Network Neuroscience Untethered: Brain-Wide Immediate Early Gene Expression for the Analysis of Functional Connectivity in Freely Behaving Animals
Abstract
Simple Summary
Abstract
1. Introduction
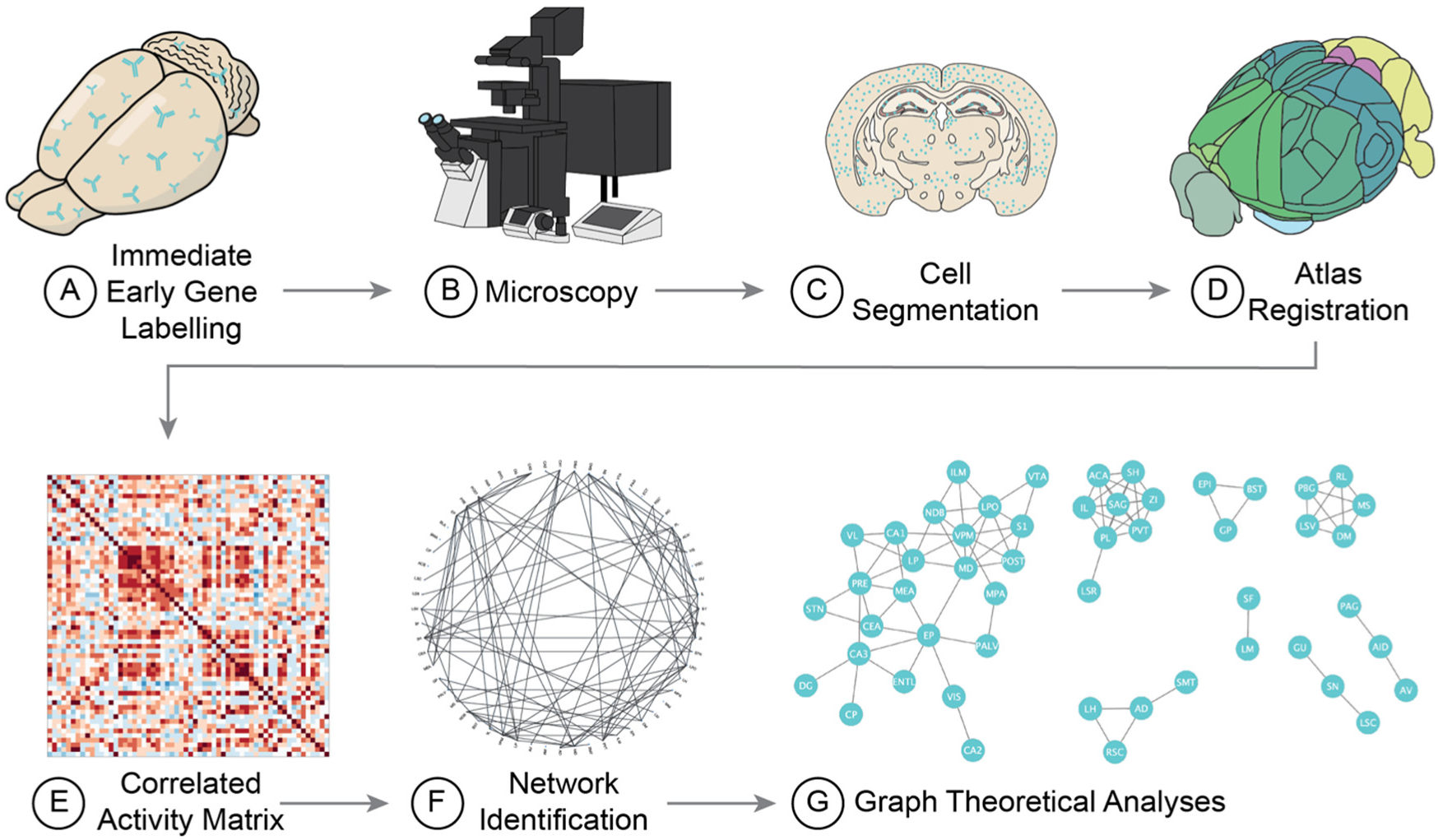
2. How to Run the Analysis
2.1. Behavioural Tasks
2.2. Histology
2.3. Imaging
2.4. Label Segmentation
2.5. Image Registration
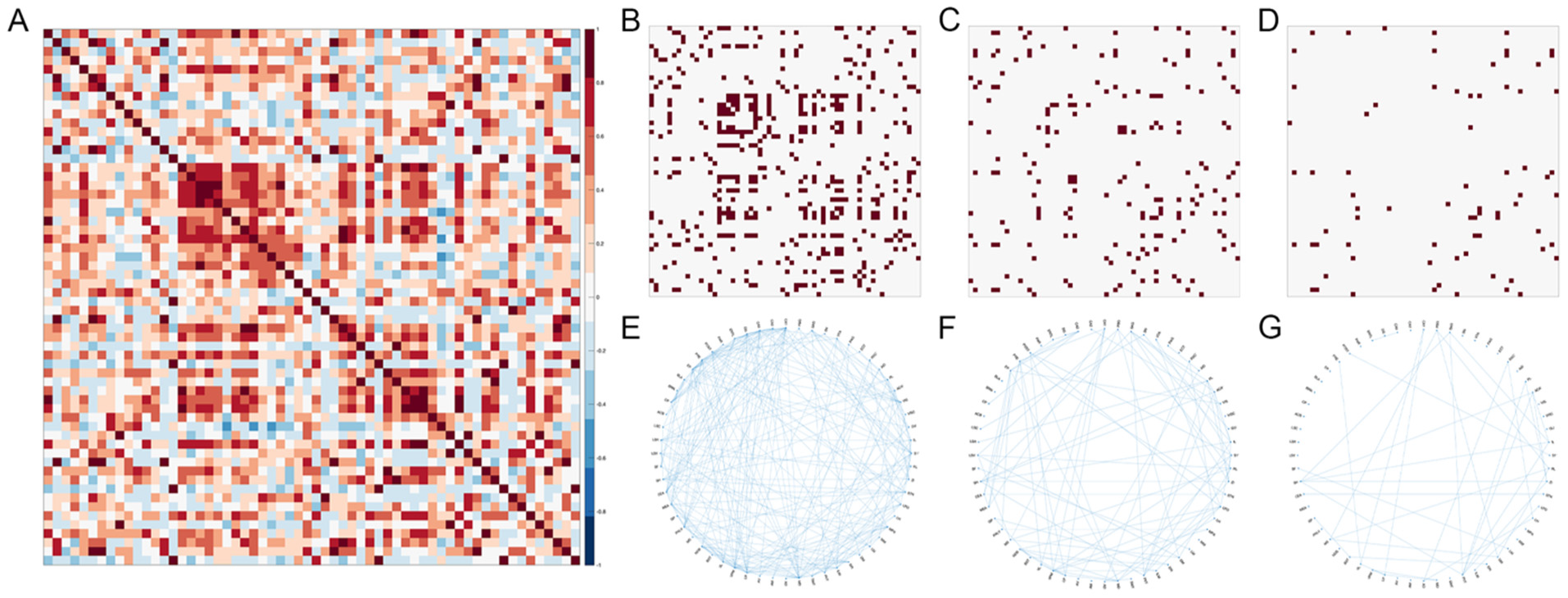
2.6. Network Analyses
3. Critical Considerations
3.1. Behavioural Tasks
3.2. Histology
3.3. Imaging
3.4. Label Segmentation
3.5. Image Registration
3.6. Network Analyses
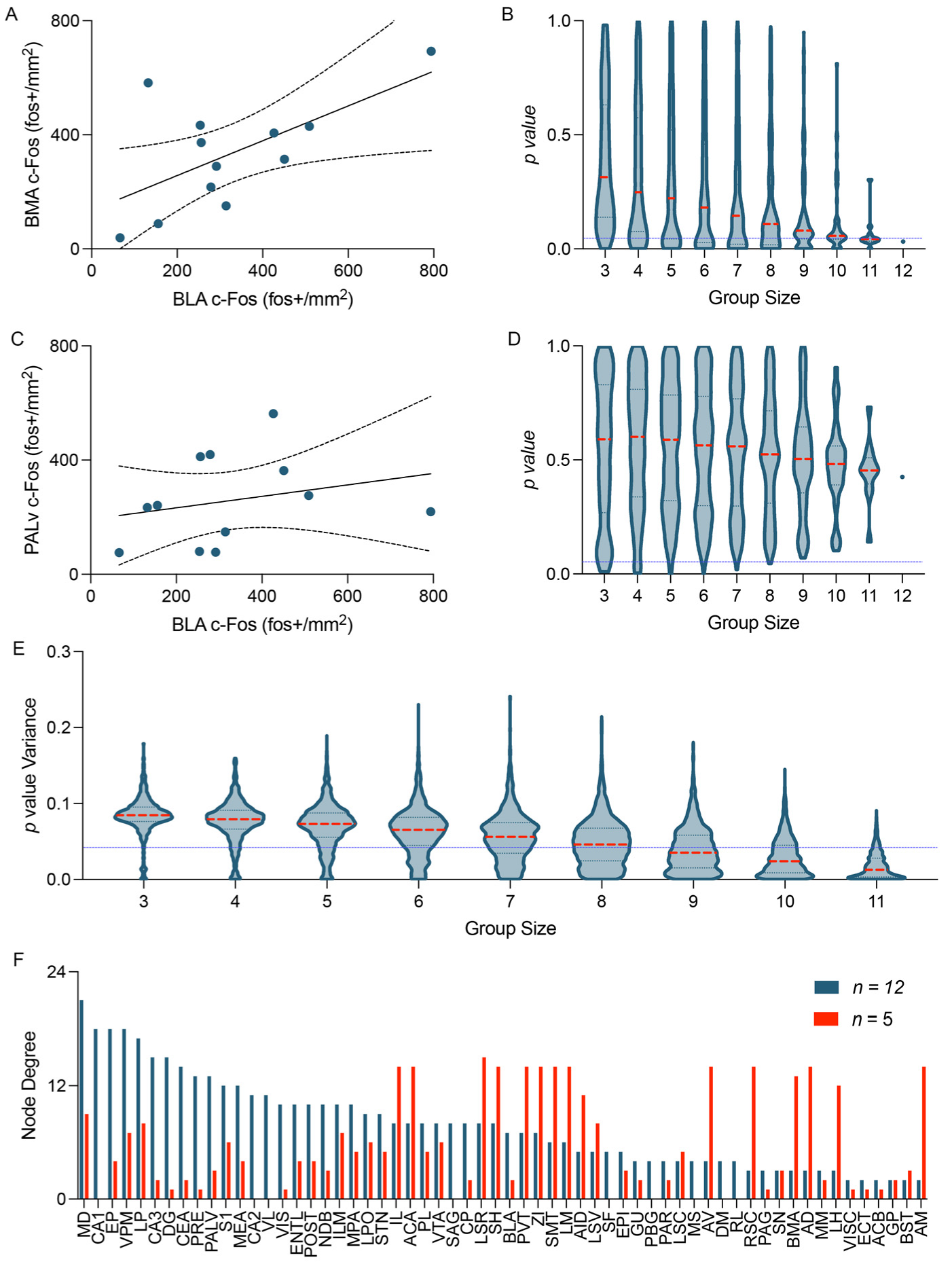
3.6.1. Group Size
3.6.2. Network Thresholding
4. Future Directions
5. Conclusions
6. Methods
6.1. Mice
6.2. Contextual Fear Conditioning
6.3. Perfusions and Histology
6.4. Brain-Wide c-Fos Quantification
6.5. Functional Connectivity Network Generation and Analysis
6.6. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, H.-J.; Friston, K. Structural and Functional Brain Networks: From Connections to Cognition. Science 2013, 342, 1238411. [Google Scholar] [CrossRef] [PubMed]
- Bandettini, P.A.; Wong, E.C.; Hinks, R.S.; Tikofsky, R.S.; Hyde, J.S. Time Course EPI of Human Brain Function during Task Activation. Magn. Reson. Med. 1992, 25, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Kwong, K.K.; Belliveau, J.W.; Chesler, D.A.; Goldberg, I.E.; Weisskoff, R.M.; Poncelet, B.P.; Kennedy, D.N.; Hoppel, B.E.; Cohen, M.S.; Turner, R. Dynamic Magnetic Resonance Imaging of Human Brain Activity during Primary Sensory Stimulation. Proc. Natl. Acad. Sci. USA 1992, 89, 5675–5679. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, S.; Lee, T.M.; Kay, A.R.; Tank, D.W. Brain Magnetic Resonance Imaging with Contrast Dependent on Blood Oxygenation. Proc. Natl. Acad. Sci. USA 1990, 87, 9868–9872. [Google Scholar] [CrossRef] [PubMed]
- Jonckers, E.; Van Audekerke, J.; De Visscher, G.; Van der Linden, A.; Verhoye, M. Functional Connectivity FMRI of the Rodent Brain: Comparison of Functional Connectivity Networks in Rat and Mouse. PLoS ONE 2011, 6, e18876. [Google Scholar] [CrossRef]
- Pawela, C.P.; Biswal, B.B.; Cho, Y.R.; Kao, D.S.; Li, R.; Jones, S.R.; Schulte, M.L.; Matloub, H.S.; Hudetz, A.G.; Hyde, J.S. Resting-State Functional Connectivity of the Rat Brain. Magn. Reson. Med. 2008, 59, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
- Klioutchnikov, A.; Wallace, D.J.; Frosz, M.H.; Zeltner, R.; Sawinski, J.; Pawlak, V.; Voit, K.-M.; Russell, P.S.J.; Kerr, J.N.D. Three-Photon Head-Mounted Microscope for Imaging Deep Cortical Layers in Freely Moving Rats. Nat. Methods 2020, 17, 509–513. [Google Scholar] [CrossRef]
- Klioutchnikov, A.; Wallace, D.J.; Sawinski, J.; Voit, K.-M.; Groemping, Y.; Kerr, J.N.D. A Three-Photon Head-Mounted Microscope for Imaging All Layers of Visual Cortex in Freely Moving Mice. Nat. Methods 2022. [Google Scholar] [CrossRef]
- Vanni, M.P.; Chan, A.W.; Balbi, M.; Silasi, G.; Murphy, T.H. Mesoscale Mapping of Mouse Cortex Reveals Frequency-Dependent Cycling between Distinct Macroscale Functional Modules. J. Neurosci. 2017, 37, 7513–7533. [Google Scholar] [CrossRef]
- Demas, J.; Manley, J.; Tejera, F.; Barber, K.; Kim, H.; Traub, F.M.; Chen, B.; Vaziri, A. High-Speed, Cortex-Wide Volumetric Recording of Neuroactivity at Cellular Resolution Using Light Beads Microscopy. Nat. Methods 2021, 18, 1103–1111. [Google Scholar] [CrossRef]
- Mohajerani, M.H.; Chan, A.W.; Mohsenvand, M.; LeDue, J.; Liu, R.; McVea, D.A.; Boyd, J.D.; Wang, Y.T.; Reimers, M.; Murphy, T.H. Spontaneous Cortical Activity Alternates between Motifs Defined by Regional Axonal Projections. Nat. Neurosci. 2013, 16, 1426–1435. [Google Scholar] [CrossRef]
- Greicius, M. Resting-State Functional Connectivity in Neuropsychiatric Disorders. Curr. Opin. Neurol. 2008, 21, 424–430. [Google Scholar] [CrossRef]
- Caviezel, M.P.; Reichert, C.F.; Sadeghi Bahmani, D.; Linnemann, C.; Liechti, C.; Bieri, O.; Borgwardt, S.; Leyhe, T.; Melcher, T. The Neural Mechanisms of Associative Memory Revisited: FMRI Evidence from Implicit Contingency Learning. Front. Psychiatry 2019, 10, 1002. [Google Scholar] [CrossRef]
- Stark, C.E.; Squire, L.R. Functional Magnetic Resonance Imaging (FMRI) Activity in the Hippocampal Region during Recognition Memory. J. Neurosci. 2000, 20, 7776–7781. [Google Scholar] [CrossRef]
- Guzowski, J.F.; Timlin, J.A.; Roysam, B.; McNaughton, B.L.; Worley, P.F.; Barnes, C.A. Mapping Behaviorally Relevant Neural Circuits with Immediate-Early Gene Expression. Curr. Opin. Neurobiol. 2005, 15, 599–606. [Google Scholar] [CrossRef]
- Stone, S.S.D.; Teixeira, C.M.; Zaslavsky, K.; Wheeler, A.L.; Martinez-Canabal, A.; Wang, A.H.; Sakaguchi, M.; Lozano, A.M.; Frankland, P.W. Functional Convergence of Developmentally and Adult-Generated Granule Cells in Dentate Gyrus Circuits Supporting Hippocampus-Dependent Memory. Hippocampus 2011, 21, 1348–1362. [Google Scholar] [CrossRef]
- Wheeler, A.L.; Teixeira, C.M.; Wang, A.H.; Xiong, X.; Kovacevic, N.; Lerch, J.P.; McIntosh, A.R.; Parkinson, J.; Frankland, P.W. Identification of a Functional Connectome for Long-Term Fear Memory in Mice. PLoS Comput. Biol. 2013, 9, e1002853. [Google Scholar] [CrossRef]
- McIntosh, A.R. Mapping Cognition to the Brain through Neural Interactions. Memory 1999, 7, 523–548. [Google Scholar] [CrossRef]
- Horwitz, B.; McIntosh, A.R.; Haxby, J.V.; Grady, C.L. Network Analysis of Brain Cognitive Function Using Metabolic and Blood Flow Data. Behav. Brain Res. 1995, 66, 187–193. [Google Scholar] [CrossRef]
- Vetere, G.; Kenney, J.W.; Tran, L.M.; Xia, F.; Steadman, P.E.; Parkinson, J.; Josselyn, S.A.; Frankland, P.W. Chemogenetic Interrogation of a Brain-Wide Fear Memory Network in Mice. Neuron 2017, 94, 363–374.e4. [Google Scholar] [CrossRef]
- Scott, G.A.; Terstege, D.J.; Vu, A.P.; Law, S.; Evans, A.; Epp, J.R. Disrupted Neurogenesis in Germ-Free Mice: Effects of Age and Sex. Front. Cell Dev. Biol. 2020, 8, 407. [Google Scholar] [CrossRef] [PubMed]
- du Plessis, K.C.; Basu, S.; Rumbell, T.H.; Lucas, E.K. Sex-Specific Neural Networks of Cued Threat Conditioning: A Pilot Study. Front. Syst. Neurosci. 2022, 16, 832484. [Google Scholar] [CrossRef]
- Silva, B.A.; Burns, A.M.; Gräff, J. A CFos Activation Map of Remote Fear Memory Attenuation. Psychopharmacology 2019, 236, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Besnard, A.; Gao, Y.; TaeWoo Kim, M.; Twarkowski, H.; Reed, A.K.; Langberg, T.; Feng, W.; Xu, X.; Saur, D.; Zweifel, L.S.; et al. Dorsolateral Septum Somatostatin Interneurons Gate Mobility to Calibrate Context-Specific Behavioral Fear Responses. Nat. Neurosci. 2019, 22, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Rosier, M.; Le Barillier, L.; Meunier, D.; El Yacoubi, M.; Malleret, G.; Salin, P.-A. Post-Learning Paradoxical Sleep Deprivation Impairs Reorganization of Limbic and Cortical Networks Associated with Consolidation of Remote Contextual Fear Memory in Mice. Sleep 2018, 41, zsy188. [Google Scholar] [CrossRef]
- Coelho, C.A.O.; Ferreira, T.L.; Kramer-Soares, J.C.; Sato, J.R.; Oliveira, M.G.M. Network Supporting Contextual Fear Learning after Dorsal Hippocampal Damage Has Increased Dependence on Retrosplenial Cortex. PLoS Comput. Biol. 2018, 14, e1006207. [Google Scholar] [CrossRef]
- Leal Santos, S.; Stackmann, M.; Muñoz Zamora, A.; Mastrodonato, A.; De Landri, A.V.; Vaughan, N.; Chen, B.K.; Lanio, M.; Denny, C.A. Propranolol Decreases Fear Expression by Modulating Fear Memory Traces. Biol. Psychiatry 2021, 89, 1150–1161. [Google Scholar] [CrossRef]
- Dos Santos Corrêa, M.; Grisanti, G.D.V.; Franciscatto, I.A.F.; Tarumoto, T.S.A.; Tiba, P.A.; Ferreira, T.L.; Fornari, R.V. Remote Contextual Fear Retrieval Engages Activity from Salience Network Regions in Rats. Neurobiol. Stress 2022, 18, 100459. [Google Scholar] [CrossRef]
- Terstege, D.J.; Durante, I.M.; Epp, J.R. Brain-Wide Neuronal Activation and Functional Connectivity Are Modulated by Prior Exposure to Repetitive Learning Episodes. Front. Behav. Neurosci. 2022, 16, 907707. [Google Scholar] [CrossRef]
- Kimbrough, A.; Lurie, D.J.; Collazo, A.; Kreifeldt, M.; Sidhu, H.; Macedo, G.C.; D’Esposito, M.; Contet, C.; George, O. Brain-Wide Functional Architecture Remodeling by Alcohol Dependence and Abstinence. Proc. Natl. Acad. Sci. USA 2020, 117, 2149–2159. [Google Scholar] [CrossRef]
- Walker, L.C.; Kastman, H.E.; Lawrence, A.J. Pattern of Neural Activation Following Yohimbine-Induced Reinstatement of Alcohol Seeking in Rats. Eur. J. Neurosci. 2020, 51, 706–720. [Google Scholar] [CrossRef]
- Borcuk, C.; Héraud, C.; Herbeaux, K.; Diringer, M.; Panzer, É.; Scuto, J.; Hashimoto, S.; Saido, T.C.; Saito, T.; Goutagny, R.; et al. Early Memory Deficits and Extensive Brain Network Disorganization in the App/MAPT Double Knock-in Mouse Model of Familial Alzheimer’s Disease. Aging Brain 2022, 2, 100042. [Google Scholar] [CrossRef]
- Worley, N.B.; Everett, S.R.; Foilb, A.R.; Christianson, J.P. Functional Networks Activated by Controllable and Uncontrollable Stress in Male and Female Rats. Neurobiol. Stress 2020, 13, 100233. [Google Scholar] [CrossRef]
- Sampedro-Piquero, P.; Álvarez-Suárez, P.; Moreno-Fernández, R.D.; García-Castro, G.; Cuesta, M.; Begega, A. Environmental Enrichment Results in Both Brain Connectivity Efficiency and Selective Improvement in Different Behavioral Tasks. Neuroscience 2018, 388, 374–383. [Google Scholar] [CrossRef]
- Bernanke, A.; Burnette, E.; Murphy, J.; Hernandez, N.; Zimmerman, S.; Walker, Q.D.; Wander, R.; Sette, S.; Reavis, Z.; Francis, R.; et al. Behavior and Fos Activation Reveal That Male and Female Rats Differentially Assess Affective Valence during CTA Learning and Expression. PLoS ONE 2021, 16, e0260577. [Google Scholar] [CrossRef]
- Ben-Ami Bartal, I.; Breton, J.M.; Sheng, H.; Long, K.L.; Chen, S.; Halliday, A.; Kenney, J.W.; Wheeler, A.L.; Frankland, P.; Shilyansky, C.; et al. Neural Correlates of Ingroup Bias for Prosociality in Rats. eLife 2021, 10, e65582. [Google Scholar] [CrossRef]
- Rogers-Carter, M.M.; Varela, J.A.; Gribbons, K.B.; Pierce, A.F.; McGoey, M.T.; Ritchey, M.; Christianson, J.P. Insular Cortex Mediates Approach and Avoidance Responses to Social Affective Stimuli. Nat. Neurosci. 2018, 21, 404–414. [Google Scholar] [CrossRef]
- Tyebji, S.; Seizova, S.; Garnham, A.L.; Hannan, A.J.; Tonkin, C.J. Impaired Social Behaviour and Molecular Mediators of Associated Neural Circuits during Chronic Toxoplasma Gondii Infection in Female Mice. Brain Behav. Immun. 2019, 80, 88–108. [Google Scholar] [CrossRef]
- Gossman, K.R.; Dykstra, B.; García, B.H.; Swopes, A.P.; Kimbrough, A.; Smith, A.S. Pair Bond-Induced Affiliation and Aggression in Male Prairie Voles Elicit Distinct Functional Connectivity in the Social Decision-Making Network. Front. Neurosci. 2021, 15, 748431. [Google Scholar] [CrossRef]
- Tanimizu, T.; Kenney, J.W.; Okano, E.; Kadoma, K.; Frankland, P.W.; Kida, S. Functional Connectivity of Multiple Brain Regions Required for the Consolidation of Social Recognition Memory. J. Neurosci. 2017, 37, 4103–4116. [Google Scholar] [CrossRef]
- Pilarzyk, K.; Klett, J.; Pena, E.A.; Porcher, L.; Smith, A.J.; Kelly, M.P. Loss of Function of Phosphodiesterase 11A4 Shows That Recent and Remote Long-Term Memories Can Be Uncoupled. Curr. Biol. 2019, 29, 2307–2321.e5. [Google Scholar] [CrossRef]
- Hodges, T.E.; Lee, G.Y.; Noh, S.H.; Galea, L.A.M. Sex and Age Differences in Cognitive Bias and Neural Activation in Response to Cognitive Bias Testing. Neurobiol. Stress 2022, 18, 100458. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Couz, M.; Conejo, N.M.; González-Pardo, H.; Arias, J.L. Functional Interactions between Dentate Gyrus, Striatum and Anterior Thalamic Nuclei on Spatial Memory Retrieval. Brain Res. 2015, 1605, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Cholvin, T.; Muller, M.A.; Cosquer, B.; Kelche, C.; Cassel, J.-C.; Pereira de Vasconcelos, A. Environmental Enrichment Enhances Systems-Level Consolidation of a Spatial Memory after Lesions of the Ventral Midline Thalamus. Neurobiol. Learn. Mem. 2017, 141, 108–123. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Fernández, R.D.; Pérez-Martín, M.; Castilla-Ortega, E.; Rosell del Valle, C.; García-Fernández, M.I.; Chun, J.; Estivill-Torrús, G.; Rodríguez de Fonseca, F.; Santín, L.J.; Pedraza, C. MaLPA1-Null Mice as an Endophenotype of Anxious Depression. Transl. Psychiatry 2017, 7, e1077. [Google Scholar] [CrossRef]
- Takeuchi, T.; Tamura, M.; Tse, D.; Kajii, Y.; Fernández, G.; Morris, R.G.M. Brain Region Networks for the Assimilation of New Associative Memory into a Schema. Mol. Brain 2022, 15, 24. [Google Scholar] [CrossRef]
- Yagi, S.; Lee, A.; Truter, N.; Galea, L.A.M. Sex Differences in Contextual Pattern Separation, Neurogenesis, and Functional Connectivity within the Limbic System. Biol. Sex Differ. 2022, 13, 42. [Google Scholar] [CrossRef]
- Morgan, J.I.; Curran, T. Stimulus-Transcription Coupling in the Nervous System: Involvement of the Inducible Proto-Oncogenes Fos and Jun. Annu. Rev. Neurosci. 1991, 14, 421–451. [Google Scholar] [CrossRef]
- Sheng, M.; Greenberg, M.E. The Regulation and Function of C-Fos and Other Immediate Early Genes in the Nervous System. Neuron 1990, 4, 477–485. [Google Scholar] [CrossRef]
- Zangenehpour, S.; Chaudhuri, A. Differential Induction and Decay Curves of C-Fos and Zif268 Revealed through Dual Activity Maps. Brain Res. Mol. Brain Res. 2002, 109, 221–225. [Google Scholar] [CrossRef]
- Kimbrough, A.; Kallupi, M.; Smith, L.C.; Simpson, S.; Collazo, A.; George, O. Characterization of the Brain Functional Architecture of Psychostimulant Withdrawal Using Single-Cell Whole-Brain Imaging. eNeuro 2021, 8, ENEURO.0208-19.2021. [Google Scholar] [CrossRef] [PubMed]
- Cruces-Solis, H.; Nissen, W.; Ferger, B.; Arban, R. Whole-Brain Signatures of Functional Connectivity after Bidirectional Modulation of the Dopaminergic System in Mice. Neuropharmacology 2020, 178, 108246. [Google Scholar] [CrossRef] [PubMed]
- Brambilla-Pisoni, C.; Muñoz-Moreno, E.; Gallego-Amaro, I.; Maldonado, R.; Ivorra, A.; Soria, G.; Ozaita, A. Auricular Transcutaneous Vagus Nerve Stimulation Acutely Modulates Brain Connectivity in Mice. Front. Cell. Neurosci. 2022, 16, 856855. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-Y.; Cho, S.-J.; Lee, S.-H.; Ryu, Y.; Jang, J.-H.; Kim, S.-N.; Park, H.-J. Peripheral ERK Modulates Acupuncture-Induced Brain Neural Activity and Its Functional Connectivity. Sci. Rep. 2021, 11, 5128. [Google Scholar] [CrossRef] [PubMed]
- Moretti, J.; Terstege, D.J.; Poh, E.Z.; Epp, J.R.; Rodger, J. Low Intensity Repetitive Transcranial Magnetic Stimulation Modulates Brain-Wide Functional Connectivity to Promote Anti-Correlated c-Fos Expression. Sci. Rep. 2022, 12, 20571. [Google Scholar] [CrossRef]
- Rinaldi, A.; De Leonibus, E.; Cifra, A.; Torromino, G.; Minicocci, E.; De Sanctis, E.; López-Pedrajas, R.M.; Oliverio, A.; Mele, A. Flexible Use of Allocentric and Egocentric Spatial Memories Activates Differential Neural Networks in Mice. Sci. Rep. 2020, 10, 11338. [Google Scholar] [CrossRef]
- Wheeler, A.L.; Creed, M.C.; Voineskos, A.N.; Nobrega, J.N. Changes in Brain Functional Connectivity after Chronic Haloperidol in Rats: A Network Analysis. Int. J. Neuropsychopharmacol. 2014, 17, 1129–1138. [Google Scholar] [CrossRef]
- Penner, M.R.; Parrish, R.R.; Hoang, L.T.; Roth, T.L.; Lubin, F.D.; Barnes, C.A. Age-Related Changes InEgr1 Transcription and DNA Methylation within the Hippocampus. Hippocampus 2016, 26, 1008–1020. [Google Scholar] [CrossRef]
- Farina, F.R.; Commins, S. Differential Expression of Immediate Early Genes Zif268 and C-Fos in the Hippocampus and Prefrontal Cortex Following Spatial Learning and Glutamate Receptor Antagonism. Behav. Brain Res. 2016, 307, 194–198. [Google Scholar] [CrossRef]
- Duclot, F.; Kabbaj, M. The Role of Early Growth Response 1 (EGR1) in Brain Plasticity and Neuropsychiatric Disorders. Front. Behav. Neurosci. 2017, 11, 35. [Google Scholar] [CrossRef]
- Rockel, J.S.; Bernier, S.M.; Leask, A. Egr-1 Inhibits the Expression of Extracellular Matrix Genes in Chondrocytes by TNFalpha-Induced MEK/ERK Signalling. Arthritis Res. Ther. 2009, 11, R8. [Google Scholar] [CrossRef]
- Kelly, M.P.; Deadwyler, S.A. Experience-Dependent Regulation of the Immediate-Early Gene Arc Differs across Brain Regions. J. Neurosci. 2003, 23, 6443–6451. [Google Scholar] [CrossRef] [PubMed]
- Rial Verde, E.M.; Lee-Osbourne, J.; Worley, P.F.; Malinow, R.; Cline, H.T. Increased Expression of the Immediate-Early Gene Arc/Arg3.1 Reduces AMPA Receptor-Mediated Synaptic Transmission. Neuron 2006, 52, 461–474. [Google Scholar] [CrossRef] [PubMed]
- Okuno, H.; Akashi, K.; Ishii, Y.; Yagishita-Kyo, N.; Suzuki, K.; Nonaka, M.; Kawashima, T.; Fujii, H.; Takemoto-Kimura, S.; Abe, M.; et al. Inverse Synaptic Tagging of Inactive Synapses via Dynamic Interaction of Arc/Arg3.1 with CaMKIIβ. Cell 2012, 149, 886–898. [Google Scholar] [CrossRef] [PubMed]
- Guzowski, J.F.; Setlow, B.; Wagner, E.K.; McGaugh, J.L. Experience-Dependent Gene Expression in the Rat Hippocampus after Spatial Learning: A Comparison of the Immediate-Early Genes Arc, c-Fos, and Zif268. J. Neurosci. 2001, 21, 5089–5098. [Google Scholar] [CrossRef]
- Barry, D.N.; Coogan, A.N.; Commins, S. The Time Course of Systems Consolidation of Spatial Memory from Recent to Remote Retention: A Comparison of the Immediate Early Genes Zif268, c-Fos and Arc. Neurobiol. Learn. Mem. 2016, 128, 46–55. [Google Scholar] [CrossRef]
- Miyashita, T.; Kubik, S.; Haghighi, N.; Steward, O.; Guzowski, J.F. Rapid Activation of Plasticity-Associated Gene Transcription in Hippocampal Neurons Provides a Mechanism for Encoding of One-Trial Experience. J. Neurosci. 2009, 29, 898–906. [Google Scholar] [CrossRef] [PubMed]
- Lonergan, M.E.; Gafford, G.M.; Jarome, T.J.; Helmstetter, F.J. Time-Dependent Expression of Arc and Zif268 after Acquisition of Fear Conditioning. Neural Plast. 2010, 2010, 139891. [Google Scholar] [CrossRef] [PubMed]
- Khodadad, A.; Adelson, P.D.; Lifshitz, J.; Thomas, T.C. The Time Course of Activity-Regulated Cytoskeletal (ARC) Gene and Protein Expression in the Whisker-Barrel Circuit Using Two Paradigms of Whisker Stimulation. Behav. Brain Res. 2015, 284, 249–256. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guzowski, J.F.; Lyford, G.L.; Stevenson, G.D.; Houston, F.P.; McGaugh, J.L.; Worley, P.F.; Barnes, C.A. Inhibition of Activity-Dependent Arc Protein Expression in the Rat Hippocampus Impairs the Maintenance of Long-Term Potentiation and the Consolidation of Long-Term Memory. J. Neurosci. 2000, 20, 3993–4001. [Google Scholar] [CrossRef]
- Nakayama, D.; Iwata, H.; Teshirogi, C.; Ikegaya, Y.; Matsuki, N.; Nomura, H. Long-Delayed Expression of the Immediate Early Gene Arc/Arg3.1 Refines Neuronal Circuits to Perpetuate Fear Memory. J. Neurosci. 2015, 35, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Bottai, D.; Guzowski, J.F.; Schwarz, M.K.; Kang, S.H.; Xiao, B.; Lanahan, A.; Worley, P.F.; Seeburg, P.H. Synaptic Activity-Induced Conversion of Intronic to Exonic Sequence in Homer 1 Immediate Early Gene Expression. J. Neurosci. 2002, 22, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.B.; Beale, H.C.; Carlisle, H.J.; Washburn, L.R. Integration of Biochemical Signalling in Spines. Nat. Rev. Neurosci. 2005, 6, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Moutin, E.; Raynaud, F.; Roger, J.; Pellegrino, E.; Homburger, V.; Bertaso, F.; Ollendorff, V.; Bockaert, J.; Fagni, L.; Perroy, J. Dynamic Remodeling of Scaffold Interactions in Dendritic Spines Controls Synaptic Excitability. J. Cell Biol. 2012, 198, 251–263. [Google Scholar] [CrossRef]
- Witharana, W.K.L.; Clark, B.J.; Trivedi, V.; Mesina, L.; McNaughton, B.L. Immediate-Early Gene Homer1a Intranuclear Transcription Focus Intensity as a Measure of Relative Neural Activation. Hippocampus 2019, 29, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Marrone, D.F.; Schaner, M.J.; McNaughton, B.L.; Worley, P.F.; Barnes, C.A. Immediate-Early Gene Expression at Rest Recapitulates Recent Experience. J. Neurosci. 2008, 28, 1030–1033. [Google Scholar] [CrossRef]
- Lin, R.; Learman, L.N.; Bangash, M.A.; Melnikova, T.; Leyder, E.; Reddy, S.C.; Naidoo, N.; Park, J.M.; Savonenko, A.; Worley, P.F. Homer1a Regulates Shank3 Expression and Underlies Behavioral Vulnerability to Stress in a Model of Phelan-McDermid Syndrome. Cell Rep. 2021, 37, 110014. [Google Scholar] [CrossRef]
- Zhang, G.-C.; Mao, L.-M.; Liu, X.-Y.; Parelkar, N.K.; Arora, A.; Yang, L.; Hains, M.; Fibuch, E.E.; Wang, J.Q. In Vivo Regulation of Homer1a Expression in the Striatum by Cocaine. Mol. Pharmacol. 2007, 71, 1148–1158. [Google Scholar] [CrossRef]
- Diering, G.H.; Nirujogi, R.S.; Roth, R.H.; Worley, P.F.; Pandey, A.; Huganir, R.L. Homer1a Drives Homeostatic Scaling-down of Excitatory Synapses during Sleep. Science 2017, 355, 511–515. [Google Scholar] [CrossRef]
- Cingolani, L.A.; Vitale, C.; Dityatev, A. Intra- and Extracellular Pillars of a Unifying Framework for Homeostatic Plasticity: A Crosstalk between Metabotropic Receptors and Extracellular Matrix. Front. Cell. Neurosci. 2019, 13, 513. [Google Scholar] [CrossRef]
- Bertaso, F.; Roussignol, G.; Worley, P.; Bockaert, J.; Fagni, L.; Ango, F. Homer1a-Dependent Crosstalk between NMDA and Metabotropic Glutamate Receptors in Mouse Neurons. PLoS ONE 2010, 5, e9755. [Google Scholar] [CrossRef]
- Spiegel, I.; Mardinly, A.R.; Gabel, H.W.; Bazinet, J.E.; Couch, C.H.; Tzeng, C.P.; Harmin, D.A.; Greenberg, M.E. Npas4 Regulates Excitatory-Inhibitory Balance within Neural Circuits through Cell-Type-Specific Gene Programs. Cell 2014, 157, 1216–1229. [Google Scholar] [CrossRef] [PubMed]
- Bloodgood, B.L.; Sharma, N.; Browne, H.A.; Trepman, A.Z.; Greenberg, M.E. The Activity-Dependent Transcription Factor NPAS4 Regulates Domain-Specific Inhibition. Nature 2013, 503, 121–125. [Google Scholar] [CrossRef]
- Coutellier, L.; Beraki, S.; Ardestani, P.M.; Saw, N.L.; Shamloo, M. Npas4: A Neuronal Transcription Factor with a Key Role in Social and Cognitive Functions Relevant to Developmental Disorders. PLoS ONE 2012, 7, e46604. [Google Scholar] [CrossRef]
- Vousden, D.A.; Epp, J.; Okuno, H.; Nieman, B.J.; van Eede, M.; Dazai, J.; Ragan, T.; Bito, H.; Frankland, P.W.; Lerch, J.P.; et al. Whole-Brain Mapping of Behaviourally Induced Neural Activation in Mice. Brain Struct. Funct. 2015, 220, 2043–2057. [Google Scholar] [CrossRef]
- Renier, N.; Wu, Z.; Simon, D.J.; Yang, J.; Ariel, P.; Tessier-Lavigne, M. IDISCO: A Simple, Rapid Method to Immunolabel Large Tissue Samples for Volume Imaging. Cell 2014, 159, 896–910. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.-H.; Rendall, S.D.; Gray, J.M. Brain-Wide Maps of Fos Expression during Fear Learning and Recall. Learn. Mem. 2017, 24, 169–181. [Google Scholar] [CrossRef]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; et al. Ilastik: Interactive Machine Learning for (Bio)Image Analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef]
- McQuin, C.; Goodman, A.; Chernyshev, V.; Kamentsky, L.; Cimini, B.A.; Karhohs, K.W.; Doan, M.; Ding, L.; Rafelski, S.M.; Thirstrup, D.; et al. CellProfiler 3.0: Next-Generation Image Processing for Biology. PLoS Biol. 2018, 16, e2005970. [Google Scholar] [CrossRef]
- Wang, A.; Zhang, Q.; Han, Y.; Megason, S.; Hormoz, S.; Mosaliganti, K.R.; Lam, J.C.K.; Li, V.O.K. A Novel Deep Learning-Based 3D Cell Segmentation Framework for Future Image-Based Disease Detection. Sci. Rep. 2022, 12, 342. [Google Scholar] [CrossRef]
- Tappan, S.J.; Eastwood, B.S.; O’Connor, N.; Wang, Q.; Ng, L.; Feng, D.; Hooks, B.M.; Gerfen, C.R.; Hof, P.R.; Schmitz, C.; et al. Automatic Navigation System for the Mouse Brain. J. Comp. Neurol. 2019, 527, 2200–2211. [Google Scholar] [CrossRef]
- Renier, N.; Adams, E.L.; Kirst, C.; Wu, Z.; Azevedo, R.; Kohl, J.; Autry, A.E.; Kadiri, L.; Umadevi Venkataraju, K.; Zhou, Y.; et al. Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell 2016, 165, 1789–1802. [Google Scholar] [CrossRef] [PubMed]
- Fürth, D.; Vaissière, T.; Tzortzi, O.; Xuan, Y.; Märtin, A.; Lazaridis, I.; Spigolon, G.; Fisone, G.; Tomer, R.; Deisseroth, K.; et al. An Interactive Framework for Whole-Brain Maps at Cellular Resolution. Nat. Neurosci. 2018, 21, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Terstege, D.J.; Oboh, D.O.; Epp, J.R. FASTMAP: Open-Source Flexible Atlas Segmentation Tool for Multi-Area Processing of Biological Images. eNeuro 2022, 9, ENEURO.0325-21.2022. [Google Scholar] [CrossRef] [PubMed]
- Mano, T.; Murata, K.; Kon, K.; Shimizu, C.; Ono, H.; Shi, S.; Yamada, R.G.; Miyamichi, K.; Susaki, E.A.; Touhara, K.; et al. CUBIC-Cloud Provides an Integrative Computational Framework toward Community-Driven Whole-Mouse-Brain Mapping. Cell Rep Methods 2021, 1, 100038. [Google Scholar] [CrossRef]
- Claudi, F.; Petrucco, L.; Tyson, A.; Branco, T.; Margrie, T.; Portugues, R. BrainGlobe Atlas API: A Common Interface for Neuroanatomical Atlases. J. Open Source Softw. 2020, 5, 2668. [Google Scholar] [CrossRef]
- Carey, H.; Pegios, M.; Martin, L.; Saleeba, C.; Turner, A.; Everett, N.; Puchades, M.; Bjaalie, J.; McMullan, S. DeepSlice: Rapid Fully Automatic Registration of Mouse Brain Imaging to a Volumetric Atlas. bioRxiv 2022. [Google Scholar] [CrossRef]
- Puchades, M.A.; Csucs, G.; Ledergerber, D.; Leergaard, T.B.; Bjaalie, J.G. Spatial Registration of Serial Microscopic Brain Images to Three-Dimensional Reference Atlases with the QuickNII Tool. PLoS ONE 2019, 14, e0216796. [Google Scholar] [CrossRef]
- Lauridsen, K.; Ly, A.; Prévost, E.D.; McNulty, C.; McGovern, D.J.; Tay, J.W.; Dragavon, J.; Root, D.H. A Semi-Automated Workflow for Brain Slice Histology Alignment, Registration, and Cell Quantification (SHARCQ). eNeuro 2022, 9, ENEURO.0483-21.2022. [Google Scholar] [CrossRef]
- Lein, E.S.; Hawrylycz, M.J.; Ao, N.; Ayres, M.; Bensinger, A.; Bernard, A.; Boe, A.F.; Boguski, M.S.; Brockway, K.S.; Byrnes, E.J.; et al. Genome-Wide Atlas of Gene Expression in the Adult Mouse Brain. Nature 2007, 445, 168–176. [Google Scholar] [CrossRef]
- Oh, S.W.; Harris, J.A.; Ng, L.; Winslow, B.; Cain, N.; Mihalas, S.; Wang, Q.; Lau, C.; Kuan, L.; Henry, A.M.; et al. A Mesoscale Connectome of the Mouse Brain. Nature 2014, 508, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Honey, C.J.; Thivierge, J.-P.; Sporns, O. Can Structure Predict Function in the Human Brain? Neuroimage 2010, 52, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Latora, V.; Marchiori, M. Efficient Behavior of Small-World Networks. Phys. Rev. Lett. 2001, 87, 198701. [Google Scholar] [CrossRef] [PubMed]
- Watts, D.J.; Strogatz, S.H. Collective Dynamics of “small-World” Networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef]
- Rubinov, M.; Sporns, O. Complex Network Measures of Brain Connectivity: Uses and Interpretations. Neuroimage 2010, 52, 1059–1069. [Google Scholar] [CrossRef]
- Achard, S.; Salvador, R.; Whitcher, B.; Suckling, J.; Bullmore, E. A Resilient, Low-Frequency, Small-World Human Brain Functional Network with Highly Connected Association Cortical Hubs. J. Neurosci. 2006, 26, 63–72. [Google Scholar] [CrossRef]
- Sporns, O.; Honey, C.J. Small Worlds inside Big Brains. Proc. Natl. Acad. Sci. USA 2006, 103, 19219–19220. [Google Scholar] [CrossRef]
- Bassett, D.S.; Meyer-Lindenberg, A.; Achard, S.; Duke, T.; Bullmore, E. Adaptive Reconfiguration of Fractal Small-World Human Brain Functional Networks. Proc. Natl. Acad. Sci. USA 2006, 103, 19518–19523. [Google Scholar] [CrossRef]
- Freeman, L.C. Centrality in Social Networks Conceptual Clarification. Soc. Netw. 1978, 1, 215–239. [Google Scholar] [CrossRef]
- Brandes, U. A Faster Algorithm for Betweenness Centrality. J. Math. Sociol. 2001, 25, 163–177. [Google Scholar] [CrossRef]
- Bullmore, E.; Sporns, O. Complex Brain Networks: Graph Theoretical Analysis of Structural and Functional Systems. Nat. Rev. Neurosci. 2009, 10, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Sporns, O. Graph Theory Methods: Applications in Brain Networks. Dialogues Clin. Neurosci. 2018, 20, 111–121. [Google Scholar] [CrossRef]
- Konganti, K.; Wang, G.; Yang, E.; Cai, J.J. SBEToolbox: A Matlab Toolbox for Biological Network Analysis. Evol. Bioinform. Online 2013, 9, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Colombo, P.J.; Brightwell, J.J.; Countryman, R.A. Cognitive Strategy-Specific Increases in Phosphorylated CAMP Response Element-Binding Protein and c-Fos in the Hippocampus and Dorsal Striatum. J. Neurosci. 2003, 23, 3547–3554. [Google Scholar] [CrossRef] [PubMed]
- Fields, R.D.; Eshete, F.; Stevens, B.; Itoh, K. Action Potential-Dependent Regulation of Gene Expression: Temporal Specificity in Ca2+, CAMP-Responsive Element Binding Proteins, and Mitogen-Activated Protein Kinase Signaling. J. Neurosci. 1997, 17, 7252–7266. [Google Scholar] [CrossRef] [PubMed]
- Speicher, M.R.; Carter, N.P. The New Cytogenetics: Blurring the Boundaries with Molecular Biology. Nat. Rev. Genet. 2005, 6, 782–792. [Google Scholar] [CrossRef]
- Guzowski, J.F.; McNaughton, B.L.; Barnes, C.A.; Worley, P.F. Environment-Specific Expression of the Immediate-Early Gene Arc in Hippocampal Neuronal Ensembles. Nat. Neurosci. 1999, 2, 1120–1124. [Google Scholar] [CrossRef]
- Gruene, T.M.; Flick, K.; Stefano, A.; Shea, S.D.; Shansky, R.M. Sexually Divergent Expression of Active and Passive Conditioned Fear Responses in Rats. eLife 2015, 4, e11352. [Google Scholar] [CrossRef]
- Colom-Lapetina, J.; Li, A.J.; Pelegrina-Perez, T.C.; Shansky, R.M. Behavioral Diversity across Classic Rodent Models Is Sex-Dependent. Front. Behav. Neurosci. 2019, 13, 45. [Google Scholar] [CrossRef]
- Earnest, D.J.; Olschowka, J.A. Circadian Regulation of C-Fos Expression in the Suprachiasmatic Pacemaker by Light. J. Biol. Rhythms 1993, 8 Suppl, S65–S71. [Google Scholar]
- Edelstein, K.; Amir, S. Non-Photic Manipulations Induce Expression of Fos Protein in the Suprachiasmatic Nucleus and Intergeniculate Leaflet in the Rat. Brain Res. 1995, 690, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Ueda, H.R.; Ertürk, A.; Chung, K.; Gradinaru, V.; Chédotal, A.; Tomancak, P.; Keller, P.J. Tissue Clearing and Its Applications in Neuroscience. Nat. Rev. Neurosci. 2020, 21, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.; Deisseroth, K. CLARITY for Mapping the Nervous System. Nat. Methods 2013, 10, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Parra-Damas, A.; Saura, C.A. Tissue Clearing and Expansion Methods for Imaging Brain Pathology in Neurodegeneration: From Circuits to Synapses and Beyond. Front. Neurosci. 2020, 14, 914. [Google Scholar] [CrossRef] [PubMed]
- Vigouroux, R.J.; Belle, M.; Chédotal, A. Neuroscience in the Third Dimension: Shedding New Light on the Brain with Tissue Clearing. Mol. Brain 2017, 10, 1–10. [Google Scholar] [CrossRef]
- Hama, H.; Hioki, H.; Namiki, K.; Hoshida, T.; Kurokawa, H.; Ishidate, F.; Kaneko, T.; Akagi, T.; Saito, T.; Saido, T.; et al. ScaleS: An Optical Clearing Palette for Biological Imaging. Nat. Neurosci. 2015, 18, 1518–1529. [Google Scholar] [CrossRef]
- Epp, J.R.; Niibori, Y.; Liz Hsiang, H.-L.; Mercaldo, V.; Deisseroth, K.; Josselyn, S.A.; Frankland, P.W. Optimization of CLARITY for Clearing Whole-Brain and Other Intact Organs. eNeuro 2015, 2, ENEURO.0022-15.2015. [Google Scholar] [CrossRef]
- Matsumoto, K.; Mitani, T.T.; Horiguchi, S.A.; Kaneshiro, J.; Murakami, T.C.; Mano, T.; Fujishima, H.; Konno, A.; Watanabe, T.M.; Hirai, H.; et al. Advanced CUBIC Tissue Clearing for Whole-Organ Cell Profiling. Nat. Protoc. 2019, 14, 3506–3537. [Google Scholar] [CrossRef]
- DeNardo, L.A.; Liu, C.D.; Allen, W.E.; Adams, E.L.; Friedmann, D.; Fu, L.; Guenthner, C.J.; Tessier-Lavigne, M.; Luo, L. Temporal Evolution of Cortical Ensembles Promoting Remote Memory Retrieval. Nat. Neurosci. 2019, 22, 460–469. [Google Scholar] [CrossRef]
- Guenthner, C.J.; Miyamichi, K.; Yang, H.H.; Heller, H.C.; Luo, L. Permanent Genetic Access to Transiently Active Neurons via TRAP: Targeted Recombination in Active Populations. Neuron 2013, 78, 773–784. [Google Scholar] [CrossRef]
- Igarashi, H.; Koizumi, K.; Kaneko, R.; Ikeda, K.; Egawa, R.; Yanagawa, Y.; Muramatsu, S.-I.; Onimaru, H.; Ishizuka, T.; Yawo, H. A Novel Reporter Rat Strain That Conditionally Expresses the Bright Red Fluorescent Protein TdTomato. PLoS ONE 2016, 11, e0155687. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Allen, W.E.; Thompson, K.R.; Tian, Q.; Hsueh, B.; Ramakrishnan, C.; Wang, A.-C.; Jennings, J.H.; Adhikari, A.; Halpern, C.H.; et al. Wiring and Molecular Features of Prefrontal Ensembles Representing Distinct Experiences. Cell 2016, 165, 1776–1788. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Ren, J.; Luo, L.; Horowitz, M. Mapping Histological Slice Sequences to the Allen Mouse Brain Atlas without 3D Reconstruction. Front. Neuroinform. 2018, 12, 93. [Google Scholar] [CrossRef] [PubMed]
- Croce, A.C.; Bottiroli, G. Lipids: Evergreen Autofluorescent Biomarkers for the Liver Functional Profiling. Eur. J. Histochem. 2017, 61, 2808. [Google Scholar] [CrossRef] [PubMed]
- Baschong, W.; Suetterlin, R.; Laeng, R.H. Control of Autofluorescence of Archival Formaldehyde-Fixed, Paraffin-Embedded Tissue in Confocal Laser Scanning Microscopy (CLSM). J. Histochem. Cytochem. 2001, 49, 1565–1572. [Google Scholar] [CrossRef]
- Billinton, N.; Knight, A.W. Seeing the Wood through the Trees: A Review of Techniques for Distinguishing Green Fluorescent Protein from Endogenous Autofluorescence. Anal. Biochem. 2001, 291, 175–197. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Kao, C.-Y.; Gore, J.C.; Ding, Z. Implicit Active Contours Driven by Local Binary Fitting Energy. In Proceedings of the 2007 IEEE Conference on Computer Vision and Pattern Recognition, Minneapolis, MN, USA, 17–22 June 2007. [Google Scholar]
- Wehrl, H.F.; Bezrukov, I.; Wiehr, S.; Lehnhoff, M.; Fuchs, K.; Mannheim, J.G.; Quintanilla-Martinez, L.; Kohlhofer, U.; Kneilling, M.; Pichler, B.J.; et al. Assessment of Murine Brain Tissue Shrinkage Caused by Different Histological Fixatives Using Magnetic Resonance and Computed Tomography Imaging. Histol. Histopathol. 2015, 30, 601–613. [Google Scholar]
- Korogod, N.; Petersen, C.C.H.; Knott, G.W. Ultrastructural Analysis of Adult Mouse Neocortex Comparing Aldehyde Perfusion with Cryo Fixation. Elife 2015, 4, e05793. [Google Scholar] [CrossRef]
- Vulders, R.C.M.; van Hoogenhuizen, R.C.; van der Giessen, E.; van der Zaag, P.J. Clearing-Induced Tisssue Shrinkage: A Novel Observation of a Thickness Size Effect. PLoS ONE 2021, 16, e0261417. [Google Scholar] [CrossRef]
- Bekkouche, B.M.B.; Fritz, H.K.M.; Rigosi, E.; O’Carroll, D.C. Comparison of Transparency and Shrinkage during Clearing of Insect Brains Using Media with Tunable Refractive Index. Front. Neuroanat. 2020, 14, 599282. [Google Scholar] [CrossRef]
- Morris, J.A.; Royall, J.J.; Bertagnolli, D.; Boe, A.F.; Burnell, J.J.; Byrnes, E.J.; Copeland, C.; Desta, T.; Fischer, S.R.; Goldy, J.; et al. Divergent and Nonuniform Gene Expression Patterns in Mouse Brain. Proc. Natl. Acad. Sci. USA 2010, 107, 19049–19054. [Google Scholar] [CrossRef] [PubMed]
- Makin, T.R.; Orban de Xivry, J.-J. Ten Common Statistical Mistakes to Watch out for When Writing or Reviewing a Manuscript. Elife 2019, 8, e58175. [Google Scholar] [CrossRef]
- Button, K.S.; Ioannidis, J.P.A.; Mokrysz, C.; Nosek, B.A.; Flint, J.; Robinson, E.S.J.; Munafò, M.R. Power Failure: Why Small Sample Size Undermines the Reliability of Neuroscience. Nat. Rev. Neurosci. 2013, 14, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Gelman, A.; Carlin, J. Beyond Power Calculations: Assessing Type S (Sign) and Type M (Magnitude) Errors. Perspect. Psychol. Sci. 2014, 9, 641–651. [Google Scholar] [CrossRef]
- Ghasemi, A.; Zahediasl, S. Normality Tests for Statistical Analysis: A Guide for Non-Statisticians. Int. J. Endocrinol. Metab. 2012, 10, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Bassett, D.S.; Bullmore, E.T. Small-World Brain Networks Revisited. Neuroscientist 2017, 23, 499–516. [Google Scholar] [CrossRef]
- Weissenbacher, A.; Kasess, C.; Gerstl, F.; Lanzenberger, R.; Moser, E.; Windischberger, C. Correlations and Anticorrelations in Resting-State Functional Connectivity MRI: A Quantitative Comparison of Preprocessing Strategies. Neuroimage 2009, 47, 1408–1416. [Google Scholar] [CrossRef]
- Murphy, K.; Birn, R.M.; Handwerker, D.A.; Jones, T.B.; Bandettini, P.A. The Impact of Global Signal Regression on Resting State Correlations: Are Anti-Correlated Networks Introduced? Neuroimage 2009, 44, 893–905. [Google Scholar] [CrossRef]
- Fox, M.D.; Snyder, A.Z.; Vincent, J.L.; Corbetta, M.; Van Essen, D.C.; Raichle, M.E. The Human Brain Is Intrinsically Organized into Dynamic, Anticorrelated Functional Networks. Proc. Natl. Acad. Sci. USA 2005, 102, 9673–9678. [Google Scholar] [CrossRef]
- Chen, G.; Chen, G.; Xie, C.; Li, S.-J. Negative Functional Connectivity and Its Dependence on the Shortest Path Length of Positive Network in the Resting-State Human Brain. Brain Connect. 2011, 1, 195–206. [Google Scholar] [CrossRef]
- Sylwestrak, E.L.; Rajasethupathy, P.; Wright, M.A.; Jaffe, A.; Deisseroth, K. Multiplexed Intact-Tissue Transcriptional Analysis at Cellular Resolution. Cell 2016, 164, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Kramer, E.E.; Steadman, P.E.; Epp, J.R.; Frankland, P.W.; Josselyn, S.A. Assessing Individual Neuronal Activity across the Intact Brain: Using Hybridization Chain Reaction (HCR) to Detect Arc MRNA Localized to the Nucleus in Volumes of Cleared Brain Tissue. Curr. Protoc. Neurosci. 2018, 84, e49. [Google Scholar] [CrossRef] [PubMed]
- Reijmers, L.; Mayford, M. Genetic Control of Active Neural Circuits. Front. Mol. Neurosci. 2009, 2, 27. [Google Scholar] [CrossRef] [PubMed]
- Reijmers, L.G.; Perkins, B.L.; Matsuo, N.; Mayford, M. Localization of a Stable Neural Correlate of Associative Memory. Science 2007, 317, 1230–1233. [Google Scholar] [CrossRef] [PubMed]
- Denny, C.A.; Kheirbek, M.A.; Alba, E.L.; Tanaka, K.F.; Brachman, R.A.; Laughman, K.B.; Tomm, N.K.; Turi, G.F.; Losonczy, A.; Hen, R. Hippocampal Memory Traces Are Differentially Modulated by Experience, Time, and Adult Neurogenesis. Neuron 2014, 83, 189–201. [Google Scholar] [CrossRef]
- Smeyne, R.J.; Schilling, K.; Robertson, L.; Luk, D.; Oberdick, J.; Curran, T.; Morgan, J.I. Fos-LacZ Transgenic Mice: Mapping Sites of Gene Induction in the Central Nervous System. Neuron 1992, 8, 13–23. [Google Scholar] [CrossRef]
- Kasof, G.M.; Mandelzys, A.; Maika, S.D.; Hammer, R.E.; Curran, T.; Morgan, J.I. Kainic Acid-Induced Neuronal Death Is Associated with DNA Damage and a Unique Immediate-Early Gene Response in c-Fos-LacZ Transgenic Rats. J. Neurosci. 1995, 15, 4238–4249. [Google Scholar] [CrossRef]
- Sakurai, K.; Zhao, S.; Takatoh, J.; Rodriguez, E.; Lu, J.; Leavitt, A.D.; Fu, M.; Han, B.-X.; Wang, F. Capturing and Manipulating Activated Neuronal Ensembles with CANE Delineates a Hypothalamic Social-Fear Circuit. Neuron 2016, 92, 739–753. [Google Scholar] [CrossRef]
- Kawashima, T.; Okuno, H.; Nonaka, M.; Adachi-Morishima, A.; Kyo, N.; Okamura, M.; Takemoto-Kimura, S.; Worley, P.F.; Bito, H. Synaptic Activity-Responsive Element in the Arc/Arg3.1 Promoter Essential for Synapse-to-Nucleus Signaling in Activated Neurons. Proc. Natl. Acad. Sci. USA 2009, 106, 316–321. [Google Scholar] [CrossRef]
- Sørensen, A.T.; Cooper, Y.A.; Baratta, M.V.; Weng, F.-J.; Zhang, Y.; Ramamoorthi, K.; Fropf, R.; LaVerriere, E.; Xue, J.; Young, A.; et al. A Robust Activity Marking System for Exploring Active Neuronal Ensembles. eLife 2016, 5, e13918. [Google Scholar] [CrossRef]
- DeNardo, L.; Luo, L. Genetic Strategies to Access Activated Neurons. Curr. Opin. Neurobiol. 2017, 45, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Bassett, D.S.; Bullmore, E.T.; Meyer-Lindenberg, A.; Apud, J.A.; Weinberger, D.R.; Coppola, R. Cognitive Fitness of Cost-Efficient Brain Functional Networks. Proc. Natl. Acad. Sci. USA 2009, 106, 11747–11752. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Sagar, S.M.; Sharp, F.R.; Curran, T. Expression of C-Fos Protein in Brain: Metabolic Mapping at the Cellular Level. Science 1988, 240, 1328–1331. [Google Scholar] [CrossRef] [PubMed]
- Gall, C.M.; Hess, U.S.; Lynch, G. Mapping Brain Networks Engaged by, and Changed by, Learning. Neurobiol. Learn. Mem. 1998, 70, 14–36. [Google Scholar] [CrossRef]
- Maleeva, N.E.; Bikbulatova, L.S.; Ivolgina, G.L.; Anokhin, K.V.; Limborskaia, S.A.; Kruglikov, R.I. Activation of the c-fos proto-oncogene in different structures of the rat brain during training and pseudoconditioning. Dokl. Akad. Nauk SSSR 1990, 314, 762–764. [Google Scholar]
- Maleeva, N.E.; Ivolgina, G.L.; Anokhin, K.V.; Limborskaia, S.A. Analysis of the expression of the c-fos proto-oncogene in the rat cerebral cortex during learning. Genetika 1989, 25, 1119–1121. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terstege, D.J.; Epp, J.R. Network Neuroscience Untethered: Brain-Wide Immediate Early Gene Expression for the Analysis of Functional Connectivity in Freely Behaving Animals. Biology 2023, 12, 34. https://doi.org/10.3390/biology12010034
Terstege DJ, Epp JR. Network Neuroscience Untethered: Brain-Wide Immediate Early Gene Expression for the Analysis of Functional Connectivity in Freely Behaving Animals. Biology. 2023; 12(1):34. https://doi.org/10.3390/biology12010034
Chicago/Turabian StyleTerstege, Dylan J., and Jonathan R. Epp. 2023. "Network Neuroscience Untethered: Brain-Wide Immediate Early Gene Expression for the Analysis of Functional Connectivity in Freely Behaving Animals" Biology 12, no. 1: 34. https://doi.org/10.3390/biology12010034
APA StyleTerstege, D. J., & Epp, J. R. (2023). Network Neuroscience Untethered: Brain-Wide Immediate Early Gene Expression for the Analysis of Functional Connectivity in Freely Behaving Animals. Biology, 12(1), 34. https://doi.org/10.3390/biology12010034





