Neurogenesis during Brittle Star Arm Regeneration Is Characterised by a Conserved Set of Key Developmental Genes
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Animal Maintenance and Collection
2.2. Immunohistochemistry
2.3. Whole-Mount In Situ Hybridisation
2.4. NanoString—nCounter Analysis
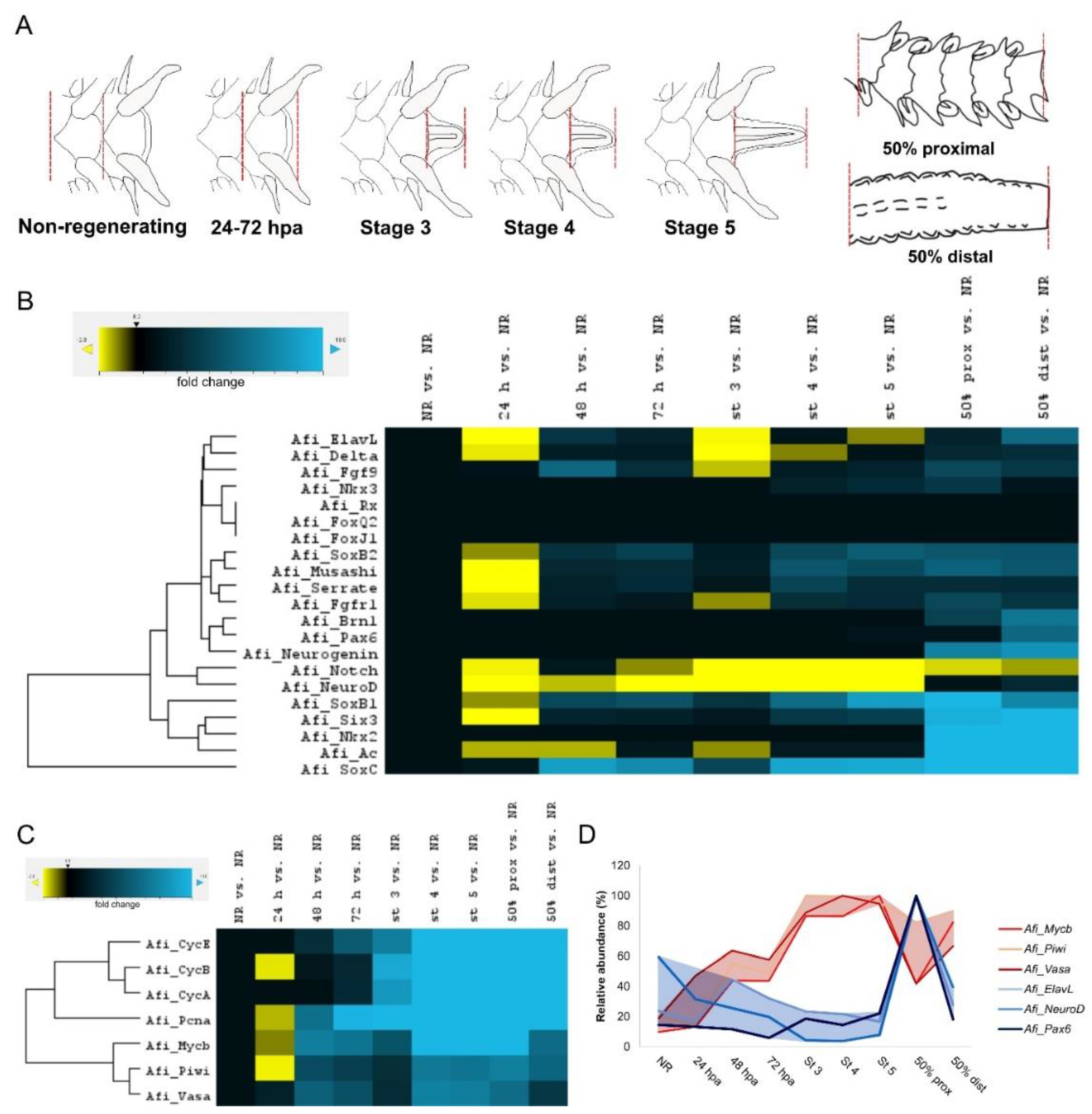
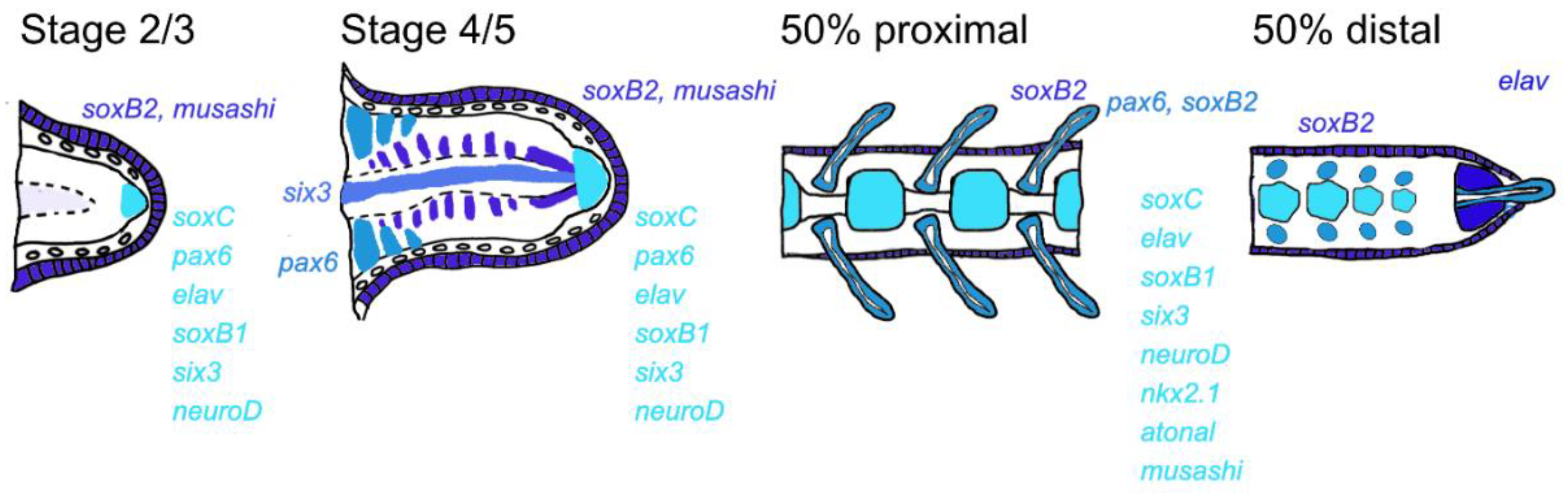
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Silver, J.; Schwab, M.E.; Popovich, P.G. Central Nervous System Regenerative Failure: Role of Oligodendrocytes, Astrocytes, and Microglia. Cold Spring Harb. Perspect Biol. 2015, 7, a020602. [Google Scholar] [CrossRef] [PubMed]
- Agata, K.; Umesono, Y. Brain regeneration from pluripotent stem cells in planarian. Phil. Trans. R. Soc. B 2008, 363, 2071–2078. [Google Scholar] [CrossRef] [PubMed]
- Diaz Quiroz, J.F.; Echeverri, K. Spinal cord regeneration: Where fish, frogs and salamanders lead the way, can we follow? Biochem. J. 2013, 451, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Zambusi, A.; Ninkovic, J. Regeneration of the central nervous system-principles from brain regeneration in adult zebrafish. World J. Stem. Cells 2020, 12, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.; Brand, M. Vertebrate brain regeneration—A community effort of fate-restricted precursor cell types. Curr. Opin. Genet. Dev. 2020, 64, 101–108. [Google Scholar] [CrossRef]
- Lust, K.; Tanaka, E.M. A Comparative Perspective on Brain Regeneration in Amphibians and Teleost Fish. Dev. Neurobiol. 2019, 79, 424–436. [Google Scholar] [CrossRef]
- Wagner, D.E.; Wang, I.E.; Reddien, P.W. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 2011, 332, 811–816. [Google Scholar] [CrossRef]
- Shin, J.; Berg, D.A.; Zhu, Y.; Shin, J.Y.; Song, J.; Bonaguidi, M.A.; Enikolopov, G.; Nauen, D.W.; Christian, K.M.; Ming, G.L.; et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 2015, 17, 360–372. [Google Scholar] [CrossRef]
- Molinaro, A.M.; Pearson, B.J. In silico lineage tracing through single cell transcriptomics identifies a neural stem cell population in planarians. Genome Biol. 2016, 17, 87. [Google Scholar] [CrossRef]
- Mollinari, C.; Zhao, J.; Lupacchini, L.; Garaci, E.; Merlo, D.; Pei, G. Transdifferentiation: A new promise for neurodegenerative diseases. Cell Death Dis. 2018, 9, 830. [Google Scholar] [CrossRef]
- Mashanov, V.S.; Zueva, O.R.; García-Arrarás, J.E. Inhibition of cell proliferation does not slow down echinoderm neural regeneration. Front. Zool. 2017, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Mashanov, V.S.; Zueva, O.R.; Heinzeller, T. Regeneration of the radial nerve cord in a holothurian: A promising new model system for studying post-traumatic recovery in the adult nervous system. Tissue Cell 2008, 40, 351–372. [Google Scholar] [CrossRef] [PubMed]
- Mashanov, V.S.; Zueva, O.R.; García-Arrarás, J.E. Transcriptomic changes during regeneration of the central nervous system in an echinoderm. BMC Genom. 2014, 15, 357. [Google Scholar] [CrossRef] [PubMed]
- Tossas, K.; Qi-Huang, S.; Cuyar, E.; García-Arrarás, J.E. Temporal and spatial analysis of enteric nervous system regeneration in the sea cucumber Holothuria glaberrima. Regeneration 2014, 1, 10–26. [Google Scholar] [CrossRef]
- San Miguel-Ruiz, J.E.; Maldonado-Soto, A.R.; García-Arrarás, J.E. Regeneration of the radial nerve cord in the sea cucumber Holothuria glaberrima. BMC Dev. Biol. 2009, 9, 3. [Google Scholar] [CrossRef]
- Burke, R.; Angerer, L.; Elphick, M.; Humphrey, G.; Yaguchi, S.; Kiyama, T.; Liang, S.; Mu, X.; Agca, C.; Klein, W.; et al. A genomic view of the sea urchin nervous system. Dev. Biol. 2006, 300, 434–460. [Google Scholar] [CrossRef]
- Garner, S.; Zysk, I.; Byrne, G.; Kramer, M.; Moller, D.; Taylor, V.; Burke, R.D. Neurogenesis in sea urchin embryos and the diversity of deuterostome neurogenic mechanisms. Development 2015, 143, 286–297. [Google Scholar] [CrossRef]
- Yankura, K.A.; Koechlein, C.S.; Cryan, A.F.; Cheatle, A.; Hinman, V.F. Gene regulatory network for neurogenesis in a sea star embryo connects broad neural specification and localized patterning. Proc. Natl. Acad. Sci. USA 2013, 110, 8591–8596. [Google Scholar] [CrossRef]
- Jarvela, A.M.C.; Yankura, K.A.; Hinman, V.F. A gene regulatory network for apical organ neurogenesis and its spatial control in sea star embryos. Development 2016, 143, 4214–4223. [Google Scholar] [CrossRef]
- Byrne, M.; Mazzone, F.; Elphick, M.R.; Thorndyke, M.C.; Cisternas, P. Expression of the neuropeptide SALMFamide-1 during regeneration of the seastar radial nerve cord following arm autotomy. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182701. [Google Scholar] [CrossRef]
- Zheng, M.; Zueva, O.; Hinman, V.F. Regeneration of the larval sea star nervous system by wounding induced respecification to the sox2 lineage. eLife 2022, 11, e72983. [Google Scholar] [CrossRef] [PubMed]
- Dylus, D.V.; Czarkwiani, A.; Stångberg, J.; Ortega-Martinez, O.; Dupont, S.; Oliveri, P. Large-scale gene expression study in the ophiuroid Amphiura filiformis provides insights into evolution of gene regulatory networks. Evodevo 2016, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Dylus, D.V.; Czarkwiani, A.; Blowes, L.M.; Elphick, M.R.; Oliveri, P. Developmental transcriptomics of the brittle star Amphiura filiformis reveals gene regulatory network rewiring in echinoderm larval skeleton evolution. Genome Biol. 2018, 19, 26. [Google Scholar] [CrossRef] [PubMed]
- Czarkwiani, A.; Dylus, D.V.; Carballo, L.; Oliveri, P. FGF signalling plays similar roles in development and regeneration of the skeleton in the brittle star Amphiura filiformis. Development 2021, 148, dev180760. [Google Scholar] [CrossRef] [PubMed]
- Czarkwiani, A.; Dylus, D.V.; Oliveri, P. Expression of skeletogenic genes during arm regeneration in the brittle star Amphiura filiformis. Gene Expr. Patterns 2013, 13, 464–472. [Google Scholar] [CrossRef]
- Czarkwiani, A.; Ferrario, C.; Dylus, D.V.; Sugni, M.; Oliveri, P. Skeletal regeneration in the brittle star Amphiura filiformis. Front. Zool. 2016, 13, 18. [Google Scholar] [CrossRef]
- Piovani, L.; Czarkwiani, A.; Ferrario, C.; Sugni, M.; Oliveri, P. Ultrastructural and molecular analysis of the origin and differentiation of cells mediating brittle star skeletal regeneration. BMC Biol. 2021, 19, 9. [Google Scholar] [CrossRef]
- Hirokawa, T.; Komatsu, M.; Nakajima, Y. Development of the nervous system in the brittle star Amphipholis kochii. Dev. Genes Evol. 2008, 218, 15–21. [Google Scholar] [CrossRef]
- Dupont, S.; Thorndyke, W.; Thorndyke, M.C.; Burke, R.D. Neural development of the brittlestar Amphiura filiformis. Dev. Genes Evol. 2009, 219, 159–166. [Google Scholar] [CrossRef]
- Zueva, O.; Khoury, M.; Heinzeller, T.; Mashanova, D.; Mashanov, V. The complex simplicity of the brittle star nervous system. Front. Zool. 2018, 15, 1. [Google Scholar] [CrossRef]
- Mashanov, V.; Whaley, L.; Davis, K.; Heinzeller, T.; Machado, D.J.; Reid, R.W.; Kofsky, J.; Janies, D. A subterminal growth zone at arm tip likely underlies life-long indeterminate growth in brittle stars. Front. Zool. 2022, 19, 15. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, T.; Hugall, A.; Thuy, B.; Moussalli, A. Phylogenomic Resolution of the Class Ophiuroidea Unlocks a Global Microfossil Record. Curr. Biol. 2014, 24, 1874–1879. [Google Scholar] [CrossRef] [PubMed]
- Thuy, B.; Stöhr, S. A new morphological phylogeny of the Ophiuroidea (Echinodermata) accords with molecular evidence and renders microfossils accessible for cladistics. PLoS ONE 2016, 11, e0156140. [Google Scholar] [CrossRef] [PubMed]
- Rentzsch, F.; Layden, M.; Manuel, M. The cellular and molecular basis of cnidarian neurogenesis. WIREs Dev. Biol. 2016, 6, e257. [Google Scholar] [CrossRef] [PubMed]
- Guth, S.I.E.; Wegner, M. Having it both ways: Sox protein function between conservation and innovation. Cell. Mol. Life Sci. 2008, 65, 3000–3018. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Hochedlinger, K. The Sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell 2013, 12, 15–30. [Google Scholar] [CrossRef]
- Powell, L.M.; Jarman, A.P. Context dependence of proneural bHLH proteins. Curr. Opin. Genet. Dev. 2008, 18, 411–417. [Google Scholar] [CrossRef]
- Simionato, E.; Ledent, V.; Richards, G.; Thomas-Chollier, M.; Kerner, P.; Coornaert, D.; Degnan, B.M.; Vervoort, M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: Insights from comparative genomics. BMC Evol. Biol. 2007, 7, 33. [Google Scholar] [CrossRef]
- Ledent, V.; Vervoort, M. The basic helix-loop-helix protein family: Comparative genomics and phylogenetic analysis. Genome Res. 2001, 11, 754–770. [Google Scholar] [CrossRef]
- Deryckere, A.; Styfhals, R.; Elagoz, A.M.; Maes, G.E.; Seuntjens, E. Identification of neural progenitor cells and their progeny reveals long distance migration in the developing octopus brain. eLife 2021, 10, e69161. [Google Scholar] [CrossRef]
- Pascale, A.; Amadio, M.; Quattrone, A. Defining a neuron: Neuronal ELAV proteins. Cell. Mol. Life Sci. 2008, 65, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Yaguchi, J.; Yaguchi, S.; Angerer, R.C.; Angerer, L.M. The sea urchin animal pole domain is a Six3-dependent neurogenic patterning center. Development 2009, 136, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- Lesser, M.P.; Carleton, K.L.; Bottger, S.A.; Barry, T.M.; Walker, C.W. Sea urchin tube feet are photosensory organs that express a rhabdomeric-like opsin and Pax6. Proc. R. Soc. B Biol. Sci. 2011, 278, 3371–3379. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Thorndyke, M.C. Growth or differentiation? Adaptive regeneration in the brittlestar Amphiura filiformis. J. Exp. Biol. 2006, 209, 3873–3881. [Google Scholar] [CrossRef]
- Biressi, A.C.M.; Zou, T.; Dupont, S.; Dahlberg, C.; Di Benedetto, C.; Bonasoro, F.; Thorndyke, M.; Carnevali, M.C. Wound healing and arm regeneration in Ophioderma longicaudum and Amphiura filiformis (Ophiuroidea, Echinodermata): Comparative morphogenesis and histogenesis. Zoomorphology 2010, 129, 1–19. [Google Scholar] [CrossRef]
- Kaneko, Y.; Sakakibara, S.-I.; Imai, T.; Suzuki, A.; Nakamura, Y.; Sawamoto, K.; Ogawa, Y.; Toyama, Y.; Miyata, T.; Okano, H. Musashi1: An evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev. Neurosci. 2000, 22, 139–153. [Google Scholar] [CrossRef]
- Farkas, J.E.; Freitas, P.D.; Bryant, D.M.; Whited, J.L.; Monaghan, J.R. Neuregulin-1 signaling is essential for nerve-dependent axolotl limb regeneration. Development 2016, 143, 2724–2731. [Google Scholar] [CrossRef]
- Kumar, A.; Brockes, J.P. Nerve dependence in tissue, organ, and appendage regeneration. Trends Neurosci. 2012, 35, 691–699. [Google Scholar] [CrossRef]
- Huet, P.M. Le rôle du système nerveux an cours de la régénération du bras chez une Etoile de mer: Asterina gibbosa Penn.(Echinoderme, Astéride). J. Embryol. Exp. Morphol. 1975, 33, 535–552. [Google Scholar]
- Sur, A.; Magie, C.R.; Seaver, E.C.; Meyer, N.P. Spatiotemporal regulation of nervous system development in the annelid Capitella teleta. Evodevo 2017, 8, 13. [Google Scholar] [CrossRef]
- Anishchenko, E.; Arnone, M.I.; D’Aniello, S. SoxB2 in sea urchin development: Implications in neurogenesis, ciliogenesis and skeletal patterning. Evodevo 2018, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Mashanov, V.; Akiona, J.; Khoury, M.; Ferrier, J.; Reid, R.; Machado, D.J.; Zueva, O.; Janies, D. Active Notch signaling is required for arm regeneration in a brittle star. PLoS ONE 2020, 15, e0232981. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, R.; Arnone, M.I. A dynamic regulatory network explains ParaHox gene control of gut patterning in the sea urchin. Development 2014, 141, 2462–2472. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, Y.; Kaneko, H.; Murray, G.; Burke, R.D. Divergent patterns of neural development in larval echinoids and asteroids. Evol. Dev. 2004, 6, 95–104. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czarkwiani, A.; Taylor, J.; Oliveri, P. Neurogenesis during Brittle Star Arm Regeneration Is Characterised by a Conserved Set of Key Developmental Genes. Biology 2022, 11, 1360. https://doi.org/10.3390/biology11091360
Czarkwiani A, Taylor J, Oliveri P. Neurogenesis during Brittle Star Arm Regeneration Is Characterised by a Conserved Set of Key Developmental Genes. Biology. 2022; 11(9):1360. https://doi.org/10.3390/biology11091360
Chicago/Turabian StyleCzarkwiani, Anna, Jack Taylor, and Paola Oliveri. 2022. "Neurogenesis during Brittle Star Arm Regeneration Is Characterised by a Conserved Set of Key Developmental Genes" Biology 11, no. 9: 1360. https://doi.org/10.3390/biology11091360
APA StyleCzarkwiani, A., Taylor, J., & Oliveri, P. (2022). Neurogenesis during Brittle Star Arm Regeneration Is Characterised by a Conserved Set of Key Developmental Genes. Biology, 11(9), 1360. https://doi.org/10.3390/biology11091360





