Cabrera, R. M., Pimienta Ramirez, P. A., Zhang, C., Ramaekers, V. T., Finnell, R. H., & Lei, Y. (2023). KDM6B Variants May Contribute to the Pathophysiology of Human Cerebral Folate Deficiency. Biology, 12(1), 74. https://doi.org/10.3390/biology12010074






