Functions of Small Non-Coding RNAs in Salmonella–Host Interactions
Abstract
Simple Summary
Abstract
1. Introduction
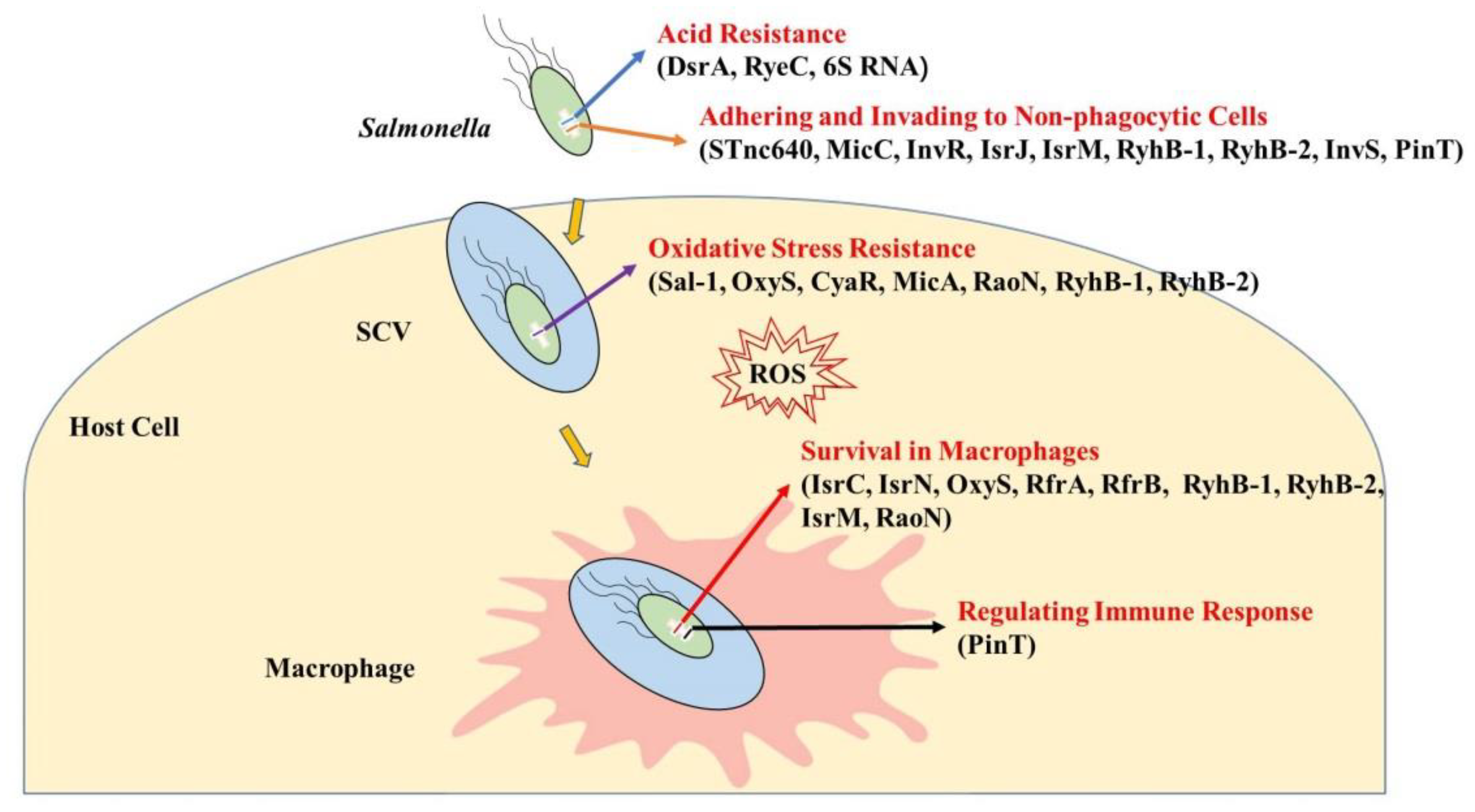
2. sRNAs’ Functions in Salmonella–Host Interactions
2.1. sRNAs Regulate Resistance to the Acidic Environment
2.2. sRNAs Regulate Adhesion to and Invasion of Non-Phagocytic Cells
2.2.1. sRNAs Regulate the Expression of a Fimbrial Subunit
2.2.2. sRNAs Regulate the Expression of Outer Membrane Proteins
2.2.3. sRNAs in SPIs and Regulation of SPI Genes Related to Invasion
2.2.4. sRNAs in OMVs
2.3. sRNAs Regulate Resistance to Oxidative Stress within Cells
2.4. sRNAs Regulate Survival in Macrophages
2.5. sRNAs Regulate the Expression of Inflammatory Cytokines in Host Cells
3. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rehman, T.; Yin, L.; Latif, M.B.; Chen, J.H.; Wang, K.Y.; Geng, Y.; Huang, X.L.; Abaidullah, M.; Guo, H.R.; Ouyang, P. Adhesive mechanism of different Salmonella fimbrial adhesins. Microb. Pathog. 2019, 137, 103748. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.D.; Ghori, N.; Falkow, S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J. Exp. Med. 1994, 180, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Bakowski, M.A.; Braun, V.; Brumell, J.H. Salmonella-containing vacuoles: Directing traffic and nesting to grow. Traffic 2008, 9, 2022–2031. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.C. Antimicrobial reactive oxygen and nitrogen species: Concepts and controversies. Nat. Rev. Microbiol. 2004, 2, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Waters, L.S.; Storz, G. Regulatory RNAs in bacteria. Cell 2009, 136, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.; Ho, J.; Liu, X.; Zhang, L.; Wong, S.H.; Chan, M.T.; Wu, W.K. Potential and use of bacterial small RNAs to combat drug resistance: A systematic review. Infect. Drug Resist. 2017, 10, 521–532. [Google Scholar] [CrossRef]
- Kwenda, S.; Gorshkov, V.; Ramesh, A.M.; Naidoo, S.; Rubagotti, E.; Birch, P.R.; Moleleki, L.N. Discovery and profiling of small RNAs responsive to stress conditions in the plant pathogen Pectobacterium atrosepticum. BMC Genom. 2016, 17, 47. [Google Scholar] [CrossRef]
- Altuvia, S.; Wagner, E.G. Switching on and off with RNA. Proc. Natl. Acad. Sci. USA 2000, 97, 9824–9826. [Google Scholar] [CrossRef]
- Chao, Y.; Vogel, J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010, 13, 24–33. [Google Scholar] [CrossRef]
- Hoe, C.H.; Raabe, C.A.; Rozhdestvensky, T.S.; Tang, T.H. Bacterial sRNAs: Regulation in stress. Int. J. Med. Microbiol. 2013, 303, 217–229. [Google Scholar] [CrossRef]
- Papenfort, K.; Vogel, J. Regulatory RNA in bacterial pathogens. Cell Host Microbe 2010, 8, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Masse, E.; Salvail, H.; Desnoyers, G.; Arguin, M. Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 2007, 10, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Rychlik, I.; Barrow, P.A. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol. Rev. 2005, 29, 1021–1040. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Zheng, K.; Liu, Z.F. Small non-coding RNAs: New insights in modulation of host immune response by intracellular bacterial pathogens. Front. Immunol. 2016, 7, 431. [Google Scholar] [CrossRef]
- Majdalani, N.; Cunning, C.; Sledjeski, D.; Elliott, T.; Gottesman, S. DsrA RNA regulates translation of RpoS message by an anti- antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 1998, 95, 12462–12467. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.; Mukherjee, M.; Nayak, R.; Dutta, R.; Suar, M. Biological and regulatory roles of acid-induced small RNA RyeC in Salmonella Typhimurium. Biochimie 2018, 150, 48–56. [Google Scholar] [CrossRef]
- Ren, J.; Sang, Y.; Qin, R.; Cui, Z.; Yao, Y.F. 6S RNA is involved in acid resistance and invasion of epithelial cells in Salmonella enterica serovar Typhimurium. Future Microbiol. 2017, 12, 1045–1057. [Google Scholar] [CrossRef]
- Althouse, C.; Patterson, S.; Fedorka-Cray, P.; Isaacson, R.E. Type 1 fimbriae of Salmonella enterica serovar Typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect. Immun. 2003, 71, 6446–6452. [Google Scholar] [CrossRef]
- Meng, X.; Meng, X.C.; Wang, J.Q.; Wang, H.; Zhu, C.H.; Ni, J.; Zhu, G.Q. Small non-coding RNA STnc640 regulates expression of fimA fimbrial gene and virulence of Salmonella enterica serovar Enteritidis. BMC Vet. Res. 2019, 15, 319. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, A.; Blyn, L.B.; Storz, G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 2004, 186, 6689–6697. [Google Scholar] [CrossRef]
- Pfeiffer, V.; Papenfort, K.; Lucchini, S.; Hinton, J.C.; Vogel, J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 2009, 16, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, V.; Sittka, A.; Tomer, R.; Tedin, K.; Brinkmann, V.; Vogel, J. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol. Microbiol. 2007, 66, 1174–1191. [Google Scholar] [CrossRef]
- Padalon-Brauch, G.; Hershberg, R.; Elgrably-Weiss, M.; Baruch, K.; Rosenshine, I.; Margalit, H.; Altuvia, S. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008, 36, 1913–1927. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Vu, G.P.; Bai, Y.; Chan, E.; Wu, R.; Yang, E.; Liu, F.Y.; Lu, S.W. A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors. PLoS Pathog. 2011, 7, e1002120. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.J.; Meng, X.; Ni, J.; He, M.P.; Chen, Y.F.; Xia, P.P.; Wang, H.; Liu, S.G.; Zhu, G.Q. Positive regulation of type III secretion effectors and virulence by RyhB paralogs in Salmonella enterica serovar Enteritidis. Vet. Res. 2021, 52, 44. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cai, X.; Wu, S.Y.; Bomjan, R.; Nakayasu, E.S.; Handler, K.; Hinton, J.C.D.; Zhou, D.G. InvS coordinates expression of PrgH and FimZ and is required for invasion of epithelial cells by Salmonella enterica serovar Typhimurium. J. Bacteriol. 2017, 199, e00824-16. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Palmer, A.D.; Vanderpool, C.K.; Slauch, J.M. The small RNA PinT contributes to PhoP-mediated regulation of the Salmonella pathogenicity island 1 type III secretion system in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2019, 201, e00312-19. [Google Scholar] [CrossRef]
- Zhao, C.H.; Zhou, Z.; Zhang, T.F.; Liu, F.Y.; Zhang, C.Y.; Zen, K.; Gu, H.W. Salmonella small RNA fragment Sal-1 facilitates bacterial survival in infected cells via suppressing iNOS induction in a microRNA manner. Sci. Rep. 2017, 7, 16979. [Google Scholar] [CrossRef]
- Briones, A.C.; Lorca, D.; Cofre, A.; Cabezas, C.E.; Kruger, G.I.; Pardo-Este, C.; Baquedano, M.S.; Salinas, C.R.; Espinoza, M.; Castro-Severyn, J.; et al. Genetic regulation of the ompX porin of Salmonella Typhimurium in response to hydrogen peroxide stress. Biol. Res. 2022, 55, 8. [Google Scholar] [CrossRef]
- Kim, S.; Lee, Y.H. Impact of small RNA RaoN on nitrosative-oxidative stress resistance and virulence of Salmonella enterica serovar Typhimurium. J. Microbiol. 2020, 58, 499–506. [Google Scholar] [CrossRef]
- Leclerc, J.M.; Dozois, C.M.; Daigle, F. Role of the Salmonella enterica serovar Typhi Fur regulator and small RNAs RfrA and RfrB in iron homeostasis and interaction with host cells. Microbiology 2013, 159, 91–602. [Google Scholar] [CrossRef]
- Penaloza, D.; Acuna, L.G.; Barros, M.J.; Nunez, P.; Montt, F.; Gil, F.; Fuentes, J.A.; Calderon, I.L. The small RNA RyhB homologs from Salmonella Typhimurium restrain the intracellular growth and modulate the SPI-1 gene expression within RAW264.7 macrophages. Microorganisms 2021, 9, 635. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Kim, S.; Helmann, J.D.; Kim, B.H.; Park, Y.K. RaoN, a small RNA encoded within Salmonella pathogenicity island-11, confers resistance to macrophage-induced stress. Microbiology 2013, 159, 1366–1378. [Google Scholar] [CrossRef] [PubMed]
- Westermann, A.J.; Forstner, K.U.; Amman, F.; Barquist, L.; Chao, Y.; Schulte, L.N.; Muller, L.; Reinhardt, R.; Stadler, P.F.; Vogel, J. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 2016, 529, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Correia Santos, S.; Bischler, T.; Westermann, A.J.; Vogel, J. MAPS integrates regulation of actin-targeting effector SteC into the virulence control network of Salmonella small RNA PinT. Cell Rep. 2021, 34, 108722. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.W.; Hall, H.K. Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 1990, 172, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Eckmann, L.; Libby, S.J.; Fang, F.C.; Okamoto, S.; Kagnoff, M.F.; Fierer, J.; Guiney, D.G. Expression of Salmonella typhimurium rpoS and rpoS-dependent genes in the intracellular environment of eukaryotic cells. Infect. Immun. 1996, 64, 4739–4743. [Google Scholar] [CrossRef]
- Ryan, D.; Ojha, U.K.; Jaiswal, S. The small RNA DsrA influences the acid tolerance response and virulence of Salmonella enterica serovar Typhimurium. Front. Microbiol. 2016, 7, 599. [Google Scholar] [CrossRef][Green Version]
- Sledjeski, D.D.; Gupta, A.; Gottesman, S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996, 15, 3993–4000. [Google Scholar] [CrossRef]
- Lease, R.A.; Smith, D.; McDonough, K.; Belfort, M. The small noncoding DsrA RNA is an acid resistance regulator in Escherichia coli. J. Bacteriol. 2004, 186, 6179–6185. [Google Scholar] [CrossRef]
- Fozo, E.M.; Kawano, M.; Fontaine, F.; Kaya, Y.; Mendieta, K.S.; Jones, K.L.; Ocampo, A.; Rudd, K.E.; Storz, G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008, 70, 1076–1093. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Sang, Y.; Ni, J.J.; Tao, J.; Lu, J.; Zhao, M.W.; Yao, Y.F. Acetylation regulates survival of Salmonella enterica serovar Typhimurium under acid stress. Appl. Environ. Microbiol. 2015, 81, 5675–5682. [Google Scholar] [CrossRef] [PubMed]
- Kanjee, U.; Houry, W.A. Mechanisms of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 2013, 67, 65–81. [Google Scholar] [CrossRef]
- Wagner, C.; Hensel, M. Adhesive mechanisms of Salmonella enterica. Adv. Exp. Med. Biol. 2011, 715, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Hara-Kaonga, B.; Pistole, T.G. OmpD but not OmpC is involved in adherence of Salmonella enterica serovar typhimurium to human cells. Can. J. Microbiol. 2004, 50, 719–727. [Google Scholar] [CrossRef]
- Hensel, M. Evolution of pathogenicity islands of Salmonella enterica. Int. J. Med. Microbiol. 2004, 294, 95–102. [Google Scholar] [CrossRef]
- Sittka, A.; Lucchini, S.; Papenfort, K.; Sharma, C.M.; Rolle, K.; Binnewies, T.T.; Hinton, J.C.; Vogel, J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008, 4, e1000163. [Google Scholar] [CrossRef]
- Li, J.; Li, N.; Ning, C.C.; Guo, Y.; Ji, C.H.; Zhu, X.Z.; Zhang, X.X.; Meng, Q.L.; Shang, Y.X.; Xiao, C.C.; et al. sRNA STnc150 is involved in virulence regulation of Salmonella Typhimurium by targeting fimA mRNA. FEMS Microbiol. Lett. 2021, 368, fnab124. [Google Scholar] [CrossRef]
- Dam, S.; Pagès, J.M.; Masi, M. Dual regulation of the small RNA MicC and the quiescent porin OmpN in response to antibiotic stress in Escherichia coli. Antibiotics 2017, 6, 33. [Google Scholar] [CrossRef]
- Wroblewska, Z.; Olejniczak, M. Hfq assists small RNAs in binding to the coding sequence of ompD mRNA and in rearranging its structure. RNA 2016, 22, 979–994. [Google Scholar] [CrossRef]
- Meng, X.; Cui, W.W.; Meng, X.C.; Wang, J.Q.; Wang, J.; Zhu, G.Q. A non-coding small RNA MicC contributes to virulence in outer membrane proteins in Salmonella Enteritidis. J. Vis. Exp. 2021, 167, e61808. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Li, J.; Meng, X.C.; Wang, H.; Wang, J.Q.; Zhu, G.Q. The non-coding small RNA InvR regulates pathogenicity of Salmonella enteritidis. Chin. J. Prev. Vet. Med. 2021, 43, 699–705. [Google Scholar] [CrossRef]
- Dos Santos, A.M.P.; Ferrari, R.G.; Conte-Junior, C.A. Virulence Factors in Salmonella Typhimurium: The Sagacity of a Bacterium. Curr. Microbiol. 2019, 76, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Chareyre, S.; Mandin, P. Bacterial iron homeostasis regulation by sRNAs. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Lou, L.X.; Zhang, P.; Piao, R.L.; Wang, Y. Salmonella pathogenicity island 1 (SPI-1) and its complex regulatory network. Front. Cell Infect. Microbiol. 2019, 9, 270. [Google Scholar] [CrossRef]
- Dos Santos, A.M.P.; Ferrari, R.G.; Conte-Junior, C.A. Type three secretion system in Salmonella Typhimurium: The key to infection. Genes Genom. 2020, 42, 495–506. [Google Scholar] [CrossRef]
- Kim, S.I.; Kim, S.; Kim, E.; Hwang, S.Y.; Yoon, H. Secretion of Salmonella pathogenicity island 1-encoded type III secretion system effectors by outer membrane vesicles in Salmonella enterica serovar Typhimurium. Front. Microbiol. 2018, 9, 2810. [Google Scholar] [CrossRef]
- McIntosh, A.; Meikle, L.M.; Ormsby, M.J.; McCormick, B.A.; Christie, J.M.; Brewer, J.M.; Roberts, M.; Wall, D.M. SipA Activation of Caspase-3 is a decisive mediator of host cell survival at early stages of Salmonella enterica serovar Typhimurium infection. Infect. Immun. 2017, 85, e00393-17. [Google Scholar] [CrossRef]
- Vonaesch, P.; Sellin, M.E.; Cardini, S.; Singh, V.; Barthel, M.; Hardt, W.D. The Salmonella Typhimurium effector protein SopE transiently localizes to the early SCV and contributes to intracellular replication. Cell Microbiol. 2014, 16, 1723–1735. [Google Scholar] [CrossRef]
- Colgan, A.M.; Kröger, C.; Diard, M.; Hardt, W.D.; Puente, J.L.; Sivasankaran, S.K.; Hokamp, K.; Hinton, J.C. The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet. 2016, 12, e1006258. [Google Scholar] [CrossRef]
- Palmer, A.D.; Kim, K.; Slauch, J.M. PhoP-mediated repression of the SPI1 type 3 secretion system in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2019, 201, e00264-19. [Google Scholar] [CrossRef] [PubMed]
- Malabirade, A.; Habier, J.; Heintz-Buschart, A.; May, P.; Godet, J.; Halder, R.; Etheridge, A.; Galas, D.; Wilmes, P.; Fritz, J.V. The RNA complement of outer membrane vesicles from Salmonella enterica Serovar Typhimurium under distinct culture conditions. Front. Microbiol. 2018, 9, 2015. [Google Scholar] [CrossRef] [PubMed]
- Mastroeni, P.; Vazquez-Torres, A.; Fang, F.C.; Xu, Y.; Khan, S.; Hormaeche, C.E.; Dougan, G. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J. Exp. Med. 2000, 192, 237–248. [Google Scholar] [CrossRef]
- Altuvia, S.; Weinstein-Fischer, D.; Zhang, A.; Postow, L.; Storz, G. A small, stable RNA induced by oxidative stress: Role as a pleiotropic regulator and antimutator. Cell 1997, 90, 43–53. [Google Scholar] [CrossRef]
- Schlosser-Silverman, E.; Elgrably-Weiss, M.; Rosenshine, I.; Kohen, R.; Altuvia, S. Characterization of Escherichia coli DNA lesions generated within J774 macrophages. J. Bacteriol. 2000, 182, 5225–5230. [Google Scholar] [CrossRef]
- Massé, E.; Gottesman, S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 4620–4625. [Google Scholar] [CrossRef]
- Calderon, I.L.; Morales, E.H.; Collao, B.; Calderon, P.F.; Chahuan, C.A.; Acuna, L.G.; Gil, F.; Saavedra, C.P. Role of Salmonella Typhimurium small RNAs RyhB-1 and RyhB-2 in the oxidative stress response. Res. Microbiol. 2014, 165, 30–40. [Google Scholar] [CrossRef]
- Steele-Mortimer, O.; Brumell, J.H.; Knodler, L.A.; Méresse, S.; Lopez, A.; Finlay, B.B. The invasion-associated type III secretion system of Salmonella enterica serovar typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol. 2002, 4, 43–54. [Google Scholar] [CrossRef]
- Srikumar, S.; Kröger, C.; Hébrard, M.; Colgan, A.; Owen, S.V.; Sivasankaran, S.K.; Cameron, A.D.; Hokamp, K.; Hinton, J.C. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog. 2015, 11, e1005262. [Google Scholar] [CrossRef]
- Medzhitov, R. Recognition of microorganisms and activation of the immune response. Nature 2007, 449, 819–826. [Google Scholar] [CrossRef]
- Zhong, J.; Kyriakis, J.M. Dissection of a signaling pathway by which pathogen-associated molecular patterns recruit the JNK and p38 MAPKs and trigger cytokine release. J. Biol. Chem. 2007, 282, 24246–24254. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, B.; Macfarlane, S.; Macfarlane, G.T. Induction of cytokine formation by human intestinal bacteria in gut epithelial cell lines. J. Appl. Microbiol. 2011, 110, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Kyrova, K.; Stepanova, H.; Rychlik, I.; Polansky, O.; Leva, L.; Sekelova, Z.; Faldyna, M.; Volf, J. The response of porcine monocyte derived macrophages and dendritic cells to Salmonella Typhimurium and lipopolysaccharide. BMC Vet. Res. 2014, 10, 244. [Google Scholar] [CrossRef]
- Kurtz, J.R.; Goggins, J.A.; McLachlan, J.B. Salmonella infection: Interplay between the bacteria and host immune system. Immunol. Lett. 2017, 190, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Garai, P.; Gnanadhas, D.P.; Chakravortty, D. Salmonella enterica serovars Typhimurium and Typhi as model organisms: Revealing paradigm of host-pathogen interactions. Virulence 2012, 3, 377–388. [Google Scholar] [CrossRef]
- Calderon, P.F.; Morales, E.H.; Acuna, L.G.; Fuentes, D.N.; Gil, F.; Porwollik, S.; McClelland, M.; Saavedra, C.P.; Calderon, I.L. The small RNA RyhB homologs from Salmonella typhimurium participate in the response to S-nitrosoglutathione-induced stress. Biochem. Biophys. Res. Commun. 2014, 450, 641–645. [Google Scholar] [CrossRef]
| Function in Infection | Serotype | sRNA | Description | Target Gene/Protein | Reference |
|---|---|---|---|---|---|
| Resisting AcidEnvironment | S. Typhimurium SB300 | DsrA | Trans-coded | rpoS | [15] |
| S. Typhimurium | RyeC | Trans-coded | ptsi | [16] | |
| S. Typhimurium | 6S RNA | Trans-coded | citGXFED, nuo operon | [17] | |
| Adhering and Invading to Non-Phagocytic Cells | S. Enteritidis 50336 | STnc640 | Trans-coded | fimA | [18,19] |
| S. Typhimurium | MicC | Trans-coded | OmpD | [20,21] | |
| S. Enteritidis | InvR | Trans-coded | OmpD | [22] | |
| S. Typhimurium | IsrJ | Trans-coded | SptP | [23] | |
| Salmonella | IsrM | Trans-coded | HilE | [24] | |
| S. Enteritidis 50336 | RyhB-1, RyhB-2 | Trans-coded | sipA, sopE | [25] | |
| S. Typhimurium | InvS | Trans-coded | PrgH, FimZ | [26] | |
| Salmonella | PinT | Trans-coded | hilA, rtsA | [27] | |
| Resisting Oxidative Stress | S. Enteritidis 2472 | Sal-1 | Trans-coded | iNOS | [28] |
| S. Typhimurium | OxyS | cis-coded | ompX | [29] | |
| S. Typhimurium | CyaR | Trans-coded | ompX | [29] | |
| S. Typhimurium | MicA | Trans-coded | ompX | [29] | |
| S. Typhimurium YK5104 | RaoN | Trans-coded | ldhA | [29] | |
| S. Typhimurium | RyhB-1, RyhB-2 | Trans-coded | - | [23,30] | |
| Survivalin Macrophages | S. Typhimurium | IsrC | cis-coded | msgA | [23] |
| S. Typhimurium | IsrN | cis-coded | STM2765 | [23] | |
| S. Typhimurium | OxyS | cis-coded | - | [23] | |
| S. Typhi | RfrA, RfrB | Trans-coded | - | [31] | |
| S. Typhimurium | RyhB-1, RyhB-2 | Trans-coded | fumA, sdhD | [32] | |
| S. Typhimurium | IsrM | Trans-coded | SopA | [24] | |
| S. Typhimurium YK5104 | RaoN | Trans-coded | - | [30,33] | |
| Regulating Inflammatory Cytokines of Hosts | S. Typhimurium | PinT | Trans-coded | IL-8, SOCS3 | [34,35] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, X.; He, M.; Xia, P.; Wang, J.; Wang, H.; Zhu, G. Functions of Small Non-Coding RNAs in Salmonella–Host Interactions. Biology 2022, 11, 1283. https://doi.org/10.3390/biology11091283
Meng X, He M, Xia P, Wang J, Wang H, Zhu G. Functions of Small Non-Coding RNAs in Salmonella–Host Interactions. Biology. 2022; 11(9):1283. https://doi.org/10.3390/biology11091283
Chicago/Turabian StyleMeng, Xia, Mengping He, Pengpeng Xia, Jinqiu Wang, Heng Wang, and Guoqiang Zhu. 2022. "Functions of Small Non-Coding RNAs in Salmonella–Host Interactions" Biology 11, no. 9: 1283. https://doi.org/10.3390/biology11091283
APA StyleMeng, X., He, M., Xia, P., Wang, J., Wang, H., & Zhu, G. (2022). Functions of Small Non-Coding RNAs in Salmonella–Host Interactions. Biology, 11(9), 1283. https://doi.org/10.3390/biology11091283







