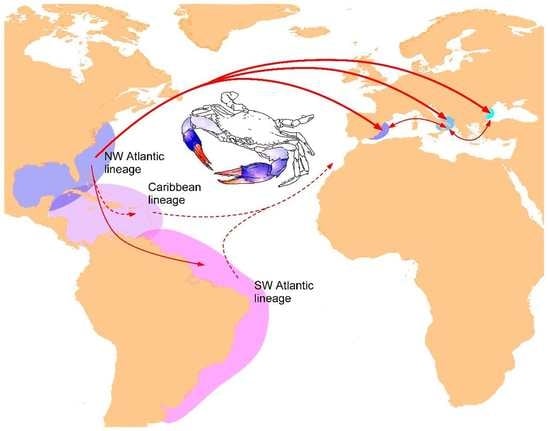
Phylogeography of the Atlantic Blue Crab Callinectes sapidus (Brachyura: Portunidae) in the Americas versus the Mediterranean Sea: Determining Origins and Genetic Connectivity of a Large-Scale Invasion
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
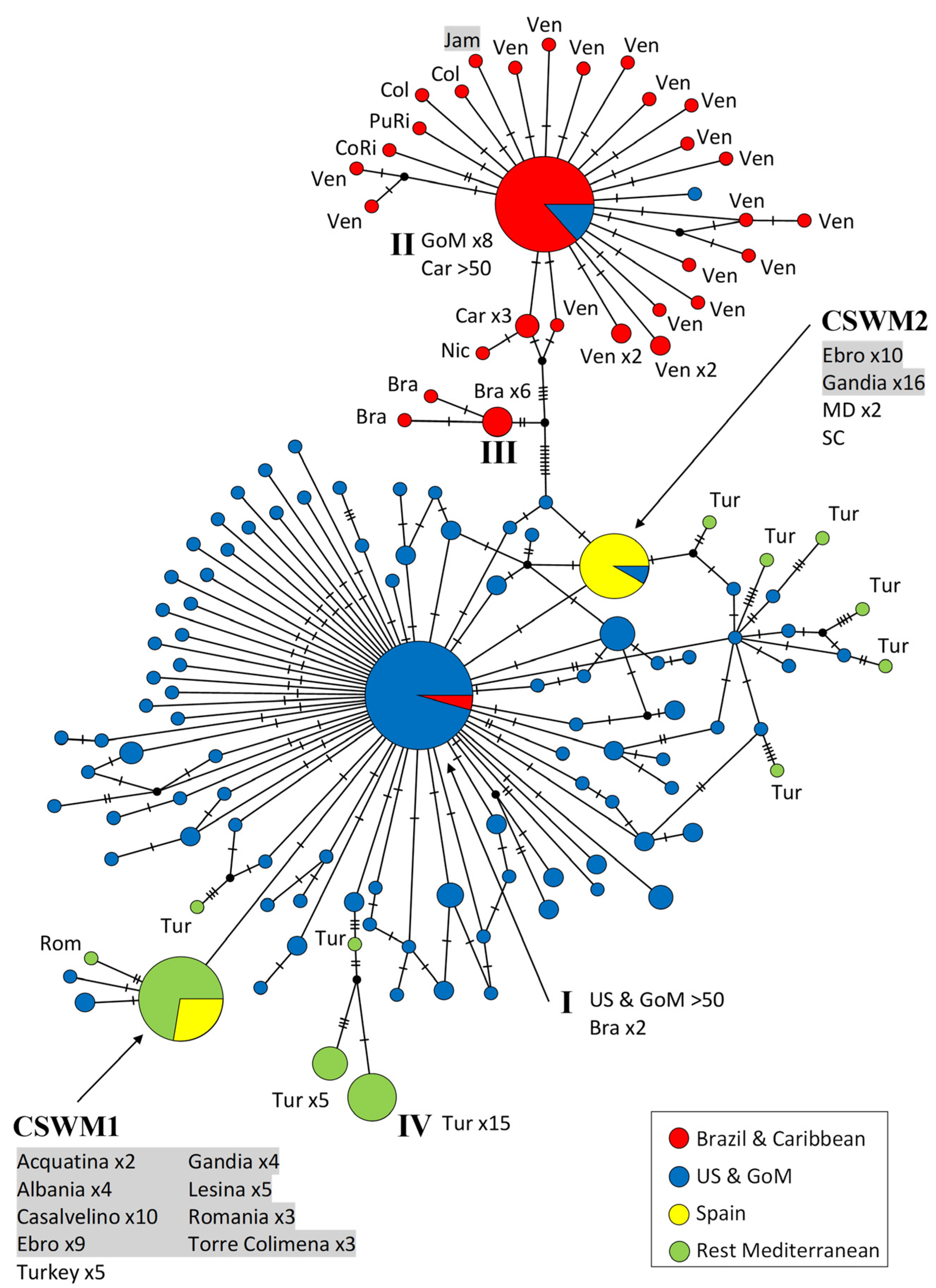
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mittermeier, R.A.; Turner, W.R.; Larsen, F.W.; Brooks, T.M.; Gascon, C. Global biodiversity conservation: The critical role of hotspots. In Biodiversity Hotspots: Distribution and Protection of Conservation Priority Areas; Zachos, F.E., Habel, J.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 3–22. [Google Scholar]
- Watson, J.E.M.; Shanahan, D.F.; DiMarco, M.; Allan, J.; Laurance, W.F.; Sanderson, E.W.; Mackey, B.; Venter, O. Catastrophic declines in wilderness areas undermine global environment targets. Curr. Biol. 2016, 26, 2929–2934. [Google Scholar] [CrossRef] [PubMed]
- Venter, O.; Sanderson, E.W.; Magrach, A.; Allan, J.R.; Beher, J.; Jones, K.R.; Possingham, H.P.; Laurance, W.F.; Wood, P.; Fekete, B.M.; et al. Global terrestrial human footprint maps for 1993 and 2009. Sci. Data 2016, 3, 160067. [Google Scholar] [CrossRef]
- Breithaupt, H. Aliens on the shores. EMBO Rep. 2003, 4, 547–550. [Google Scholar] [CrossRef] [PubMed]
- Mainka, S.A.; Howard, G.W. Climate change and invasive species: Double jeopardy. Integr. Zool. 2010, 5, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Doherty, T.S.; Glen, A.S.; Nimmo, D.G.; Ritchie, E.G.; Dickman, C.R. Invasive predators and global biodiversity loss. Proc. Natl. Acad. Sci. USA 2016, 113, 11261–11265. [Google Scholar] [CrossRef]
- Doherty, T.S.; Davis, R.A.; van Etten, E.J.B.; Algar, D.; Collier, N.; Dickman, C.R.; Edwards, G.; Masters, P.; Palmer, R.; Robinson, S. A continental-scale analysis of feral cat diet in Australia. J. Biogeogr. 2015, 42, 964–975. [Google Scholar] [CrossRef]
- Harris, D.B.; Macdonald, D.W. Interference competition between introduced black rats and endemic Galápagos rice rats. Ecology 2007, 88, 2330–2344. [Google Scholar] [CrossRef]
- Wyatt, K.B.; Campos, P.F.; Gilbert, M.T.P.; Kolokotronis, S.-O.; Hynes, W.H.; DeSalle, R.; Daszak, P.; MacPhee, R.D.E.; Greenwood, A.D. Historical mammal extinction on Christmas Island (Indian Ocean) correlates with introduced infectious disease. PLoS ONE 2008, 3, e3602. [Google Scholar] [CrossRef]
- Raitsos, D.E.; Beaugrand, G.; Georgopoulos, D.; Zenetos, A.; Pancucci-Papadopoulou, A.M.; Theocharis, A.; Papathanassiou, E. Global climate change amplifies the entry of tropical species into the Eastern Mediterranean Sea. Limnol. Oceanogr. 2010, 55, 1478–1484. [Google Scholar] [CrossRef]
- Occhipinti-Ambrogi, A.; Marchini, A.; Cantone, G.; Castelli, A.; Chimenz, C.; Cormaci, M.; Froglia, C.; Furnari, G.; Gambi, M.; Giaccone, G.; et al. Alien species along the Italian coasts: An overview. Biol. Invasions 2011, 13, 215–237. [Google Scholar] [CrossRef]
- Zenetos, A.; Gofas, S.; Morri, C.; Rosso, A.; Violanti, D.; Garcia Raso, J.E.; Cinar, M.E.; Almogi-Labin, A.; Ates, A.S.; Azzurro, E.; et al. Alien species in the Mediterranean Sea by 2012. A contribution to the application of European Union’s Marine Strategy Framework Directive (MSFD). Part 2. Introduction trends and pathways. Mediterr. Mar. Sci. 2012, 13, 328–352. [Google Scholar] [CrossRef]
- Coll, M.; Piroddi, C.; Steenbeek, J.; Kaschner, K.; Lasram, F.B.R.; Aguzzi, J.; Ballesteros, E.; Bianchi, C.N.; Corbera, J.; Dailianis, T. The biodiversity of the Mediterranean Sea: Estimates, patterns, and threats. PLoS ONE 2010, 5, e11842. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, G.M.; Carlton, J.T.; Grosholz, E.D.; Hines, A.H. Global invasions of marine and estuarine habitats by non-indigenous species: Mechanisms, extent, and consequences. Am. Zool. 1997, 37, 621–632. [Google Scholar] [CrossRef]
- Tsirintanis, K.; Azzurro, E.; Crocetta, F.; Dimiza, M.; Froglia, C.; Gerovasileiou, V.; Langeneck, J.; Mancinelli, G.; Rosso, A.; Stern, N.; et al. Bioinvasion impacts on biodiversity, ecosystem services, and human health in the Mediterranean Sea. Aquat. Invasions 2022, 17, 308–352. [Google Scholar] [CrossRef]
- Olenin, S.; Elliott, M.; Bysveen, I.; Culverhouse, P.F.; Daunys, D.; Dubelaar, G.B.J.; Gollasch, S.; Goulletquer, P.; Jelmert, A.; Kantor, Y.; et al. Recommendations on methods for the detection and control of biological pollution in marine coastal waters. Mar. Pollut. Bull. 2011, 62, 2598–2604. [Google Scholar] [CrossRef]
- Streftaris, N.; Zenetos, A. Alien marine species in the Mediterranean—The 100 ‘Worst Invasives’ and their impact. Mediterr. Mar. Sci. 2006, 7, 87–117. [Google Scholar] [CrossRef]
- Galil, B. A sea under siege—Alien species in the Mediterranean. Biol. Invasions 2000, 2, 177–186. [Google Scholar] [CrossRef]
- Galil, B.S. The alien crustaceans in the Mediterranean Sea: An historical review. In In the Wrong Place—Alien Marine Crustaceans: Distribution, Biology and Impacts; Galil, B.S., Clark, P.F., Carlton, J.T., Eds.; Invading Nature—Springer Series in Invasion Ecology; Springer: Berlin/Heidelberg, Germany, 2011; Volume 6, pp. 377–401. [Google Scholar]
- Zenetos, A.; Galanidi, M. Mediterranean non indigenous species at the start of the 2020s: Recent changes. Mar. Biodivers. Rec. 2020, 13, 10. [Google Scholar] [CrossRef]
- Williams, A.B. The swimming crabs of the genus Callinectes (Decapoda: Portunidae). Fish. Bull. 1974, 72, 685–798. [Google Scholar]
- Nehring, S. Invasion history and success of the American blue crab Callinectes sapidus in European and adjacent waters. In In the Wrong Place—Alien Marine Crustaceans: Distribution, Biology and Impacts; Galil, B.S., Clark, P.F., Carlton, J.T., Eds.; Invading Nature—Springer Series in Invasion Ecology; Springer: Berlin/Heidelberg, Germany, 2011; Volume 6, pp. 607–624. [Google Scholar]
- Millikin, M.R.; Williams, A.B. Synopsis of biological data on blue crab, Callinectes sapidus Rathbun. In FAO Fisheries Synopsis; FAO: Rome, Italy, 1984; Volume 38, pp. 1–39. [Google Scholar]
- Hines, A.H. Ecology of juvenile and adult blue crabs. In The Blue Crab: Callinectes sapidus; Kennedy, V.S., Cronin, L.E., Eds.; Maryland Sea Grant College: College Park, MD, USA, 2007; pp. 565–654. [Google Scholar]
- Mancinelli, G.; Bardelli, R.; Zenetos, A. A global occurrence database of the Atlantic blue crab Callinectes sapidus. Sci. Data 2021, 8, 111. [Google Scholar] [CrossRef]
- Scalici, M.; Chiesa, S.; Mancinelli, G.; Rontani, P.M.; Voccia, A.; Nonnis Marzano, F. Euryhaline aliens invading Italian inland waters: The case of the Atlantic blue crab Callinectes sapidus Rathbun, 1896. Appl. Sci. 2022, 12, 4666. [Google Scholar] [CrossRef]
- González-Ortegón, E.; Jenkins, S.; Galil, B.S.; Drake, P.; Cuesta, J.A. Accelerated invasion of decapod crustaceans in the southernmost point of the Atlantic coast of Europe: A non-natives’ hot spot? Biol. Invasions 2020, 22, 3487–3492. [Google Scholar] [CrossRef]
- Bouvier, E.L. Sur un Callinectes sapidus M. Rathbun trouvé à Rocheford. Bull. Mus. D’histoire Nat. Paris 1901, 7, 16–17. [Google Scholar]
- Giordani Soika, A. Il Neptunus pelagicus (L.) nell’alto Adriatico. Natura 1951, 42, 18–20. [Google Scholar]
- González-Ortegón, E.; Berger, S.; Encarnação, J.; Chairi, H.; Morais, P.; Teodósio, M.A.; Oliva-Paterna, F.J.; Schubart, C.D.; Cuesta, J.A. Free pass through the pillars of Hercules? Genetic and historical insights into the recent expansion of the Atlantic blue crab Callinectes sapidus to the West and the East of the Strait of Gibraltar. Front. Mar. Sci. 2022, 9, 1121. [Google Scholar] [CrossRef]
- Schubart, C.D. Mitochondrial DNA and decapod phylogenies: The importance of pseudogenes and primer optimization. In Decapod Crustacean Phylogenetics; Martin, J.W., Crandall, K.A., Felder, D.L., Eds.; Taylor & Francis/CRC Press: Boca Raton, FL, USA, 2009; Volume 18, pp. 47–65. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; pp. 95–98. [Google Scholar]
- Clement, M.; Snell, Q.; Walker, P.; Posada, D.; Crandall, K. TCS: Estimating gene genealogies. In Parallel and Distributed Processing Symposium, International; IEEE Computer Society: Washington, DC, USA, 2002; p. 184. [Google Scholar]
- Hijmans, R.J.; Guarino, L.; Cruz, M.; Rojas, E. Computer tools for spatial analysis of plant genetic resources data: 1. DIVA-GIS. Plant Genet. Resour. Newsl. 2001, 127, 15–19. [Google Scholar]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Knowles, L.L.; Futuyma, D.J.; Eanes, W.F.; Rannala, B. Insights into speciation mode from historical demography in the phytophagous beetle Ophraella. Evolution 1999, 53, 1846–1856. [Google Scholar] [PubMed]
- Hoffman, J.I.; Grant, S.M.; Forcada, J.; Phillips, C.D. Bayesian inference of a historical bottleneck in a heavily exploited marine mammal. Mol. Ecol. 2011, 20, 3989–4008. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.A.; Cestari Dumont, L.F.; Meira dos Santos, C.R.; D’Incao, F.; Weiss, S.; Froufe, E. Two distinct mtDNA lineages of the blue crab reveal large-scale population structure in its native Atlantic distribution. Estuar. Coast. Shelf Sci. 2017, 197, 45–53. [Google Scholar] [CrossRef]
- Santos, C.R.M.; D’Incao, F. Crustáceos no cerrito Ariano Souza, Rio Grande, Rio Grande do Sul e distribuição de Callinectes sapidus (Brachyura, Portunidae). Iheringia Série Zool. 2004, 94, 73–76. [Google Scholar] [CrossRef]
- Windsor, A.M.; Moore, M.K.; Warner, K.A.; Stadig, S.R.; Deeds, J.R. Evaluation of variation within the barcode region of Cytochrome c Oxidase I (COI) for the detection of commercial Callinectes sapidus Rathbun, 1896 (blue crab) products of non-US origin. PeerJ 2019, 7, e7827. [Google Scholar] [CrossRef]
- Suguío, K.; Martin, L.; Bittencourt, A.C.S.P.; Dominguez, J.M.L.; Flexor, J.-M.; de Azevedo, A.E.G. Flutuações do nível relativo do mar durante o Quaternário Superior ao longo do litoral brasileiro e suas implicações na sedimentação costeira. Rev. Bras. Geocienc. 1985, 15, 273–286. [Google Scholar] [CrossRef]
- McMillen-Jackson, A.L.; Bert, T.M.; Steele, P. Population genetics of the blue crab Callinectes sapidus: Modest population structuring in a background of high gene flow. Mar. Biol. 1994, 118, 53–65. [Google Scholar] [CrossRef]
- Yednock, B.K.; Neigel, J.E. An investigation of genetic population structure in blue crabs, Callinectes sapidus, using nuclear gene sequences. Mar. Biol. 2014, 161, 871–886. [Google Scholar] [CrossRef]
- Feng, X.; Williams, E.P.; Place, A.R. High genetic diversity and implications for determining population structure in the blue crab Callinectes sapidus. J. Shellfish Res. 2017, 36, 231–242. [Google Scholar] [CrossRef]
- Macedo, D.; Caballero, I.; Mateos, M.; Leblois, R.; McCay, S.; Hurtado, L.A. Population genetics and historical demographic inferences of the blue crab Callinectes sapidus in the US based on microsatellites. PeerJ 2019, 7, e7780. [Google Scholar] [CrossRef] [PubMed]
- García-Castellanos, D.; Estrada, F.; Jiménez-Munt, I.; Gorini, C.; Fernàndez, M.; Vergés, J.; De Vicente, R. Catastrophic flood of the Mediterranean after the Messinian Salinity Crisis. Nature 2009, 462, 778–781. [Google Scholar] [CrossRef] [PubMed]
- Cordero, D.; Peña, J.B.; Saavedra, C. Phylogeographic analysis of introns and mitochondrial DNA in the clam Ruditapes decussatus uncovers the effects of Pleistocene glaciations and endogenous barriers to gene flow. Mol. Phylogen. Evol. 2014, 71, 274–287. [Google Scholar] [CrossRef] [PubMed]
- Kordos, L.M.; Burton, R.S. Genetic differentiation of Texas Gulf Coast populations of the blue crab Callinectes sapidus. Mar. Biol. 1993, 117, 227–233. [Google Scholar] [CrossRef]
- Plough, L.V. Population genomic analysis of the blue crab Callinectes sapidus using genotyping-by-sequencing. J. Shellfish Res. 2017, 36, 249–261. [Google Scholar] [CrossRef]
- Clayton, S.; Dutkiewicz, S.; Jahn, O.; Follows, M.J. Dispersal, eddies, and the diversity of marine phytoplankton. Limnol. Oceanogr. Fluids Environ. 2013, 3, 182–197. [Google Scholar] [CrossRef]
- McMillen-Jackson, A.L.; Bert, T.M. Mitochondrial DNA variation and population genetic structure of the blue crab Callinectes sapidus in the eastern United States. Mar. Biol. 2004, 145, 769–777. [Google Scholar] [CrossRef]
- Lacerda, A.L.F.; Kersanach, R.; Cortinhas, M.C.S.; Prata, P.F.S.; Dumont, L.F.C.; Proietti, M.C.; Maggioni, R.; D’Incao, F. High connectivity among blue crab (Callinectes sapidus) populations in the Western South Atlantic. PLoS ONE 2016, 11, e0153124. [Google Scholar] [CrossRef]
- Piola, A.R.; Campos, E.J.D.; Matano, R.P.; Möller, O.O., Jr.; Palma, E.D. The influence of the Plata River discharge on the western South Atlantic Shelf. Geophys. Res. Lett. 2005, 32, 1–4. [Google Scholar] [CrossRef]
- Prakash, M. Fundamentals of Gene Evolution; Discovery Publishing House: New Delhi, India, 2007; p. 328. [Google Scholar]
- Darling, J.A.; Bagley, M.J.; Roman, J.O.E.; Tepolt, C.K.; Geller, J.B. Genetic patterns across multiple introductions of the globally invasive crab genus Carcinus. Mol. Ecol. 2008, 17, 4992–5007. [Google Scholar] [CrossRef]
- Zitari-Chatti, R.; Chatti, N.; Fulgione, D.; Caiazza, I.; Aprea, G.; Elouaer, A.; Said, K.; Capriglione, T. Mitochondrial DNA variation in the caramote prawn Penaeus (Melicertus) kerathurus across a transition zone in the Mediterranean Sea. Genetica 2009, 136, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Deli, T.; Chatti, N.; Said, K.; Schubart, C.D. Concordant patterns of mtDNA and nuclear phylogeographic structure reveal a Pleistocene vicariant event in the green crab Carcinus aestuarii across the Siculo-Tunisian Strait. Medit. Mar. Sci. 2016, 17, 533–551. [Google Scholar] [CrossRef][Green Version]
- Serra, I.A.; Innocenti, A.M.; Di Maida, G.; Calvo, S.; Migliaccio, M.; Zambianchi, E.; Pizzigalli, C.; Arnaud-Haond, S.; Duarte, C.M.; Serrao, E.A.; et al. Genetic structure in the Mediterranean seagrass Posidonia oceanica: Disentangling past vicariance events from contemporary patterns of gene flow. Mol. Ecol. 2010, 19, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Georgiadis, C.; Georgiadis, G. Zur Kenntnis der Crustacea Decapoda des Golfes von Thessaloniki. Crustaceana 1974, 26, 239–248. [Google Scholar] [CrossRef]
- Onofri, V.; Dulčić, J.; Conides, A.; Matić-Skoko, S.; Glamuzina, B. The occurrence of the blue crab, Callinectes sapidus Rathbun, 1896 (Decapoda, Brachyura, Portunidae) in the eastern Adriatic (Croatian coast). Crustaceana 2008, 81, 403–409. [Google Scholar]
- Öztürk, R.Ç.; Terzi, Y.; Feyzioğlu, A.M.; Şahin, A.; Aydın, M. Genetic characterization of the invasive Blue crab, Callinectes sapidus (Rathbun, 1896), in the Black Sea. Reg. Stud. Mar. Sci. 2020, 39, 101412. [Google Scholar] [CrossRef]
- Neumann, G. Ocean Currents; Elsevier: Amsterdam, The Netherlands, 2014; 352p. [Google Scholar]
- Astraldi, M.; Bianchi, C.N.; Gasparini, G.P.; Morri, C. Climatic fluctuations, current variability and marine species distribution-a case-study in the Ligurian sea (north-west Mediterranean). Oceanol. Acta 1995, 18, 139–149. [Google Scholar]
- Hines, A.H.; Johnson, E.G.; Darnell, M.Z.; Rittschof, D.; Miller, T.J.; Bauer, L.J.; Rodgers, P.; Aguilar, R. Predicting effects of climate change on blue crabs in Chesapeake Bay. In Biology and Management of Exploited Crab Populations under Climate Change; Kruse, G.H., Eckert, G.L., Foy, R.J., Lipcius, R.N., Sainte-Marie, B., Stram, D.L., Woodby, D., Eds.; University of Alaska: Fairbanks, AK, USA, 2010; pp. 109–127. [Google Scholar]
- Epifanio, C.E. Transport of crab larvae between estuaries and the continental shelf. In Coastal-Offshore Ecosystem Interactions; Springer: Berlin/Heidelberg, Germany, 1988; pp. 291–305. [Google Scholar]
- Morgan, S.G.; Zimmer-Faust, R.K.; Heck, K.L., Jr.; Coen, L.D. Population regulation of blue crabs Callinectes sapidus in the northern Gulf of Mexico: Postlarval supply. Mar. Ecol. Prog. Ser. 1996, 133, 73–88. [Google Scholar] [CrossRef]
- Forward, R.B.; Rittschof, D. Photoresponses of crab megalopae in offshore and estuarine waters: Implications for transport. J. Exp. Mar. Biol. Ecol. 1994, 182, 183–192. [Google Scholar] [CrossRef]
- Sulkin, S.; Van Heukelem, W.; Kelly, P.; Van Heukelem, L. The behavioral basis of larval recruitment in the crab Callinectes sapidus Rathbun: A laboratory investigation of ontogenetic changes in geotaxis and barokinesis. Biol. Bull. 1980, 159, 402–417. [Google Scholar] [CrossRef]



| Sampling Location | Extraction Number | Coordinates | N |
|---|---|---|---|
| Albania: Kavaja | T598 | 41.18, 19.48 | 4 |
| Brazil: Ubatuba | L30 | −23.44, −45.06 | 1 |
| Italy: Acquatina | T603 | 40.44, 18.24 | 2 |
| Italy: Casalvelino | T675 | 40.17, 15.12 | 10 |
| Italy: Lesina | T602 | 41.89, 15.45 | 5 |
| Italy: Torre Colimena | T603 | 40.29, 17.74 | 3 |
| Jamaica: Belmont | T589 | 18.16, −78.03 | 2 |
| Louisiana: Isles Dernieres | L5 | 29.06, −90.81 | 1 |
| Mexico: Tampico | T602-7 | 22.26, −97.78 | 1 |
| Romania: Constanta | C1-C3 | 44.18, 28.68 | 3 |
| Spain: Ebro | T650, T652 | 40.63, 0.74 | 19 |
| Spain: Gandía | T649 | 38.99, −0.15 | 20 |
| Total | 71 |
| Virginia | Maryland | Florida | South Carolina | Mexico | South America | |
|---|---|---|---|---|---|---|
| Virginia | -- | 0.025 | 0.015 | −0.008 | 0.003 | 0.075 ** |
| Maryland | 0.021 | -- | 0.024 * | 0.003 | 0.035 * | 0.062 ** |
| Florida | −0.029 | 0.047 * | -- | 0.006 | 0.01 | 0.064 ** |
| South Carolina | −0.018 | 0.013 | −0.002 | -- | 0.00 | 0.062 ** |
| Mexico | −0.027 | 0.027 | −0.027 | −0.007 | -- | 0.054 * |
| South America | 0.631 *** | 0.649 *** | 0.602 *** | 0.632 *** | 0.607 *** | -- |
| Virginia | Gandía | Ebro | Tyrrhenian | Eastern Basin | Turkey | |
|---|---|---|---|---|---|---|
| Virginia | -- | 0.358 *** | 0.258 *** | 0.441 *** | 0.464 *** | 0.307 *** |
| Gandía | 0.283 *** | -- | 0.109 | 0.731 *** | 0.718 *** | 0.620 *** |
| Ebro | 0.219 *** | 0.109 | -- | 0.416 * | 0.418 *** | 0.523 *** |
| Tyrrhenian | 0.450 *** | 0.731 *** | 0.416 * | -- | −0.036 | 0.736 *** |
| Eastern Basin | 0.519 *** | 0.763 *** | 0.476 *** | −0.036 | -- | 0.732 *** |
| Turkey | 0.766 *** | 0.867 *** | 0.844 *** | 0.89 *** | 0.91 *** | -- |
| Population | N | Nh | h | π | K | Tajima’s D | Fu’s FS |
| Native sampling sites | |||||||
| Virginia | 18 | 15 | 0.961 ± 0.039 | 0.0048 ± 0.0007 | 3.091 | −2.030 | −11.774 |
| Maryland | 19 | 14 | 0.953 ± 0.036 | 0.0037 ± 0.0004 | 2.385 | −1.660 | −10.898 |
| Florida | 12 | 12 | 1.000 ± 0.034 | 0.0053 ± 0.0005 | 3.393 | −1.723 | −10.5 |
| South Carolina | 18 | 16 | 0.987 ± 0.023 | 0.0045 ± 0.0005 | 2.895 | −1.957 | −15.081 |
| Mexico | 10 | 9 | 0.978 ± 0.054 | 0.0038 ± 0.0005 | 2.422 | −1.044 | −6.487 |
| South America | 17 | 10 | 0.875 ± 0.07 | 0.0147 ± 0.0033 | 9.382 | 0.381 | 0.46 |
| Overall data | 94 | 61 | 0.979 ± 0.007 | 0.0112 ± 0.0015 | 7.162 | −1.628 | −24.923 |
| Invaded sampling sites | |||||||
| Gandía | 20 | 2 | 0.337 ± 0.110 | 0.0015 ± 0.0005 | 1.01 | 0.52 | 2.973 |
| Ebro | 19 | 2 | 0.526 ± 0.040 | 0.0024 ± 0.0001 | 1.578 | 2.282 | 4.317 |
| Tyrrhenian | 10 | 1 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 | 0.00 | 0.00 |
| Eastern Basin | 18 | 2 | 0.111 ± 0.096 | 0.0001 ± 0.0001 | 0.111 | −1.164 | −0.794 |
| Turkey | 18 | 2 | 0.425 ± 0.099 | 0.0033 ± 0.0007 | 2.124 | 1.459 | 5.388 |
| Overall data | 85 | 5 | 0.666 ± 0.031 | 0.0078 ± 0.0008 | 5.026 | 1.393 | 9.783 |
| Genetic Diversity Indices | Statistical Assessment of Difference (One-Way ANOVA) between Native and Invaded Sampling Sites | |||
|---|---|---|---|---|
| Mean Squares between Groups | Mean Squares within Groups | F | p | |
| Haplotype diversity (h) | 1.257 | 0.022 | 56.12 | *** |
| Nucleotide diversity () | 0.00006 | 0.00001 | 5.417 | * |
| Mean nucleotide differences (K) | 23.953 | 4.425 | 5.413 | * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schubart, C.D.; Deli, T.; Mancinelli, G.; Cilenti, L.; Gil Fernández, A.; Falco, S.; Berger, S. Phylogeography of the Atlantic Blue Crab Callinectes sapidus (Brachyura: Portunidae) in the Americas versus the Mediterranean Sea: Determining Origins and Genetic Connectivity of a Large-Scale Invasion. Biology 2023, 12, 35. https://doi.org/10.3390/biology12010035
Schubart CD, Deli T, Mancinelli G, Cilenti L, Gil Fernández A, Falco S, Berger S. Phylogeography of the Atlantic Blue Crab Callinectes sapidus (Brachyura: Portunidae) in the Americas versus the Mediterranean Sea: Determining Origins and Genetic Connectivity of a Large-Scale Invasion. Biology. 2023; 12(1):35. https://doi.org/10.3390/biology12010035
Chicago/Turabian StyleSchubart, Christoph D., Temim Deli, Giorgio Mancinelli, Lucrezia Cilenti, Alberto Gil Fernández, Silvia Falco, and Selina Berger. 2023. "Phylogeography of the Atlantic Blue Crab Callinectes sapidus (Brachyura: Portunidae) in the Americas versus the Mediterranean Sea: Determining Origins and Genetic Connectivity of a Large-Scale Invasion" Biology 12, no. 1: 35. https://doi.org/10.3390/biology12010035
APA StyleSchubart, C. D., Deli, T., Mancinelli, G., Cilenti, L., Gil Fernández, A., Falco, S., & Berger, S. (2023). Phylogeography of the Atlantic Blue Crab Callinectes sapidus (Brachyura: Portunidae) in the Americas versus the Mediterranean Sea: Determining Origins and Genetic Connectivity of a Large-Scale Invasion. Biology, 12(1), 35. https://doi.org/10.3390/biology12010035