Simple Summary
Fungi can produce many types of secondary metabolites, including mycotoxins. Poisonous mushrooms and mycotoxins that cause food spoilage have been known for a very long time. For example, Aspergillus flavus, which can grow on grains and nuts, produces highly toxic substances called Aflatoxins. Despite their menace to other living organisms, mycotoxins can be used for medicinal purposes, i.e., as antibiotics, growth-promoting compounds, and other kinds of drugs. These and other secondary metabolites produced by plant-pathogenic fungi may cause host plants to display disease symptoms and may play a substantial role in disease progression. Therefore, the identification and characterization of the genes involved in their biosynthesis are essential for understanding the molecular mechanism involved in their biosynthetic pathways and further promoting sustainable knowledge-based crop production.
Abstract
Polyketides are structurally diverse and physiologically active secondary metabolites produced by many organisms, including fungi. The biosynthesis of polyketides from acyl-CoA thioesters is catalyzed by polyketide synthases, PKSs. Polyketides play roles including in cell protection against oxidative stress, non-constitutive (toxic) roles in cell membranes, and promoting the survival of the host organisms. The genus Verticillium comprises many species that affect a wide range of organisms including plants, insects, and other fungi. Many are known as causal agents of Verticillium wilt diseases in plants. In this study, a comparative genomics approach involving several Verticillium species led us to evaluate the potential of Verticillium species for producing polyketides and to identify putative polyketide biosynthesis gene clusters. The next step was to characterize them and predict the types of polyketide compounds they might produce. We used publicly available sequences from ten species of Verticillium including V. dahliae, V. longisporum, V. nonalfalfae, V. alfalfae, V. nubilum, V. zaregamsianum, V. klebahnii, V. tricorpus, V. isaacii, and V. albo-atrum to identify and characterize PKS gene clusters by utilizing a range of bioinformatic and phylogenetic approaches. We found 32 putative PKS genes and possible clusters in the genomes of Verticillium species. All the clusters appear to be complete and functional. In addition, at least five clusters including putative DHN-melanin-, cytochalasin-, fusarielien-, fujikurin-, and lijiquinone-like compounds may belong to the active PKS repertoire of Verticillium. These results will pave the way for further functional studies to understand the role of these clusters.
1. Introduction
Verticillium spp. are ascomycete fungi with an extensive host range of more than 350 plant species [1,2]. Out of ten main plant-associated species, six cause vascular wilts in crops [3], with V. dahliae and V. albo-atrum being two of the most destructive, responsible for billions of dollars in annual losses worldwide [1,4]. Among these, V. dahliae is the most ubiquitous species and the primary cause of Verticillium wilt, especially in temperate and subtropical climates [1].
Controlling pathogenic Verticillium species is difficult due to their melanized resting structures known as microsclerotia, which serve as the primary inoculum and survive in the soil for years [5]. In addition, Verticillium sp. populations are highly diverse, with variable and complex pathogenicity due to frequent chromosomal rearrangements and the involvement of many pathogenicity-related genes [6]. The latter have been studied in V. dahliae using genome, transcriptome, proteome, and T-DNA mutant libraries [7], as well as using functional analysis of multifunctional genes involved in fungal growth and pathogenicity [8]. Previous studies have revealed that V. dahliae contains code for around 530 transcription factors classified into 42 families that play an essential role in host–pathogen interactions [9]. For example, a V. dahliae T-DNA mutant lost pathogenicity in cotton when the expression of a flanking gene encoding cytochrome P450 monooxygenase (P450, VdCYP1) was considerably suppressed [10]. Pathogenicity has also been linked to melanin production, which is regulated by the cluster-specific transcription factor VdCmr1 and two other genes within the cluster encoding a polyketide synthase (VdPKS1) and a laccase (VdLac1) [11]. In a study, using a vector-free split-marker recombination method, the ExoPG gene was deleted in V. dahliae. The results showed that ExoPG was expressed more in the highly aggressive isolate than in the weakly aggressive isolate in response to potato leaf and stem extracts [12]. The function of the isochorismatase hydrolase (ICSH1) gene in V. dahlia’s pathogenicity was investigated by Zhu et al. [13], who identified that VdICSH1 was up-regulated in V. dahliae after induction with extracts from potato tissues. Furthermore, Zhu et al. [14] compared in vitro expression of NADPH oxidase (NoxA) in highly and weakly aggressive isolates of V. dahliae after elicitation with extracts from different potato tissues and found that NoxA expression was induced more in the weakly aggressive isolate than in the highly aggressive isolate in response to leaf and stem extracts.
Fungi produce hundreds of low-molecular-weight and structurally diverse compounds called secondary metabolites or natural products (SMs and NPs, respectively) that function as toxins, antibiotics, pigments, and more [15]. SMs can be categorized into three groups: polyketides derived from acyl-CoAs, terpenes produced from acyl-CoAs, and small peptides derived from amino acids [16]. SMs such as non-ribosomal peptides, terpenes [17], alkaloids, phenols, and polyketides [18], although not considered essential for fungal growth or reproduction [19], have a wide range of functions [20]. For example, SMs are known to play a crucial role in the infection process and disease establishment [21] of numerous pathogenic fungi and in enhancing competitiveness against other microbes [22]. Ascomycete genomes contain an average of 10 non-ribosomal protein synthases (NRPS), 16 polyketide synthases (PKS), and four tryptophan synthetases (TS) and dimethylallyl tryptophan synthetases (DMATS), which is more than the secondary metabolism-related genes identified in basidiomycetes, archeo-ascomycetes, chytridiomycetes, hemi-ascomycetes, and zygomycetes [23]. Due to their high biological activity, SMs are widely used in pharmaceutical research and industry [24,25], i.e., penicillin (a β-lactam antibiotic), lovastatin (a cholesterol-lowering drug) [26], many other antibacterial compounds, and even anti-cancer drugs [27].
Advances in genomics have helped us understand fungal SMs and their biosynthesis at the whole genome scale [28]. For example, genes involved in the biosynthesis of SMs are clustered together, forming what is known as an SM cluster [29]. Moreover, the complete genome sequence of Penicillium expansum provided new insights into secondary metabolism biosynthetic gene clusters in fungi, especially those responsible for patulin and citrinin biosynthesis [30]. Genes in clusters are not constitutively expressed, and formerly actively expressed genes can become transcriptionally inactive with repeated culturing [31]. In addition, antiSMASH (a freely available online tool to investigate the presence and diversity of secondary metabolite biosynthesis gene clusters) analyses of fungal genomes showed that ascomycete biotrophs show a lower rate of SM and CAZyme-related gene gain and loss events in the genome [32]. Taylor et al. [33] predicted SM biosynthetic gene clusters in the genome of Sclerotinia sclerotiorum and analyzed their gene expression during the infection of Brassica napus and reported 80 putative SM clusters. They also found that clusters are homologous to those in the closely related plant pathogen Botrytis cinerea to produce carotenoids, hydroxamate siderophores, dihydroxynaphthalene melanin, and botcinic acid [29].
Polyketide-derived metabolites have accounted for most fungal SMs discovered thus far [34]. Polyketides have a variety of structures, ranging from single aromatic-ring compounds such as orsellinic acid, to multi-ring compounds such as aflatoxins and linear structures with an amine group, such as in fumonisin. Polyketides are synthesized from short-chain carboxylic molecules (i.e., acyl-coenzyme A [CoA]) in a process such as fatty acid biosynthesis, which is catalyzed by large multi-domain enzymes called polyketide synthases (PKSs) [17]. PKSs in fungi are large, multi-domain enzymes that act iteratively to produce the polyketide chain [35]. Fungal polyketides have remarkable biological activities and include some beneficial molecules used in medicine and agriculture. Lovastatin, for example, is used to lower cholesterol, chaetoviridin is an antibacterial agent, and echinocandins and azoxystrobin are antifungal agents. On the other hand, some fungal polyketides such as aflatoxin, patulin, zearalenone, and fumonisin B1 are harmful mycotoxins that contaminate foods, cost farmers billions of dollars each year, and threaten human, animal and plant health [36].
Recent advances in biosynthetic gene clusters have revealed several fungal PKSs that require the action of collaborating enzymes to synthesize the carbon backbone [36]. Fungal PKSs are divided into three groups based on their domain content, activity, and phylogenetic relationships: non-reducing PKSs (NR-PKSs), reducing PKSs (R-PKSs), and partially reducing PKSs (PR-PKSs) [22,37]. NR-PKSs generally synthesize aromatic polyketides and consist of starter unit-specific acyltransferase (SAT), keto synthase (KS), extender unit acyltransferase (MAT), product template (PT), and acyl-carrier protein (ACP) domains. R-PKSs generally synthesize polyketides that consist of a fully saturated carbon chain. R-PKSs contain KS, acyltransferase (AT), dehydratase (DH), methyltransferase (ME), enoyl reductase (ER), keto reductase (KR), and ACP domains. PR-PKSs synthesize aromatic polyketides and share some R-PKS domains such as KS, AT, and KR, the presence or absence of DH and ER domains varies, and they always lack an ME domain. Polyketide products can be modified by other enzymes encoded by genes that cluster together with the PKS gene, such as methyltransferases, oxidoreductases, and transcription factors [38]. Based on their proximity to PKS and similar biosynthetic genes for secondary metabolites, commonly found putative genes can be predicted to be involved in the cluster. The size of polyketide biosynthetic clusters can be small, such as the 18 kb bikaverin cluster in Fusarium fujikuroi, or larger, as in the 82 kb aflatoxin biosynthetic cluster in Aspergillus parasiticus [39,40].
Gene distance and conserved domains can identify putative loci involved in the prediction of SMs. Furthermore, phylogenetic and comparative genomic analyses are instrumental in identifying gene clusters involved in the production of SMs by making predictions of identical or related compounds that have been characterized in other fungi [41].
The main objective of this study was to identify putative PKS genes and their gene clusters in the different species of Verticillium genomes and characterize them at the structural and phylogenetic levels by using bioinformatic predictions to identify PKS clusters and the compounds they may produce, with a possible role in pathogenicity.
2. Materials and Methods
2.1. Full Genome Sequences
Full genome sequences of ten Verticillium species were obtained from the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov). The full descriptions and accession numbers of the Verticillium genomes examined in this research are displayed in Table 1.

Table 1.
Isolates numbers and genome sequence information for the species used in this study.
2.2. Prediction, Annotation, and Identification of Polyketide Synthase Genes and Gene Clusters
Putative PKS genes and related clusters in the Verticillium genomes were predicted using the fungal version of antiSMASH 6.0 known as fungiSMASH at the Secondary Metabolite Bioinformatics Portal (http://www.secondarymetabolites.org [47]) using the FASTA file of the genome sequence assembly as an input. Based on synchronized genome information, antiSMASH predicts which genes might be a part of the biosynthetic cluster. A BLASTp search was conducted against the non-redundant protein sequences in the NCBI database for each PKS or hybrid polyketide synthase-non-ribosomal peptide synthetase (PKS-NRPS) gene identified in Verticillium genomes. Furthermore, manual annotation in the CLC genomics workbench version 22 (CLCBio, Aarhus, Denmark) was performed to confirm the borders of each identified PKS cluster. For this purpose, the PKS-related ketosynthase (KS) domain in the full genome sequences of Verticillium species was searched using BLASTn as well as BLASTx searches against the whole genome in the CLC genomics workbench. Verticillium genomes with sequences of genes previously described to be needed for the PKS biosynthesis cluster were also enquired about. These included transporters, dehydrogenases, transcription factors, cytochrome oxidases, acyltransferases, as well as oxidoreductases. The regions 30 kb upstream and downstream of the predicted PKS genes were then annotated using the WebAUGUSTUS Service (http://bioinf.uni-greifswald.de/webaugustus/ [48]) with the default program parameters based on the V. longisporum gene model as the closest available genome to Verticillium. The results from both manual and fungiSMASH analyses were evaluated to obtain conclusions about the presence and absence of PKS gene clusters in the Verticillium genomes. The conservation of predicted PKS clusters among different Verticillium species was evaluated according to the PKS enzyme conservation with BLASTp on identified protein databases (identity > 50% and query coverage > 50%).
2.3. Prediction of Verticillium PKS Domains
InterPro Scan tool (https://www.ebi.ac.uk/interpro/ [49]), MOTIF search (http://www.genome.jp/tools/motif/) and NCBI’s Conserved Domain Database [50] were used to identify the domain architecture of each PKS or hybrid PKS-NRPS protein sequence. In addition, the core genes present in the identified clusters were examined for the presence of PKS domains representative of the PKS biosynthesis enzymes using the online PKS-NRPS analysis tool (http://nrps.igs.umaryland.edu/nrps [51]). A putative gene was confirmed as PKS if it contained at least the necessary PKS domains (i.e., KS, AT and ACP) [52]. The results of this analysis were also used to predict the PKS module architecture. Moreover, the PKS genes were categorized based on the presence or absence of reducing (i.e., KR, DH or ER domains) or non-reducing (i.e., SAT, TE, PT and MET) domains.
2.4. PKS Cluster Comparison Analysis of Verticillium and Other Fungi
The recovered contigs assumed to be PKS clusters were aligned with sequences of known PKS clusters publicly accessible from other annotated genomes of filamentous fungi. Preservation of synteny of clusters between Verticillium genomes and other known clusters from other fungal species investigated by BlastP search in GenBank (E-value cutoff 1e-5, query coverage > 50%, and identity > 35%) and affirmed by manual verification of potential homologous clusters and flanking sequences. Verticillium PKS clusters were considered homologous to other known fungal PKS clusters when they shared more than 50% sequence similarity, conjointly when their domain architecture showed high resemblance.
2.5. Phylogenetic Analysis
Evolutionary analyses were conducted in MEGA X [53]. For this purpose, the amino acid sequences of predicted PKS genes (in terms of PKS-III) or their KS domains alone (For NR-PKS-I and R-PKS-I sequences) were used to construct phylogenetic trees. Blastp with the Verticillium sequences was used to enquiry JGI via MycoCosm [54] as well as NCBI’s protein database. The top blast hit protein sequences identified with an E value ≤ 10−5 were used in the analysis. In this analysis, 145 Verticillium PKSs (116 R-PKSs, 19 NR-PKSs and 10 PKS-III) and 157 PKSs from other Ascomycetes (128 R-PKSs, 21 NR-PKSs and 8 PKS-III) were used in total. The sequences were then subjected to an online version of MAFFT (multiple alignments using fast Fourier transform) [55] using the iterative refinement method of E-INS-I for alignment which is normally used for sequences with multiple conserved domains. Aligned sequences were observed in BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html; accessed on 12 March 2022) and manually trimmed by removing non-aligned sequences. In each case, the best evolutionary model was anticipated using ProtTest [56]. The Verticillium PKS evolutionary history was constructed using the maximum likelihood as well as neighbor joining methods with 1000 bootstrap replicates. In all cases, the tree with the highest log-likelihood was plotted (Figure 1 and Figure 2, and Supplementary Figure S1). The initial tree(s) for the heuristic search were obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the appropriate models, and then selecting the topology with a superior log-likelihood value. All positions with less than 95% site coverage were eliminated, i.e., fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position (partial deletion option).

Figure 1.
Maximum likelihood tree of 116 and 128 Verticillium and other fungi beta-ketoacyl synthase (KS) domains of the reducing PKS-I gene sequences, respectively, that were used in this study. The 26 main clades recovered for the Verticillium species are labeled from VRPKS-I-1 to VRPKS-I-26. Similar groups were obtained with the neighbor-joining analysis. GenBank accession numbers for each of these sequences are provided. In this analysis, we used 1000 bootstrap repeats as indicated in the internodes.
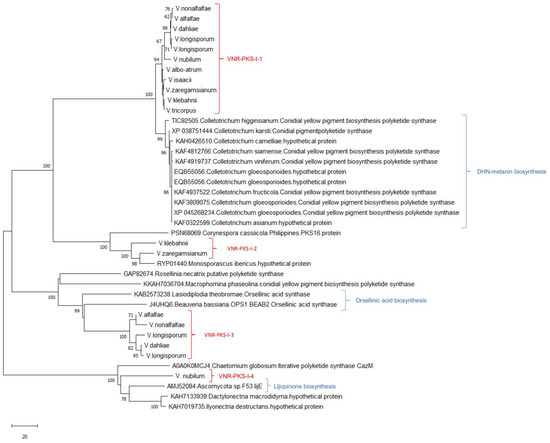
Figure 2.
Neighbor-joining tree inferred from nucleotide sequences of non-reducing PKS coding regions of Verticillium species as well as NR-PKS sequences obtained from other Ascomycetes. GenBank accession numbers for each sequence are provided. Similar groups were obtained with the maximum likelihood analysis. In this analysis, we used 1000 bootstrap repeats as indicated in the internodes.
3. Results
3.1. Prediction of Verticillium PKS Domains
Discovering domain architecture in each predicted PKS gene would help determine if all essential domains involved in the functionality of PKS genes are present or not. It would also help to understand whether the final PKS products are reduced or non-reduced. Domain analysis was performed for all the anticipated PKS and PKS-NRPS protein sequences using the PKS-NRPS webpage and motif finder. Since VNR-PKS-I-1 to VNR-PKS-I-4 did not comprise reducing domains, they are most likely non-reducing PKS enzymes. On the other hand, based on all optional domains expected to be present in all highly reducing PKS enzymes, including KR, DH, and ER domains, VRPKS-I-1 to VRPKS-I-26 are likely to be of the reducing type of PKSs enzymes (Figure 3, Table 2 and Table 3). The only exception was VRPKS-I-10, with a domain organization of KS-AT-KR-ACP that was found only in V. nubilum. Based on the domain architecture of VRPKS-I-10, this enzyme is likely to be a partially reducing PKS, as it comprises the KR domain but not the complete DH or ER domains. In the VRPKS-I-5 clade, the PKS enzyme in V. albo-atrum, V. zaregamsianum, V. isaacii, and V. klebahnii comprised of a KS-AT-DH-MET-ER-KR-C-A-ACP domain organization. The VRPKS-I-5 enzyme in V. tricorpus showed an extra NAD_binding_4 domain at the end of the protein sequence. The same unusual domain (NAD_binding_4) was also observed in VRPKS-I-12, where PKS enzymes in V. isaacii, V. klebahnii and V. tricorpus contained a KS-AT-DH-MET-ER-KR-ACP-C-A-ACP-NAD_binding_4 domain architecture, whereas the NAD_binding_4 domain was missing in the V. zaregamsianum and Verticillium albo-atrum VRPKS-I-12 enzyme. Genomic analyses on the PKS genes and gene clusters of Verticillium species revealed that, in total, a minimum of 10 (V. nonalfalfae and V. dahliae) and a maximum of 19 (V. klebahnii) PKSs or hybrid PKS-NRPS biosynthetic gene clusters were present in the genomes of different Verticillium species.

Figure 3.
Predicted PKS and PKS-NRPS biosynthetic genes in different Verticillium species.

Table 2.
Comparison of the size (in amino acid residues) and domain structure of the Verticillium NR-PKS-I proteins with that of the top BLASTp hit in the NCBI database.

Table 3.
Comparison of the size and domain structure of the Verticillium R-PKS-I proteins with top BLASTp hits in the NCBI database.
3.2. Comparison of Verticillium PKS Cluster with Those of Other Fungi
According to the phylogenetic analyses, 19 PKS core enzymes (VRPKS-I-1, VRPKS-I-2, VRPKS-I-3, VRPKS-I-6, VRPKS-I-7, VRPKS-I-8, VRPKS-I-13, VRPKS-I-16, VRPKS-I-17, VRPKS-I-21, VRPKS-I-22, VRPKS-I-23, VRPKS-I-24, VRPKS-I-25, VRPKS-I-26, VNR-PKS-I-1, VNR-PKS-I-2, VNR-PKS-I-3, and VNR-PKS-I-4), showed high resemblance to PKS enzymes from previously known gene clusters. Therefore, comparative genomic approaches were used to predict the probable final products of these secondary metabolite biosynthetic clusters. To this end, flank genes were checked, in addition to the core genes of identified Verticillium clusters, and compared to the previously known clusters from other fungal species. These comparisons revealed five interesting PKS-related clusters that displayed previously characterized orthologous gene clusters in other Ascomycetes putatively associated with the biosynthesis of a cytochalasan-like compound (VRPKS-I-3), a fusarielien-like compound (VRPKS-I-7), a fujikurin-like compound (VRPKS-I-8), a melanin compound (VNR-PKS-I-1), and a lijiquinone-like compound (VNR-PKS-I-4). A detailed explanation of the genes proposed for each of the identified Verticillium PKS clusters is shown in Supplementary Table S1.
3.2.1. Clusters Common among All Examined Verticillium Genomes
The VNR-PKS-I-1 gene and cluster highly resembled those from other fungi that produce dihydroxynaphthalene (DHN) melanin (Figure 4A,B and Supplementary Tables S1 and S2). A 1, 3, 8-trihydroxynaphthalene reductase and a cmr1 transcription factor were contained within the Verticillium VNR-PKS-I-1 cluster. Homologs of SCD1 and THR1 reductase coding genes were also found in all Verticillium species at different locations rather than the contigs in which the initial PKS cluster was found. Interestingly, the melanin biosynthesis cluster in V. albo-atrum, V. isaacii, V. zaregamsianum, V. klebahnii and V. tricorpus was found next to VRPKS-I-12, which is a PKS-NRPs cluster. Remarkably, the whole VRPKS-I-12 cluster was missing in other Verticillium species (Figure 4B, Supplementary Table S1).
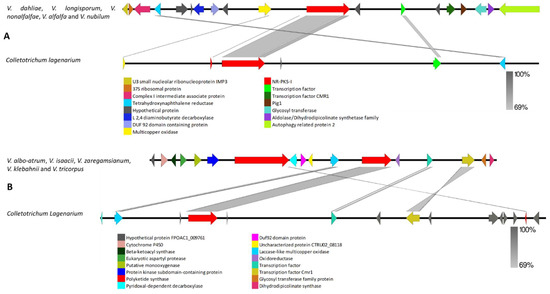
Figure 4.
Comparison of putative cluster genes for Verticillium VNR-PKS-I-1 and the DHN melanin-producing PKS. (A) Putative VNR-PKS-I-1 cluster of V. dahliae, V. longisporum, V. nonalfalfae, V. alfalfae and V. nubilum compared to Colletotrichum lagenarium DHN melanin cluster (B) Putative biosynthetic cluster for V. albo-atrum, V. isaacii, V. zaregamsianum, V. klebahnii and V. tricorpus gene compared to C. lagenarium DHN melanin cluster. The arrowheads indicate the direction of transcription and the types of genes common to the two clusters being compared are displayed in the same color. Shaded lines represent similarities between nucleotide sequences.
Orthologs for the PKS-III backbone gene were found in all examined Verticillium genomes. A highly conserved single copy of this cluster was also observed in all the Verticillium genomes (Figure 5). This cluster is composed of an oxidoreductase, a feruloyl esterase, a fungal-specific transcription factor, a BFR2 protein, a PKS-III, a fructose bisphosphate aldolase, a specific exonuclease, a mitochondrial two oxoglutarate carrier protein, a hypothetical protein, a splicing factor spf30, an iron–sulphur protein, a cell division control protein, and a short-chain dehydrogenase. This cluster thus has all the required gene inventories of a functional PKS cluster. The genes in this cluster exhibited high similarity to the orthologues present in a range of ascomycetes, including Colletotrichum higginsianum, Colletotrichum incanum, and Sodiomyces alkalinus (Figure 5, Supplementary Tables S1 and S2).
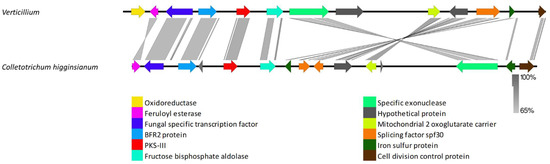
Figure 5.
Comparison of Verticillium genes flanking PKS-III that are closely related homologs of PKS-III gene cluster from Colletotrichum higginsianum. The arrowheads indicate the direction of transcription and the types of genes common to the two clusters being compared are displayed in the same color. Shaded lines represent similarities between nucleotide sequences. This highly conserved cluster was found in all Verticillium genomes examined.
Notably, clusters VRPKS-I-17, VRPKS-I-19, VRPKS-I-23, and VRPKS-I-26 are also found in all Verticillium genomes. However, they did not show any similarities in terms of gene content to the known PKS clusters from other fungi.
3.2.2. Clusters Missing in a Few Genomes but Present in Other Species
The VRPKS-I-8 cluster was found only in the pathogenic species, i.e., V. dahliae and V. longisporum, and displayed a high resemblance to the fujikurin synthase gene (FfuPks19) of F. fujikuroi and showed significant similarities (39–85% protein identity) to genes in the fujikurin biosynthesis cluster in protein–protein comparisons (Figure 6, Supplementary Table S1). The FfuPks19 showed a domain organization of KS-AT-DH-ER-KR. The VRPKS-I-8 has a similar domain organization (Table 2). The fujikurin biosynthesis cluster contains five genes, including a highly reducing PKS (CCT72377), an enoyl reductase domain-containing protein (CCT72378), a hydrolase domain-containing protein (CCT72379), a putative transcription factor (CCT72381), and a putative transporter (CCT72380). Nevertheless, homologs genes of those found in the F. fujikuroi fujikurin biosynthesis cluster were found in V. dahlia and V. longisporum (Figure 6). Hydrolase, transcription factor, and transporter genes were found on different contigs of the V. longisporum genome. This could be due to assembly problems. In this case, the sequences were assembled to the reference genome to obtain the whole cluster in one contig. Interestingly, two more genes designated as cytochrome P450 and MFS transporter were found in the VRPKS-I-8 cluster in V. dahliae and V. longisporum, which were not identified in the FfuPks19 cluster (Figure 6).
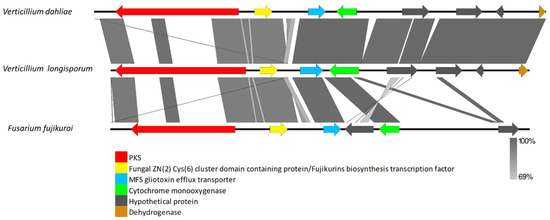
Figure 6.
Comparison of V. dahliae and V. longisporum genes flanking VRPKS-I-8 PKSs that are closely related homologs of fujikurin synthase gene cluster from Fusarium fujikuroi. The arrowheads indicate the direction of transcription and the types of genes common to the two clusters being compared are displayed in the same color. Shaded lines represent similarities between nucleotide sequences.
The VRPKS-I-20 was found only in V. isaacii, V. klebahnii, and V. tricorpus and showed high protein sequence similarity to an unknown PKS identified in Penicillium arizonense (XP_022486633). The VRPKS-I-20 protein exhibited the same domain architecture (KS-AT-DH-MET-ER-KR-ACP) as PKS identified in Penicillium arizonense. Synteny comparison between VRPKS-I-20 locus in V. isaacii, V. klebahnii and V. tricorpus genomes and Penicillium arizonense reveals extensive conservation (Figure 7). Besides PKS, the cluster comprised of some putative cluster genes, including a mannan endo-1,4-beta-mannosidase coding gene displaying 82% identity with the orthologue present in Plectosphaerella plurivora (KAH6695781), a cytochrome P450 exhibiting 75 % identity with the orthologue present in Colletotrichum fioriniae (EXF83899), a hypothetical protein exhibiting 72% identity with the orthologue present in Penicillium arizonense (XP_022486531), another cytochrome P450 displaying 65% identity with the orthologue present in Penicillium arizonense (XP_022486670), and an oxidoreductase protein exhibiting 73% identity with the orthologue present in Talaromyces rugulosus (XP_035348268).
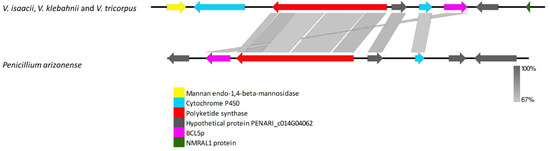
Figure 7.
Comparison of putative cluster genes for VRPKS-I-20 locus found in the genomes of V. isaacii, V. klebahnii and V. tricorpus and an unknown PKS cluster from Penicillium arizonense. The arrowheads indicate the direction of transcription and the types of genes common between the two clusters being compared are displayed in the same color. Shaded lines represent similarities between nucleotide sequences.
The VRPKS-I-6 core gene clustered with burnettramic acid biosynthesis PKS from Aspergillus burnettii and other Aspergillus PKS genes, including A. avenaceus hypothetical protein (KAE8150730) and A. alliaceus burnettramic acids biosynthesis cluster protein A (KAE8392342) (Figure 1). The genes flank to the core gene also showed high similarity in protein sequences to the A. alliaceus cluster (Figure 8, Supplementary Table S1). VRPKS-I-6 cluster found in V. zaregamsianum, V. isaacii, V. klebahnii and V. tricorpus comprised of a PKS-NRPS displaying 67% identity with the orthologue present in A. avenaceus (KAE8150730), a hypothetical protein is exhibiting 64% identity with the orthologue present in Didymosphaeria enalia (KAF2268082), a hydrolase exhibiting 58% identity with the orthologue present in Clohesyomyces aquaticus (ORY13587), a cytochrome P450 oxygenase displaying 66% identity with the orthologue present in A. avenaceus (KAE8150736), a dehydrogenase displaying 77% identity with the orthologue present in A. avenaceus (KAE8150737), and another cytochrome P450 oxygenase exhibiting 65% identity with the orthologue found in A. avenaceus (KAE8150739). VRPKS-I-6 (KS-AT-DH-ER-KR-ACP-C-A-ACP) exhibited the same domain organization as that of burnettramic acid biosynthesis PKS from A. burnettii and A. avenaceus (KS-AT-DH-MET-ER-KR-ACP-C-A-ACP) except that it lacks an MT domain (Table 3). Hence, methyl groups may be missing from the final product of the VRPKS-I-6 gene cluster.
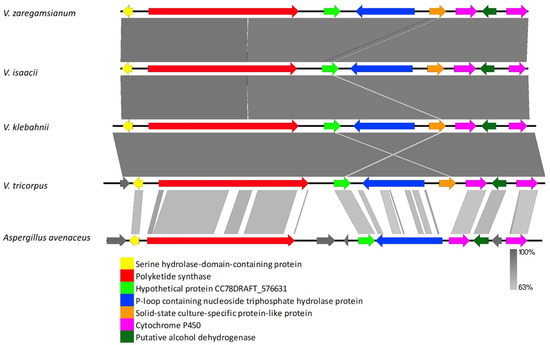
Figure 8.
Comparison of putative cluster genes for VRPKS-I-6 cluster found in V. zaregamsianum, V. isaacii, V. klebahnii and V. tricorpus and burnettramic acid biosynthesis PKS cluster from Aspergillus avenaceus. The arrowheads indicate the direction of transcription and the types of genes common to the two clusters being compared are displayed in the same color. Shaded lines represent similarities between nucleotide sequences.
VRPKS-I-21 was found in V. zaregamsianum, V. klebahnii and V. nubilum and grouped in a clade with a good bootstrap value to the alternapyrone biosynthesis gene alt5 protein sequence from Alternaria alternata and lovastatin diketide synthase LovF 2 from Colletotrichum chlorophyte (Figure 1). The A. alternata Alt5 protein showed the following domain organization: KS-AT-DH-MT-ER-KR-ACP. The VRPKS-I-21 showed the same domain architecture, except it lacks an ACP domain. In the VRPKS-I-21 biosynthetic cluster, VRPKS-I-21 is sitting next to some putative cluster-related genes such as NADH-ubiquinone oxidoreductase, cytochrome p450 domain-containing protein, and drug resistance protein, but not the oxidase or the other two cytochrome P450 proteins that may be part of the alternapyrone biosynthesis gene cluster (Supplementary Figure S2).
VRPKS-I-22 was found only in pathogenic V. dahliae, V. longisporum, V. alfalfae, and V. nonalfalfae and showed a high resemblance in protein sequence to the fusaric acid gene cluster core gene, PKS6 (Figure 1, Supplementary Table S2). VRPKS-I-22 exhibited the same domain organization (KS-AT-DH-MET-ER-KR-ACP) as that of fusaric acid biosynthesis PKS from F. mexicanum (KAF5539484) and F. miscanthi (ALQ32877). However, the rest of the genes in the VRPKS-I-22 locus did not show any similarities to the genes needed for fusaric acid production. The VRPKS-I-22 cluster is comprised of a sulfate permease, displaying 51% identity with the orthologue present in Trematosphaeria pertusa (XP_033684343); a hydrolase, exhibiting 62% identity with the orthologue present in Monosporascus sp. (RYP55738); two hypothetical proteins, exhibiting 60% and 79% identity with the orthologues present in Fusarium graminearum (CZS74879) and Colletotrichum musicola (KAF6840530), respectively; an MSF-1 protein, exhibiting 85% identity with the orthologue present in Sodiomyces alkalinus (XP_028463299); a methionine aminipeptidase, displaying 82% identity with the orthologue present in Colletotrichum sidae (TEA17142); a peroxiredoxin family protein, displaying 67% identity with the orthologue present in Sodiomyces alkalinus (XP_028463297); and an amino acid aminotransferase, exhibiting 75% identity with the orthologue present in Sodiomyces alkalinus (XP_028463295). On the other hand, the five putative fusaric acid cluster genes encode proteins with resemblance to a homoserine-O-acyltransferase, a hypothetical protein, a serine hydrolase, an aspartate kinase, and a polyketide synthase (Supplementary Figure S3).
3.2.3. Species-Specific Clusters
The VNR-PKS-I-4 gene cluster was found only in V. nubilum. Protein sequences of V. nubilum VNR-PKS-I-4 genes revealed a ~48 kb cluster, comprised of 12 open reading frames with close resemblance to the lijiquinone biosynthesis cluster found in Muyocopron atromaculans (Figure 9, Supplementary Tables S1 and S2). The protein sequences of the V. nubilum VNR-PKS-I-4 genes showed high similarity to putative hydroxylase exhibiting 78−81% identity with the orthologue present in M. atromaculans (XP_007816049) and Dactylonectria macrodidyma (KAH7133940), a hypothetical protein exhibiting 49% identity with the orthologue present in Hypoxylon sp (OTB08973), an R-PKS (VRPKS-I-15) displaying 61−68% identity with the orthologue present in Dactylonectria macrodidyma (KAH7133936) and Fusarium fujikuroi (KLP04343), a short chain dehydrogenase exhibiting 66% identity with the orthologue present in Ilyonectria destructans (KAH7019726), a serine hydrolase exhibiting 54−61% identity with the orthologue present in M. atromaculans (PVH94008) and Dactylonectria macrodidyma (KAH7133931), a putative short-chain dehydrogenase exhibiting 74% identity with the orthologue found in M. atromaculans (XP_007816049), another putative hydrolase displaying 74% identity with the orthologue present in Dactylonectria macrodidyma (KAH7133934), a putative oxidase exhibiting 71% identity with the orthologue present in M. atromaculans (KEQ59244), an NR-PKS (VNR-PKS-I-4) exhibiting 68−61% identity with the orthologue present in M. atromaculans (AMJ52084), Pseudogymnoascus destructans (XP_024319559) and Dactylonectria macrodidyma (KAH7133936), an inorganic phosphate transport displaying 42% identity with the orthologue present in Scedosporium apiospermum (XP_016645996), a putative O-methyltransferase exhibiting 86% identity with the orthologue present in Dactylonectria macrodidyma (KAH7133929) and a putative ariadne-like RING finger protein R811 exhibiting 56% identity with the orthologue present in Colletotrichum siamense (XP_024319559) (Supplementary Table S1). The M. atromaculans R-PKS found in the lijiquinone biosynthesis cluster showed a KS-AT-DH-MT-ER-KR-ACP domain organization. V. nubilum VRPKS-I-15 showed the same domain architecture but lacked an ACP domain. The domain organization of the second PKS in the cluster, VNR-PKS-I-4 (KS-AT-ACP-MT-R) and lijiquinone biosynthesis NR-PKS (SAT-KS-AT-PT-ACP-MET-R) was somewhat different, as the SAT and PT domains were missing in VNR-PKS-I-4.
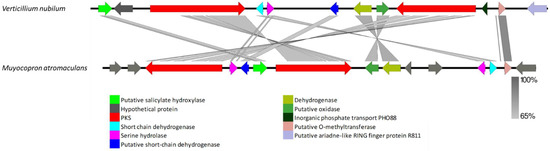
Figure 9.
Comparison of putative cluster genes for Verticillium nubilum VNR-PKS-I-4 and the lijiquinone biosynthesis cluster of Muyocopron atromaculans. The arrowheads indicate the direction of transcription and the types of genes common to the two clusters being compared are displayed in the same color. Shaded lines represent similarities between nucleotide sequences.
VRPKS-I-3, a PKS-NRPS cluster, was found only in the V. albo-atrum genome and grouped in a clade with a good bootstrap value to the cytochalasans biosynthesis PKS sequences, with the most significant similarity to pyrichalasin H synthase protein from Pyricularia oryzae (A0A4P8WAE5) (Figure 1). The fungiSMASH could not predict a product for the V. albo-atrum VRPKS-I-3 cluster, but it likely allows for the formation of cytochalasans in this fungus. VRPKS-I-3 had a KS-AT-DH-ER-KR-C-A-ACP domain organization, identical to the domain organization of pyrichalasin H synthase protein. Both V. albo-atrum VRPKS-I-3 and P. oryzae cytochalasan clusters were predicted to have high similarity in terms of protein sequences and gene order (Figure 10, Supplementary Table S1).
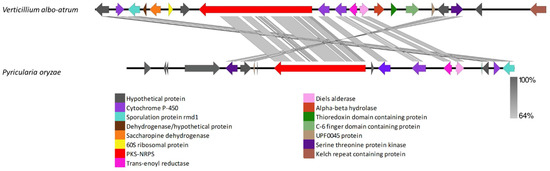
Figure 10.
Comparison of putative cluster genes for Verticillium albo-atrum VRPKS-I-3 and P. oryzae cytochalasan clusters. The arrowheads indicate the direction of transcription and the types of genes common to the two clusters being compared are displayed in the same color. Shaded lines represent similarities between nucleotide sequences.
VRPKS-I-7 was found only in one copy in V. zaregamsianum and grouped in a clade with the fusarielin biosynthesis gene PKS-9 protein sequence from Fusarium pseudograminearum. VRPKS-I-7 showed a high protein sequence similarity of 82% with the F. pseudograminearumas PKS9 fusarielin biosynthesis (XP 009262583) protein with an identical PKS domains sequence of KS-AT-DH-MET-ER-KR. In addition to a highly reducing PKS gene, the cluster contains a protein with a thioesterase (FSL2) domain (FGSG_10463) exhibiting 61% sequence identity with the orthologue present in V. zaregamsianum VRPKS-I-7 cluster, an epimerase (FSL3)-containing domain protein (FGSG_17368) exhibiting 55% identity with the orthologue present in VRPKS-I-7 cluster, a cytochrome P450 oxygenase (FSL4) protein (FGSG_10461) exhibiting 57% identity with the orthologue present in VRPKS-I-7 cluster, an enoyl reductase (FSL5) protein (FGSG_17367) with no homologs found in V. zaregamsianum VRPKS-I-7 cluster, and a transcription factor (FSL7) protein (FGSG_10458) exhibiting 85% identity with the orthologue present in VRPKS-I-7 cluster. The fusarielin biosynthesis gene cluster also contains a hypothetical protein (FGSG_10459) with an unknown role in the fusarielin biosynthesis pathway exhibiting 85% identity with the orthologue present in the VRPKS-I-7 cluster (Figure 11, Supplementary Table S1).
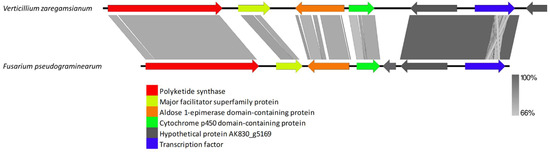
Figure 11.
Comparison of putative cluster genes for Verticillium zaregamsianum VRPKS-I-7 cluster and fusarielin biosynthesis gene cluster from Fusarium pseudograminearum. The arrowheads indicate the direction of transcription and the types of genes common to the two clusters being compared are displayed in the same color. Shaded lines represent similarities between nucleotide sequences.
None of the other Verticillium R-PKS clusters showed similarities to previously identified or unknown clusters from other fungi. However, based on their gene content and cluster-related genes such as transporters, cytochrome P-450, hydrolases, dehydrogenases, and transcription factors, some of these Verticillium R-PKS clusters such as VRPKS-I-1, VRPKS-I-9, VRPKS-I-17 and VRPKS-I-25 are likely to represent a functional PKS biosynthesis cluster.
3.3. Missing PKS Gene Clusters in Closely Related Species
To determine which PKS or PKS-NRPS genes/clusters were present in different Verticillium species but absent in closely related species or vice versa, we examined the distribution patterns of these genes. Such distribution patterns were observed in seven PKS or PKS-NRPS genes. For instance, VRPKS-I-2 was found in V. albo-atrum, V. isaacii, V. tricorpus and V. klebahnii, but not in V. zaregamsianum. The presence of the VRPKS-I-8 gene (likely involved in fujikurine biosynthesis) in V. dahliae and V. longisporum and its absence in closely related species (V. alfalfae and V. nonalfalfae) is also in congruence with gene gain/loss events. However, more analysis is needed to confirm this statement. In another example, VRPKS-I-11 was absent in V. dahliae but was present in V. longisporum, V. alfalfae and V. nonalfalfae. In V. dahliae, the second PKS-NRPS gene along with seven other genes next to VRPKS-I-11 was missing (Figure 12). The VRPKS-I-9 cluster was lost in V. zaregamsianum, but intact homologs of this cluster were observed in V. isaacii, V. tricorpus and V. klebahnii. Interestingly, the whole VRPKS-I-9 cluster was also observed in V. alfalfa and V. nonalfalfae. Homologs of the VRPKS-I-18 cluster were found in V. isaacii, V. zaregamsianum and V. klebahnii, but not in V. tricorpus. Such PKS gene gain/loss event was also found in VRPKS-I-21 homologs. The VRPKS-I-21 gene cluster was observed in V. zaregamsianum and V. klebahnii but was absent in V. isaacii and V. tricorpus. It is also worth noting that the homolog of the VRPKS-I-21 cluster was found in V. nubilum, a distantly related species.
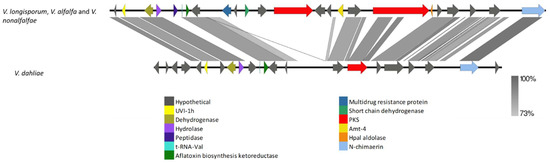
Figure 12.
Comparison of VRPKS-I-11 gene cluster among pathogenic strains of Verticillium. VRPKS-I-11 was absent in V. dahliae but was present in V. longisporum, V. alfalfae, and V. nonalfalfae.
3.4. Phylogenetic Analysis
The NR-PKS phylogenetic tree predicted five Verticillium clades, labeled as VNR-PKS-I-1 to VNR-PKS-I-4. The NR-PKS phylogenetic analysis confirmed that these predicted VNR-PKSs were present in all Verticillium genomes clustered with previously described enzymes. The presence of VNR-PKSs in different Verticillium species is illustrated in Figure 2 and Figure 3. The VNR-PKS-I-1 sequences were found in all Verticillium genomes and grouped with melanin biosynthesis PKSs from different Colletotrichum species. VNR-PKS-I-2 was only found in V. zaregamsianum, and V. klebahnii clustered with grayanic acid (GRA) synthase which is involved in the production of orcinol depsidone specific to in Cladonia grayi [57]. VNR-PKS-I-3 was detected in the genomes of V. dahliae, V. longisporum, V. alfalfae and V. nonalfalfae, forming a distinct and well-supported clade with orsellinic acid synthase, OpS1 which is responsible for producing oosporein toxin synthesized by entomopathogenic fungus and Beauveria bassiana [58]. VNR-PKS-I-4 was found only in V. nubilum and clustered with the azaphilone polyketide synthesis coding gene.
Phylogenetic analysis of the KS domain of the predicted R-PKS-I protein sequences separated Verticillium species into 26 well-supported clades, labeled from VRPKS-I-1 to VRPKS-I-26. The presence of VRPKSs in different Verticillium species is illustrated in Figure 1 and Figure 3. The VRPKS-I-1 clade consists of sequences from V. dahliae, V. zaregamsianum, V. alfalfa, and V. longisporum. The VRPKS-I-2 clade included sequences from the closely related species V. isaacii, V. klebahnii, V. tricorpus, and V. albo-atrum. The VRPKS-I-3 clade was observed only in the V. albo-atrum genome. The VRPKS-I-4, VRPKS-I-10, and VRPKS-I-15 clades contained sequences found only in V. nubilum. The VRPKS-I-5, VRPKS-I-12, VRPKS-I-16, and VRPKS-I-25 clades consist of sequences from V. isaacii, V. klebahnii, V. tricorpus, V. zaregamsianum, and V. albo-atrum. The VRPKS-I-6, VRPKS-I-13, and VRPKS-I-14 clades included KS sequences from V. isaacii, V. klebahnii, and V. tricorpus, V. zaregamsianum. The VRPKS-I-7 clade was observed only in the V. zaregamsianum genome, while the VRPKS-I-8 clade was detected only in the V. dahliae and V. longisporum genomes. The VRPKS-I-9 clade included sequences from V. isaacii, V. klebahnii, V. tricorpus, V. albo-atrum, V. alfalfae, and V. nonalfalfae. The VRPKS-I-11 and VRPKS-I-22 clades were comprised of KS sequences from V. dahliae, V. nonalfalfae, V. alfalfae, and V. longisporum. The VRPKS-I-17, VRPKS-I-19, VRPKS-I-23, and VRPKS-I-26 clades were found in all Verticillium genomes. The VRPKS-I-18 clade consists of KS sequences from V. isaacii, V. klebahnii, and V. zaregamsianum. The VRPKS-I-20 clade included sequences from V. isaacii, V. klebahnii, and V. tricorpus. Finally, the VRPKS-I-21 clade contained KS sequences from V. klebahnii, V. zaregamsianum, and V. nubilum.
The HR-PKS phylogenetic analysis showed that 15 out of the 26 predicted Verticillium HR-PKSs (VRPKS-I-1, VRPKS-I-2, VRPKS-I-3, VRPKS-I-6, VRPKS-I-7, VRPKS-I-8, VRPKS-I-13, VRPKS-I-16, VRPKS-I-17, VRPKS-I-21, VRPKS-I-22, VRPKS-I-23, VRPKS-I-24, VRPKS-I-25, and VRPKS-I-26) grouped with previously described enzymes. VRPKS-I-1 and VRPKS-I-2 grouped with oxaleimides biosynthesis-related genes, poxE from Penicillium oxalicum, and fusaridione A synthase and fsds from Fusarium heterosporum. VRPKS-I-3 clustered with the clade that contains ffsA, which is involved in cytochalasin production. VRPKS-I-6 was grouped in a clade that contains burnettramic acids biosynthesis cluster protein-coding A. VRPKS-I-7 and VRPKS-I-8 were grouped with fusarielin toxin biosynthesis gene PKS-9 from Fusarium pseudograminearum and fujikurin synthase coding gene from F. fujikuroi, respectively. VRPKS-I-13 was clustered with sdnO, involved in sordarin biosynthesis, a glycoside antibiotic with the ability of protein inhibition in fungi produced by Sordaria araneosa [59] and an unknown PKS gene from Penicillium sp. VRPKS-I-16 and VRPKS-I-17 were grouped with the T-toxin synthase PKS2 from Cochliobolus heterostrophus and the reducing polyketide synthase from Colletotrichum species which is also involved in T-toxin biosynthesis [60]. VRPKS-I-21 clustered in a clade containing the alt5 PKS gene. VRPKS-I-22 and VRPKS-I-23 clustered with fusaric acid synthase coding gene FUB1 from Fusarium species and phenolpthiocerol synthesis polyketide synthase ppsA from Sodiomyces alkalinus, respectively [61,62]. VRPKS-I-23 was clustered with FUB1 from the Colletotrichum species. VRPKS-I-24 was found only in V. albo-atrum and grouped with a PKS gene involved in trichothecene biosynthesis. VRPKS-I-25 and VRPKS-I-26 grouped with fungal maleidrides, scytalidin scyPKS from Stachybotrys chartarum, and fumagillin synthase from Aspergillus fumigatus [63]. VRPKS-I-26 also clustered with the phaseopelide biosynthesis coding gene amplA, a fungal aliphatic macrolide from Arthrinium phaeospermum [64]. The remaining R-PKS genes from Verticillium species did not show any resemblance to known PKS cluster-related genes. Hence, 15 out of 26 R-PKS genes were grouped with previously known PKS genes from other Ascomycetes, allowing us to further investigate their putative functions. Verticillium R-PKS-I phylogeny analysis proposed independent and multiple evolutionary origins for the PKS gene in the Verticillium genomes.
4. Discussion
4.1. PKS Identification
Although polyketides have been claimed to be pathogenicity factors in fungi, our knowledge of PKSs in different members of Verticillium is very limited. In this study, using ten publicly available Verticillium genome sequences, we found that different species of Verticillium differ in PKS-related genes/gene clusters and possible secondary metabolite production. For example, V. klebahnii and V. tricorpus showed much higher PKS clusters than V. alfalfae and V. dahliae. The genome analysis of V. dahliae has already revealed secondary metabolite genes and gene clusters [21]. Similar research was conducted by Hansen et al. [65], in which they compared PKSs and non-ribosomal peptide synthetases (NRPSs) from ten different Fusarium species and found 52 NRPS and 52 PKS orthology groups.
In this study, we identified a minimum of 10 (V. nonalfalfae and V. dahliae) and a maximum of 19 PKS or hybrid PKS-NRPS biosynthetic gene clusters in the Verticillium genomes and predicted products synthesized by these gene clusters. All the Verticillium PKS and hybrid PKS-NRPS genes encode enzymes that include all the required PKS domains. With each Verticillium PKS or hybrid PKS-NRPS, we were able to determine which genes would be part of biosynthetic clusters. Besides phylogenetic analysis, we also searched for homologs of the identified clusters in other fungi. Based on phylogenetic and genomic data and chemical structure considerations, we propose that five PKSs in the VNR-PKS-I-1, VNR-PKS-I-4, VRPKS-I-3, VRPKS-I-7, and VRPKS-I-8 clades are involved in synthesizing known products that include melanin, lijiquinone, cytochalasin, fusarielin and fujikurin, respectively.
We showed that most PKS genes found in Verticillium species are in predicted SM biosynthetic clusters, suggesting that the increase in PKS gene copy number specifically involves genes related to secondary metabolite production. Fatema et al. [66] reported similar findings in C. rosea and stated that most PKS genes were in predicted SM biosynthetic clusters. In the present study, the PKSs were divided into three (VNR-PKSs, VRPKSs and PKS-III) groups and further divided into 32 well-supported PKS gene clades. Based on the domain structures in each clade, we speculate that PKS homologs within each of the 32 R-PKS clades are responsible for the synthesis of the same polyketide structure that is different from polyketides synthesized by PKSs in other clades.
Homologs of the PKS clades VRPKS-I-17, VRPKS-I-19, VRPKS-I-23, and VRPKS-I-26 were detected in all Verticillium species. The groupings among the sequences in these clades largely resemble the Verticillium taxonomic expectations. This finding is consistent with studies conducted by Brown and Proctor [22], who identified 488 PKS genes in 31 Fusarium species and stated multiple and disparate patterns of distribution. The occurrence of the PKS genes in all species has been documented in previous studies [67,68]. Phylogenetic and comparative analyses of PKS homologs using different species of fungi can help find new SMs and understand SM biosynthesis pathways [69]. The discontinuous distribution of SM biosynthetic genes in fungi has been attributed to multiple genetic processes such as gene gain or loss [70] and horizontal gene transfer (HGT) [71] affecting single genes or gene clusters, providing explanations for the diversity and taxonomic distribution of SM gene clusters across fungal species [72].
4.2. Clusters Common among All Examined Verticillium Genomes
The VNR-PKS-I-1 sequences were found in all Verticillium genomes and grouped with PKSs from different Colletotrichum species that have been associated with DHN melanin biosynthesis [73]. This conserved gene cluster was found in all Verticillium genomes examined. Melanin biosynthesis requires SCD1 and THR1 reductase. In addition, a 1,3,8-trihydroxynaphthalene reductase and a cmr1 transcription factor are required for DHN fungal melanin biosynthesis [73] and are contained within the Verticillium VNR-PKS-I-1 cluster. Shi-Kunne et al. [21] identified 11 potential PKS genes/clusters in the V. dahliae genome, including loci that can be implicated in DHN-melanin and fujikurin production. It has been proven that the DHN melanin biosynthesis pathway in other fungi also requires scytalone dehydratase SCD1 and THR1 reductase coding genes in order to function [73]. In V. dahlia, mutation of either the VNR-PKS-I-1 or transcription factor VdCmr1, resulted in the production of transparent transformants that also reduced the responses of V. dahliae to stress-related factors [11]. In another study, Li et al. [74] screened the effect of VdPKS9 (VR-PKS-I-17 in our study) deletions in the virulent strain of V. dahliae and found that VdPKS9 negatively controls the VdPKS1 (VNR-PKS-I-1)-associated melanin pathway.
The analysis has putatively linked type III PKSs found in all Verticillium genomes with a similar PKS-III gene in other ascomycetes. This cluster was conserved in all genomes examined in this study and showed high similarity in gene order and sequence with several Colletotrichum strains. Type III PKSs produce SMs with various biological activities, including antimicrobial activity. Despite extensive research in plants and bacteria, only a few type III PKSs from fungi have been identified [75]. Type III PKSs, also known as the chalcone synthase-like PKSs, were initially found in plants and then in bacteria. Later, with progress in genomics analysis, four type III PKS genes (CsyA, CsyB, CsyC, and CsyD) were identified in Aspergillus oryzae and type III PKS genes in other fungi [76]. Phylogenetic analyses of type III PKSs from plants, bacteria and fungi showed a unique origin for the fungal clade [77,78]. Sayari et al. [79] examined 20 genomes of Ceratocystidaceae and reported that the PKS III-containing gene cluster is highly conserved and present in all 20 of the genomes examined. PKS III genes encode polyketide pyrones and resorcinols [80].
4.3. Species-Specific Clusters
VRPKS-I-3 is clustered with the clade that contains ffsA, which plays a role in cytochalasan production. Cytochalasins or cytochalasans are a large family of structurally diverse macromolecular fungal polyketide synthase non-ribosomal peptide synthetase (PKS-NRPS) hybrid secondary metabolites [81]. Cytochalasins produced by Aspergillus flavipes and the pyrichalasin H synthase coding gene are important toxins produced by Pyricularia oryzae that show substantial cytotoxicity [82]. Cytochalasins, such as cytochalasin E produced by Aspergillus clavatus, are polyketide-amino acid hybrid molecules that belong to the cytochalasan family of fungal secondary metabolites [83]. Generally, to form a cytochalasin biosynthesis cluster, four genes are essential, including PKS-NRPS, enoyl reductase, putative Diels−Alderase (pDA), and α/β hydrolase (HYD) genes [84]. All four of these genes were present in V. albo-atrum VRPKS-I-3 cluster.
In the phylogenetic analysis, VNR-PKS-I-4, which is found only in the genome of V. nubilum, clustered with the azaphilone polyketide synthesis coding gene lijiquinone and Muyocopron and chaetoviridin (caz) biosynthesis-related PKS from Chaetomium globosum [85,86]. Based on the cluster comparison analyses, it can be anticipated that the final product of the VNR-PKS-I-4 gene cluster, which was found only in V. nubilum, is an antifungal compound in the azaphilone group. This is because the comparison analysis revealed that the genes flank to the VNR-PKS-I-4 gene showed high protein sequence similarity to the previously known azaphilone polyketide lijiquinone cluster identified in Chinese yew fungal endophyte M. atromaculans [85].
The VRPKS-I-7 gene cluster, found only in the genome of V. zaregamsianum, was putatively linked with the fusarielin biosynthesis cluster of Fusarium pseudograminearuum. Fusarielins are decalin core polyketides produced by several species of Fusarium and Aspergillus. Sieber et al. [87] showed that the genes of the metabolites aurofusarin and fusarielin are conserved in the closely related F. pseudograminearum but were absent in other Fusarium species. It has also been proven that the fusarielin biosynthesis cluster of F. pseudograminearuum was obtained through HGT. Likewise, in the current analysis, the genes for this cluster were only found in V. zaregamsianum and were not present in other genomes examined. Droce et al. [88] have proposed that the fusarielin biosynthesis gene cluster of F. pseudograminearumas is composed of seven genes. The comparison analysis showed that six out of seven genes needed for the biosynthesis of fusarielin are also present in the VRPKS-I-7 cluster. The VRPKS-I-7 core gene also matched with A. avenaceus (58.44%), Aspergillus wentii (58.29%), Sarocladium implicatum (78.19%), Whalleya microplaca (57.63%), Penicillium nordicum (56.06%), and Penicillium camemberti (56.15%). Therefore, the VRPKS-I-7 cluster might be gained through HGT from distantly related species, although more investigation is required to prove it.
4.4. Clusters Missing in a Few Genomes but Present in Others
The VPKS-8 gene cluster was putatively linked with the fujikurin biosynthesis cluster identified in Fusarium fujikuroi. This cluster is found only in highly pathogenic Verticillium species, including V. dahliae and V. longisporum. Fujikurins are natural products isolated from the fungus F. fujikuroi. It has been reported that the PKS-19 gene controls fusarielin toxin biosynthesis in F. pseudograminearum and the fujikurin synthase coding gene in F. fujikuroi [88,89]. Fujikurins and fujikurin-like compounds can play important roles in host–pathogen interactions in several pathogens, including some economically significant plant pathogenic fungi [90]. The core gene of the VPKS-8 gene cluster showed a high similarity to the PKS-19 gene from F. fujikuroi. Moreover, the fujikurin biosynthesis cluster contains five genes [89]. The homologs of all these five genes were also found in V. dahliae and V. longisporum. Since this cluster has been found only in highly pathogenic species of Verticillium, we hypothesize that the putative fujikurin-like compound gene cluster plays a significant role in pathogen infection and lifecycle, as in Fusarium.
4.5. Clusters Found in Distantly Related Fungi
Some of the Verticillium R-PKS-I clusters found in this study were also found in distantly related fungi. For example, from the phylogenetic analysis results, the VRPKS-I-1 was grouped with oxaleimides biosynthesis-related genes, such as poxE from P. oxalicum [91,92]. In another example, VRPKS-I-20 was found only in V. isaacii, V. klebahnii, and V. tricorpus and showed high protein sequence similarity to an unknown PKS identified in P. arizonense (XP_022486633) [93]. As mentioned earlier, the VRPKS-I-6 is also grouped in a clade that contains burnettramic acids biosynthesis cluster protein-coding A, an exceptional PKS-NRPS-derived bolaamphiphilic pyrrolizidinedione with antifungal activity from A. burnettii and A. alliaceus [94]. VRPKS-I-21 also clustered in a clade containing alt5, a PKS gene involved in the production of decaketide compound, alternapyrone, from Alternaria solani [95].
Large-scale genomic and functional comparisons elucidated the various proposed mechanisms that might explain the evolution of host adaptation in Verticillium species, such as enrichment for transposable elements, extensive chromosomal rearrangements, additional dispensable chromosomes, a high proportion of positively selected genes, horizontal gene transfer, and gene gain or loss [96]. Phylogenetically distant or unrelated species can exchange genetic material via horizontal gene transfer (HGT) which could explain why some of the above-mentioned clusters are present in Verticillium genomes. The horizontally transferred genes can benefit the recipient organism by boosting its ability to withstand environmental stress, colonize hosts, access nutrients, or gain an evolutionary advantage. However, more research is needed to validate this hypothesis.
4.6. Comparison of the Number of PKS Genes in Pathogenic and Non-Pathogenic Verticillium Strains
In this study, we found a greater number of PKS clusters in non-pathogenic/weakly aggressive species of Verticillium compared to the pathogenic ones. Pathogens and plants are evolving in a perpetual arms race, resulting in the relatively fast evolution of effectors and resistance (R)- and susceptibility (S)-related proteins. In general, effectors are called virulence factors if they promote virulence. On the contrary, effectors that are recognized by R proteins become avirulence (Avr) factors (Avr proteins). These avirulence and virulence factors, determining resistance and susceptibility of plants, respectively, are identified in many plants’ pathogenic fungi [97,98]. Plants have transferred Ave1 to Verticillium through HGT. Ave1 is a secreted protein recognized by Ve1 in tomatoes, and Ave1 expression is induced during host colonization [99]. The findings show that pathogenic Verticillium species such as V. dahliae, V. longisporum, V. albo-atrum, V. alfalfae, and V. nonalfalfae have fewer polyketide clusters than non-pathogenic and weekly aggressive species such as V. tricorpus, V. zaregamsianum, V. nubilum, V. isaacii, and V. klebahnii. Although it has been reported that the polyketide synthase VdPKS9 plays an important role in melanin biosynthesis and hyphal growth to promote V. dahlia’s virulence, it has also been reported that mutation of either the transcription factor VdCmr1 or the VdPKS1 did not reduce virulence and both genes were not required for pathogenesis on tobacco and lettuce [11]. These results concur with the idea that to maintain their virulence, pathogenic fungi tend to lose genes that might be targeted or recognized by plants.
4.7. Missing PKS Gene Clusters in Closely Related Species
When it comes to genetic variation, fungi have long been thought to be limited because they reproduce asexually. Notably, adaptive evolution occurs regardless of the absence of meiotic recombination in asexual organisms. The different mechanisms of adaptive evolution can lead to genetic divergence over long periods, which may eventually lead to the emergence of new species [100]. Multiple evolutionary processes, including horizontal transfer and loss/deletion of the genes, have been proposed to contribute to such discontinuity in Verticillium genomes. In the current study, such distribution patterns were observed in seven PKS or PKS-NRPS genes. For instance, VRPKS-I-2 was found in V. albo-atrum, V. isaacii, V. tricorpus, and V. klebahnii, but not in V. zaregamsianum. On the other hand, V. zaregamsianum obtained a VRPKS-I-2 gene cluster which has also been found in distantly related species to V. zaregamsianum, including V. dahliae, V. longisporum, V. alfalfa, and V. nonalfalfae. The presence of the VRPKS-I-8 gene (likely involved in fujikurine biosynthesis) in V. dahliae and V. longisporum and its absence in closely related species (V. alfalfae and V. nonalfalfae) is also in congruence with gene gain/loss events. Genome sequencing projects have revealed large and frequent changes between species in the size of gene families. Numerous morphological, physiological, behavioral, and genetic differences between species can be traced back to these changes. They are also thought to be responsible for much of the natural genetic and genomic diversity that exists. Gene duplications and losses have also been found to contribute to long-term differences in the size of gene families between species [101]. Gene loss was frequently overlooked as an evolutionary driver in the past, mainly because it was associated with the elimination of redundant gene duplicates that had no obvious functional consequences [102]. Recent genomic data, on the other hand, suggest that gene loss may be a major cause of phenotypic diversity and fungal habitat change [103]. For example, gains, losses, or changes in the function of TRI genes have contributed to the structural diversity of trichothecenes, a family of toxic secondary metabolites produced by various Aspergillus, Fusarium, Trichoderma, and Trichothecium fungal species [104]. Comparative genomic analysis of V. dahliae strains revealed extensive large-scale genomic rearrangements, likely facilitated by incorrect double-stranded break repair, which led to lineage-specific genomic regions that are enriched for planta-expressed effector genes encoding secreted proteins that aid in host colonization [105,106].
4.8. PKS-NRPS Cluster Comparison among Different Verticillium Strains
PKS-NRPSs are formed because of the fusion of PKS and NRPS domains and are considered among the most important groups of clusters due to their structural complexity [107]. In this study, a total of nine PKS-NRPS genes were found. The VRPKS-I-1 cluster was found in V. dahliae, V. alfalfae, V. longisporum, and V. zaregamsianum. The analyses could not link this cluster to any previously known cluster. However, it has been proven that the nag 1 gene identified in this cluster is involved in the pathogenicity of V. dahliae [108]. Functional analysis of nag 1 by dsRNA-mediated gene silencing showed that the lack of nag 1 exponentially reduced conidial production and fungal growth. In addition, the Nag1-silenced mutants displayed noticeable deficiencies in fungal virulence and microsclerotia formation. In alignment with a decrease in microsclerotia formation, melanin biosynthesis and transcripts of genes involved in melanin formation were significantly decreased in Nag1-silenced mutants. Hence, although more functional analysis is needed, it seems that the VRPKS-I-1 cluster plays the same roles in V. alfalfae, V. longisporum, and V. zaregamsianum. Interestingly, when we searched for the nag 1 gene in these clusters, we could find it in the VRPKS-I-1 cluster of V. dahliae, V. alfalfae, and V. longisporum. However, in V. zaregamsianum, this gene was found in a different contig, and it seems that it is not part of the same cluster.
5. Conclusions
The genome comparison approach has provided a significant understanding of fungal pathogens’ evolution. In this study, we used a combination of phylogenetic and genomic approaches to identify 32 putative PKS genes and possible clusters in the genomes of Verticillium members. All the identified clusters seem to be complete and hence, possibly functional. We anticipate that at least five clusters, including those for putative DHN-melanin-, cytochalasin-, fusarielien-, fujikurin-, and lijiquinone-like compounds, may belong to the active PKS repertoire of Verticillium. Further functional analyses will shed light on the role of the identified clusters. Our findings provide a solid framework for further research into the functionality of polyketide biosynthesis gene clusters in this commercially significant fungal genus.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology11091252/s1, Figure S1: Neighbor-joining tree inferred from nucleotide sequences of polyketide synthase III coding regions of Verticillium species as well as PKS III sequences obtained from other Ascomycetes. GenBank accession numbers for each sequence are provided. Similar groups were obtained with the maximum likelihood analysis. In this analysis, we used 1000 bootstrap repeats as indicated at the internodes. Figure S2: Comparison of putative cluster genes for Verticillium VRPKS-I-21 found in V. zaregamsianum, V. klebahnii, and V. nubilum and the alternapyrone biosynthesis gene cluster of Alternaria alternata. The arrowheads indicate direction of transcription and types of genes common to the two clusters being compared are displayed in the same color. Shaded lines represent similarities between nucleotide sequences. Figure S3: Comparison of putative cluster genes for VRPKS-I-22 locus found in the genomes of V. dahliae, V. longisporum, V. alfalfa, and V. nonalfalfae and the fusaric acid gene cluster. The arrowheads indicate direction of transcription and types of genes common to the two clusters being compared are displayed in the same color. Shaded lines represent similarities between nucleotide sequences. Table S1: The tables below show the predicted polyketide synthase (PKS) gene clusters in all examined Verticillium genomes. Table S2: The tables below show the top 10 tBLASTn hits NCBI’s non-redundant nucleotide database for each of the Verticillium PKS genes examined in this study. The BLAST scores (E-value, percent sequence identity, and coverage) and the accession number of the top hits are indicated.
Author Contributions
Conceptualization, M.S. and F.D.; methodology, M.S. and P.K.R.; validation, M.S. and F.D.; formal analysis, M.S.; investigation, M.S.; resources, F.D.; data curation, M.S., A.D., P.K.R.; writing—original draft preparation, M.S., M.E.-S. and A.D.; writing—review and editing, M.S, M.E.-S., A.D. and F.D.; supervision, F.D.; project administration, F.D; funding acquisition, F.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a Discovery grant to F. Daayf from the Natural Sciences and Engineering Research Council of Canada.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The study did not report any data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pegg, G.F.; Brady, B.L. Verticillium Wilts; CABI: Wallingford, UK, 2002. [Google Scholar]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Barbara, D.J.; Clewes, E. Plant pathogenic Verticillium species: How many of them are there? Mol. Plant Pathol. 2003, 4, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Rowe, R.C.; Powelson, M.L. Potato early dying: Management challenges in a changing production environment. Plant Dis. 2002, 86, 1184–1193. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, S. Longevity of the Verticillium wilt fungus in the laboratory and field. Phytopathology 1955, 45, 180–181. [Google Scholar]
- Song, R.; Li, J.; Xie, C.; Jian, W.; Yang, X. An overview of the molecular genetics of plant resistance to the Verticillium wilt pathogen Verticillium dahliae. Int. J. Mol. Sci. 2020, 21, 1120. [Google Scholar] [CrossRef]
- Wu, L.; Du, G.; Bao, R.; Li, Z.; Gong, Y.; Liu, F. De novo assembly and discovery of genes involved in the response of Solanum sisymbriifolium to Verticillium dahlia. Physiol. Mol. Biol. Plants 2019, 25, 1009–1027. [Google Scholar] [CrossRef]
- Luo, X.; Xie, C.; Dong, J.; Yang, X.; Sui, A. Interactions between Verticillium dahliae and its host: Vegetative growth, pathogenicity, plant immunity. Appl. Microbiol. Biotechnol. 2014, 98, 6921–6932. [Google Scholar] [CrossRef]
- Depotter, J.R.; Shi-Kunne, X.; Missonnier, H.; Liu, T.; Faino, L.; van den Berg, G.C.; Wood, T.A.; Zhang, B.; Jacques, A.; Seidl, M.F. Dynamic virulence-related regions of the plant pathogenic fungus Verticillium dahliae display enhanced sequence conservation. Mol. Ecol. 2019, 28, 3482–3495. [Google Scholar] [CrossRef]
- Zhang, D.-D.; Wang, X.-Y.; Chen, J.-Y.; Kong, Z.-Q.; Gui, Y.-J.; Li, N.-Y.; Bao, Y.-M.; Dai, X.-F. Identification and characterization of a pathogenicity-related gene VdCYP1 from Verticillium dahliae. Sci. Rep. 2016, 6, 27979. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, X.; Fang, Y.; Anchieta, A.; Goldman, P.H.; Hernandez, G.; Klosterman, S.J. Transcription factor VdCmr1 is required for pigment production, protection from UV irradiation, and regulates expression of melanin biosynthetic genes in Verticillium dahliae. Microbiology 2018, 164, 685. [Google Scholar] [CrossRef]
- Zhu, X.; Sayari, M.; Daayf, F. Role of Exopolygalacturonase-Related Genes in Potato-Verticillium dahliae Interaction. Pathogens 2021, 10, 642. [Google Scholar] [CrossRef]
- Zhu, X.; Soliman, A.; Islam, M.R.; Adam, L.R.; Daayf, F. Verticillium dahliae’s isochorismatase hydrolase is a virulence factor that contributes to interference with potato’s salicylate and jasmonate defense signaling. Front. Plant Sci. 2017, 8, 399. [Google Scholar] [CrossRef]
- Zhu, X.; Sayari, M.; Islam, M.R.; Daayf, F. NOXA Is Important for Verticillium dahliae’s Penetration Ability and Virulence. J. Fungi 2021, 7, 814. [Google Scholar] [CrossRef]
- Vining, L.C. Functions of secondary metabolites. Annu. Rev. Microbiol. 1990, 44, 395–427. [Google Scholar] [CrossRef]
- Mosunova, O.; Navarro-Muñoz, J.C.; Collemare, J. The biosynthesis of fungal secondary metabolites: From fundamentals to biotechnological applications. Ref. Modul. Life Sci. 2021, 2, 458–476. [Google Scholar]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism—From biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef]
- Awad, N.E.; Kassem, H.A.; Hamed, M.A.; El-Feky, A.M.; Elnaggar, M.A.; Mahmoud, K.; Ali, M.A. Isolation and characterization of the bioactive metabolites from the soil derived fungus Trichoderma viride. Mycology 2018, 9, 70–80. [Google Scholar] [CrossRef]
- Cole, R.J.; Schweikert, M.A.; Jarvis, B.B. Handbook of Secondary Fungal Metabolites; Gulf Professional Publishing: Houston, TX, USA, 2003; Volume 3. [Google Scholar]
- Scharf, D.H.; Heinekamp, T.; Brakhage, A.A. Human and plant fungal pathogens: The role of secondary metabolites. PLoS Pathog. 2014, 10, e1003859. [Google Scholar] [CrossRef]
- Shi-Kunne, X.; Jové, R.d.P.; Depotter, J.R.; Ebert, M.K.; Seidl, M.F.; Thomma, B.P. In silico prediction and characterisation of secondary metabolite clusters in the plant pathogenic fungus Verticillium dahliae. FEMS Microbiol. Lett. 2019, 366, fnz081. [Google Scholar] [CrossRef]
- Brown, D.W.; Proctor, R.H. Insights into natural products biosynthesis from analysis of 490 polyketide synthases from Fusarium. Fungal Genet. Biol. 2016, 89, 37–51. [Google Scholar] [CrossRef]
- Collemare, J.; Billard, A.; Böhnert, H.U.; Lebrun, M.-H. Biosynthesis of secondary metabolites in the rice blast fungus Magnaporthe grisea: The role of hybrid PKS-NRPS in pathogenicity. Mycol. Res. 2008, 112, 207–215. [Google Scholar] [CrossRef]
- Keller, N.P. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol. 2015, 11, 671–677. [Google Scholar] [CrossRef]
- Dias, D.A.; Urban, S.; Roessner, U. A historical overview of natural products in drug discovery. Metabolites 2012, 2, 303–336. [Google Scholar] [CrossRef]
- Boruta, T. Uncovering the repertoire of fungal secondary metabolites: From Fleming’s laboratory to the International Space Station. Bioengineered 2018, 9, 12–16. [Google Scholar] [CrossRef]
- Lobanovska, M.; Pilla, G. Focus: Drug development: Penicillin’s discovery and antibiotic resistance: Lessons for the future? Yale J. Biol. Med. 2017, 90, 135. [Google Scholar]
- Medema, M.H.; Fischbach, M.A. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015, 11, 639–648. [Google Scholar] [CrossRef]
- Wiemann, P.; Keller, N.P. Strategies for mining fungal natural products. J. Ind. Microbiol. Biotechnol. 2014, 41, 301–313. [Google Scholar] [CrossRef]
- Ballester, A.-R.; Marcet-Houben, M.; Levin, E.; Sela, N.; Selma-Lázaro, C.; Carmona, L.; Wisniewski, M.; Droby, S.; González-Candelas, L.; Gabaldón, T. Genome, transcriptome, and functional analyses of Penicillium expansum provide new insights into secondary metabolism and pathogenicity. Mol. Plant-Microbe Interact. 2015, 28, 232–248. [Google Scholar] [CrossRef]
- Hoffmeister, D.; Keller, N.P. Natural products of filamentous fungi: Enzymes, genes, and their regulation. Nat. Prod. Rep. 2007, 24, 393–416. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Yan, J.; Guo, M.; Xu, L.; Hou, L.; Zou, Q. Comparative genome analysis of plant ascomycete fungal pathogens with different lifestyles reveals distinctive virulence strategies. BMC Genom. 2022, 23, 34. [Google Scholar] [CrossRef]
- Graham-Taylor, C.; Kamphuis, L.G.; Derbyshire, M.C. A detailed in silico analysis of secondary metabolite biosynthesis clusters in the genome of the broad host range plant pathogenic fungus Sclerotinia sclerotiorum. BMC Genom. 2020, 21, 7. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Noar, R.D.; Daub, M.E. Bioinformatics prediction of polyketide synthase gene clusters from Mycosphaerella fijiensis. PLoS ONE 2016, 11, e0158471. [Google Scholar] [CrossRef] [PubMed]
- Skellam, E. Biosynthesis of fungal polyketides by collaborating and trans-acting enzymes. Nat. Prod. Rep. 2022, 39, 754–783. [Google Scholar] [CrossRef]
- Cox, R.J.; Simpson, T.J. Fungal type I polyketide synthases. Methods Enzymol. 2009, 459, 49–78. [Google Scholar]
- Khaldi, N.; Seifuddin, F.T.; Turner, G.; Haft, D.; Nierman, W.C.; Wolfe, K.H.; Fedorova, N.D. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 2010, 47, 736–741. [Google Scholar] [CrossRef]
- Wiemann, P.; Willmann, A.; Straeten, M.; Kleigrewe, K.; Beyer, M.; Humpf, H.U.; Tudzynski, B. Biosynthesis of the red pigment bikaverin in Fusarium fujikuroi: Genes, their function and regulation. Mol. Microbiol. 2009, 72, 931–946. [Google Scholar] [CrossRef]
- Yu, J.; Bhatnagar, D.; Cleveland, T.E. Completed sequence of aflatoxin pathway gene cluster in Aspergillus parasiticus. FEBS Lett. 2004, 564, 126–130. [Google Scholar] [CrossRef]
- Cairns, T.; Meyer, V. In silico prediction and characterization of secondary metabolite biosynthetic gene clusters in the wheat pathogen Zymoseptoria tritici. BMC Genom. 2017, 18, 631. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Subbarao, K.V.; Kang, S.; Veronese, P.; Gold, S.E.; Thomma, B.P.; Chen, Z.; Henrissat, B.; Lee, Y.H.; Park, J.; et al. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 2011, 7, e1002137. [Google Scholar] [CrossRef]
- Shi-Kunne, X.; Faino, L.; van den Berg, G.C.M.; Thomma, B.; Seidl, M.F. Evolution within the fungal genus Verticillium is characterized by chromosomal rearrangement and gene loss. Environ. Microbiol. 2018, 20, 1362–1373. [Google Scholar] [CrossRef]
- Kasson, M.T.; Kasson, L.R.; Wickert, K.L.; Davis, D.D.; Stajich, J.E. Genome sequence of a lethal vascular wilt fungus, Verticillium nonalfalfae, a biological control used against the invasive Ailanthus altissima. Microbiol. Resour. Announc. 2019, 8, e01619-18. [Google Scholar] [CrossRef]
- Harting, R.; Starke, J.; Kusch, H.; Pöggeler, S.; Maurus, I.; Schlüter, R.; Landesfeind, M.; Bulla, I.; Nowrousian, M.; de Jonge, R. A 20-kb lineage-specific genomic region tames virulence in pathogenic amphidiploid Verticillium longisporum. Mol. Plant Pathol. 2021, 22, 939–953. [Google Scholar] [CrossRef]
- Seidl, M.F.; Faino, L.; Shi-Kunne, X.; van den Berg, G.C.; Bolton, M.D.; Thomma, B.P. The genome of the saprophytic fungus Verticillium tricorpus reveals a complex effector repertoire resembling that of its pathogenic relatives. Mol. Plant-Microbe Interact. 2015, 28, 362–373. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, W465–W467. [Google Scholar] [CrossRef]
- Zdobnov, E.M.; Apweiler, R. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010, 39, D225–D229. [Google Scholar] [CrossRef]
- Bachmann, B.O.; Ravel, J. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009, 458, 181–217. [Google Scholar]
- Cox, R.J. Polyketides, proteins and genes in fungi: Programmed nano-machines begin to reveal their secrets. Org. Biomol. Chem. 2007, 5, 2010–2026. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.i.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef]
- Armaleo, D.; Sun, X.; Culberson, C. Insights from the first putative biosynthetic gene cluster for a lichen depside and depsidone. Mycologia 2011, 103, 741–754. [Google Scholar] [CrossRef]
- Feng, P.; Shang, Y.; Cen, K.; Wang, C. Fungal biosynthesis of the bibenzoquinone oosporein to evade insect immunity. Proc. Natl. Acad. Sci. USA 2015, 112, 11365–11370. [Google Scholar] [CrossRef]
- Kudo, F.; Matsuura, Y.; Hayashi, T.; Fukushima, M.; Eguchi, T. Genome mining of the sordarin biosynthetic gene cluster from Sordaria araneosa Cain ATCC 36386: Characterization of cycloaraneosene synthase and GDP-6-deoxyaltrose transferase. J. Antibiot. 2016, 69, 541–548. [Google Scholar] [CrossRef]
- Inderbitzin, P.; Asvarak, T.; Turgeon, B.G. Six new genes required for production of T-toxin, a polyketide determinant of high virulence of Cochliobolus heterostrophus to maize. Mol. Plant-Microbe Interact. 2010, 23, 458–472. [Google Scholar] [CrossRef]
- Brown, D.W.; Lee, S.-H.; Kim, L.-H.; Ryu, J.-G.; Lee, S.; Seo, Y.; Kim, Y.H.; Busman, M.; Yun, S.-H.; Proctor, R.H. Identification of a 12-gene fusaric acid biosynthetic gene cluster in Fusarium species through comparative and functional genomics. Mol. Plant-Microbe Interact. 2015, 28, 319–332. [Google Scholar] [CrossRef]
- Grum-Grzhimaylo, A.A.; Falkoski, D.L.; van den Heuvel, J.; Valero-Jiménez, C.A.; Min, B.; Choi, I.G.; Lipzen, A.; Daum, C.G.; Aanen, D.K.; Tsang, A. The obligate alkalophilic soda-lake fungus Sodiomyces alkalinus has shifted to a protein diet. Mol. Ecol. 2018, 27, 4808–4819. [Google Scholar] [CrossRef]
- Lin, H.-C.; Chooi, Y.-H.; Dhingra, S.; Xu, W.; Calvo, A.M.; Tang, Y. The fumagillin biosynthetic gene cluster in Aspergillus fumigatus encodes a cryptic terpene cyclase involved in the formation of β-trans-bergamotene. J. Am. Chem. Soc. 2013, 135, 4616–4619. [Google Scholar] [CrossRef] [PubMed]
- Morishita, Y.; Zhang, H.; Taniguchi, T.; Mori, K.; Asai, T. The discovery of fungal polyene macrolides via a postgenomic approach reveals a polyketide macrocyclization by trans-acting thioesterase in fungi. Org. Lett. 2019, 21, 4788–4792. [Google Scholar] [CrossRef] [PubMed]
- Hansen, F.T.; Gardiner, D.M.; Lysøe, E.; Fuertes, P.R.; Tudzynski, B.; Wiemann, P.; Sondergaard, T.E.; Giese, H.; Brodersen, D.E.; Sørensen, J.L. An update to polyketide synthase and non-ribosomal synthetase genes and nomenclature in Fusarium. Fungal Genet. Biol. 2015, 75, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Fatema, U.; Broberg, A.; Jensen, D.F.; Karlsson, M.; Dubey, M. Functional analysis of polyketide synthase genes in the biocontrol fungus Clonostachys rosea. Sci. Rep. 2018, 8, 15009. [Google Scholar] [CrossRef]
- Brown, D.W.; Butchko, R.A.; Baker, S.E.; Proctor, R.H. Phylogenomic and functional domain analysis of polyketide synthases in Fusarium. Fungal Biol. 2012, 116, 318–331. [Google Scholar] [CrossRef]
- Wiemann, P.; Sieber, C.M.; Von Bargen, K.W.; Studt, L.; Niehaus, E.-M.; Espino, J.J.; Huss, K.; Michielse, C.B.; Albermann, S.; Wagner, D. Deciphering the cryptic genome: Genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013, 9, e1003475. [Google Scholar] [CrossRef]
- Throckmorton, K.; Wiemann, P.; Keller, N.P. Evolution of chemical diversity in a group of non-reduced polyketide gene clusters: Using phylogenetics to inform the search for novel fungal natural products. Toxins 2015, 7, 3572–3607. [Google Scholar] [CrossRef]
- Campbell, M.A.; Rokas, A.; Slot, J.C. Horizontal transfer and death of a fungal secondary metabolic gene cluster. Genome Biol. Evol. 2012, 4, 289–293. [Google Scholar] [CrossRef]
- Reynolds, H.T.; Slot, J.C.; Divon, H.H.; Lysøe, E.; Proctor, R.H.; Brown, D.W. Differential retention of gene functions in a secondary metabolite cluster. Mol. Biol. Evol. 2017, 34, 2002–2015. [Google Scholar] [CrossRef]
- Lind, A.L.; Wisecaver, J.H.; Lameiras, C.; Wiemann, P.; Palmer, J.M.; Keller, N.P.; Rodrigues, F.; Goldman, G.H.; Rokas, A. Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLoS Biol. 2017, 15, e2003583. [Google Scholar] [CrossRef]
- Tsuji, G.; Kenmochi, Y.; Takano, Y.; Sweigard, J.; Farrall, L.; Furusawa, I.; Horino, O.; Kubo, Y. Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and Pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn (II) 2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner. Mol. Microbiol. 2000, 38, 940–954. [Google Scholar]
- Li, H.; Wang, D.; Zhang, D.-D.; Geng, Q.; Li, J.-J.; Sheng, R.-C.; Xue, H.-S.; Zhu, H.; Kong, Z.-Q.; Dai, X.-F. A polyketide synthase from Verticillium dahliae modulates melanin biosynthesis and hyphal growth to promote virulence. BMC Biol. 2022, 20, 125. [Google Scholar] [CrossRef]
- Navarro-Muñoz, J.C.; Collemare, J. Evolutionary histories of type III polyketide synthases in fungi. Front. Microbiol. 2020, 3018. [Google Scholar] [CrossRef]
- Seshime, Y.; Juvvadi, P.R.; Fujii, I.; Kitamoto, K. Discovery of a novel superfamily of type III polyketide synthases in Aspergillus oryzae. Biochem. Biophys. Res. Commun. 2005, 331, 253–260. [Google Scholar] [CrossRef]
- Shimizu, Y.; Ogata, H.; Goto, S. Type III polyketide synthases: Functional classification and phylogenomics. ChemBioChem 2017, 18, 50–65. [Google Scholar] [CrossRef]
- Yan, H.; Sun, L.; Huang, J.; Qiu, Y.; Xu, F.; Yan, R.; Zhu, D.; Wang, W.; Zhan, J. Identification and heterologous reconstitution of a 5-alk (en) ylresorcinol synthase from endophytic fungus Shiraia sp. Slf14. J. Microbiol. 2018, 56, 805–812. [Google Scholar] [CrossRef]
- Sayari, M.; Steenkamp, E.T.; van der Nest, M.A.; Wingfield, B.D. Diversity and evolution of polyketide biosynthesis gene clusters in the Ceratocystidaceae. Fungal Biol. 2018, 122, 856–866. [Google Scholar] [CrossRef]
- Hashimoto, M.; Nonaka, T.; Fujii, I. Fungal type III polyketide synthases. Nat. Prod. Rep. 2014, 31, 1306–1317. [Google Scholar] [CrossRef]
- Skellam, E. The biosynthesis of cytochalasans. Nat. Prod. Rep. 2017, 34, 1252–1263. [Google Scholar] [CrossRef]
- Heard, S.C.; Wu, G.; Winter, J.M. Discovery and characterization of a cytochalasan biosynthetic cluster from the marine-derived fungus Aspergillus flavipes CNL-338. J. Antibiot. 2020, 73, 803–807. [Google Scholar] [CrossRef]
- Qiao, K.; Chooi, Y.-H.; Tang, Y. Identification and engineering of the cytochalasin gene cluster from Aspergillus clavatus NRRL 1. Metab. Eng. 2011, 13, 723–732. [Google Scholar] [CrossRef]
- Wang, C.; Hantke, V.; Cox, R.J.; Skellam, E. Targeted gene inactivations expose silent cytochalasans in Magnaporthe grisea NI980. Org. Lett. 2019, 21, 4163–4167. [Google Scholar] [CrossRef]
- Cain, J.W.; Miller, K.I.; Kalaitzis, J.A.; Chau, R.; Neilan, B.A. Genome mining of a fungal endophyte of Taxus yunnanensis (Chinese yew) leads to the discovery of a novel azaphilone polyketide, lijiquinone. Microb. Biotechnol. 2020, 13, 1415–1427. [Google Scholar] [CrossRef]
- Sato, M.; Winter, J.M.; Kishimoto, S.; Noguchi, H.; Tang, Y.; Watanabe, K. Combinatorial generation of chemical diversity by redox enzymes in chaetoviridin biosynthesis. Org. Lett. 2016, 18, 1446–1449. [Google Scholar] [CrossRef]
- Sieber, C.M.; Lee, W.; Wong, P.; Münsterkötter, M.; Mewes, H.-W.; Schmeitzl, C.; Varga, E.; Berthiller, F.; Adam, G.; Güldener, U. The Fusarium graminearum genome reveals more secondary metabolite gene clusters and hints of horizontal gene transfer. PLoS ONE 2014, 9, e110311. [Google Scholar]
- Droce, A.; Saei, W.; Jørgensen, S.H.; Wimmer, R.; Giese, H.; Wollenberg, R.D.; Sondergaard, T.E.; Sørensen, J.L. Functional analysis of the fusarielin biosynthetic gene cluster. Molecules 2016, 21, 1710. [Google Scholar] [CrossRef]
- Von Bargen, K.W.; Niehaus, E.-M.; Krug, I.; Bergander, K.; Würthwein, E.-U.; Tudzynski, B.; Humpf, H.-U. Isolation and structure elucidation of fujikurins A–D: Products of the PKS19 gene cluster in Fusarium fujikuroi. J. Nat. Prod. 2015, 78, 1809–1815. [Google Scholar] [CrossRef]
- Sbaraini, N.; Andreis, F.C.; Thompson, C.E.; Guedes, R.L.; Junges, Â.; Campos, T.; Staats, C.C.; Vainstein, M.H.; Ribeiro de Vasconcelos, A.T.; Schrank, A. Genome-Wide Analysis of Secondary Metabolite Gene Clusters in O phiostoma ulmi and Ophiostoma novo-ulmi Reveals a Fujikurin-Like Gene Cluster with a Putative Role in Infection. Front. Microbiol. 2017, 8, 1063. [Google Scholar] [CrossRef] [PubMed]
- Kakule, T.B.; Sardar, D.; Lin, Z.; Schmidt, E.W. Two related pyrrolidinedione synthetase loci in Fusarium heterosporum ATCC 74349 produce divergent metabolites. ACS Chem. Biol. 2013, 8, 1549–1557. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Dander, J.E.; Sato, C.; Hung, Y.-S.; Gao, S.-S.; Tang, M.-C.; Hang, L.; Winter, J.M.; Garg, N.K.; Watanabe, K. Collaborative biosynthesis of maleimide-and succinimide-containing natural products by fungal polyketide megasynthases. J. Am. Chem. Soc. 2017, 139, 5317–5320. [Google Scholar] [CrossRef] [PubMed]
- Grijseels, S.; Nielsen, J.C.; Randelovic, M.; Nielsen, J.; Nielsen, K.F.; Workman, M.; Frisvad, J.C. Penicillium arizonense, a new, genome sequenced fungal species, reveals a high chemical diversity in secreted metabolites. Sci. Rep. 2016, 6, 35112. [Google Scholar] [CrossRef]
- Li, H.; Gilchrist, C.L.; Lacey, H.J.; Crombie, A.; Vuong, D.; Pitt, J.I.; Lacey, E.; Chooi, Y.-H.; Piggott, A.M. Discovery and heterologous biosynthesis of the burnettramic acids: Rare PKS-NRPS-derived bolaamphiphilic pyrrolizidinediones from an Australian fungus, Aspergillus burnettii. Org. Lett. 2019, 21, 1287–1291. [Google Scholar] [CrossRef]
- Fujii, I.; Yoshida, N.; Shimomaki, S.; Oikawa, H.; Ebizuka, Y. An iterative type I polyketide synthase PKSN catalyzes synthesis of the decaketide alternapyrone with regio-specific octa-methylation. Chem. Biol. 2005, 12, 1301–1309. [Google Scholar] [CrossRef]
- Chen, J.Y.; Liu, C.; Gui, Y.J.; Si, K.W.; Zhang, D.D.; Wang, J.; Short, D.P.; Huang, J.Q.; Li, N.Y.; Liang, Y. Comparative genomics reveals cotton-specific virulence factors in flexible genomic regions in Verticillium dahliae and evidence of horizontal gene transfer from Fusarium. New Phytol. 2018, 217, 756–770. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Li, J.; Cornelissen, B.; Rep, M. Host-specificity factors in plant pathogenic fungi. Fungal Genet. Biol. 2020, 144, 103447. [Google Scholar] [CrossRef]
- De Jonge, R.; Peter van Esse, H.; Maruthachalam, K.; Bolton, M.D.; Santhanam, P.; Saber, M.K.; Zhang, Z.; Usami, T.; Lievens, B.; Subbarao, K.V. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 5110–5115. [Google Scholar] [CrossRef]
- Flot, J.-F.; Hespeels, B.; Li, X.; Noel, B.; Arkhipova, I.; Danchin, E.G.; Hejnol, A.; Henrissat, B.; Koszul, R.; Aury, J.-M. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 2013, 500, 453–457. [Google Scholar] [CrossRef]
- Han, M.V.; Thomas, G.W.; Lugo-Martinez, J.; Hahn, M.W. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 2013, 30, 1987–1997. [Google Scholar] [CrossRef]
- Olson, M.V. When less is more: Gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 1999, 64, 18–23. [Google Scholar] [CrossRef]
- Albalat, R.; Cañestro, C. Evolution by gene loss. Nat. Rev. Genet. 2016, 17, 379–391. [Google Scholar] [CrossRef]
- Brown, D.W.; Villani, A.; Susca, A.; Moretti, A.; Hao, G.; Kim, H.-S.; Proctor, R.H.; McCormick, S.P. Gain and loss of a transcription factor that regulates late trichothecene biosynthetic pathway genes in Fusarium. Fungal Genet. Biol. 2020, 136, 103317. [Google Scholar] [CrossRef]
- De Jonge, R.; Bolton, M.D.; Kombrink, A.; van den Berg, G.C.; Yadeta, K.A.; Thomma, B.P. Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 2013, 23, 1271–1282. [Google Scholar] [CrossRef]
- Faino, L.; Seidl, M.F.; Shi-Kunne, X.; Pauper, M.; van den Berg, G.C.; Wittenberg, A.H.; Thomma, B.P. Transposons passively and actively contribute to evolution of the two-speed genome of a fungal pathogen. Genome Res. 2016, 26, 1091–1100. [Google Scholar] [CrossRef]
- Boettger, D.; Hertweck, C. Molecular diversity sculpted by fungal PKS–NRPS hybrids. ChemBioChem 2013, 14, 28–42. [Google Scholar] [CrossRef]
- Xiong, D.; Wang, Y.; Tian, C. A novel gene from a secondary metabolism gene cluster is required for microsclerotia formation and virulence in Verticillium dahliae. Phytopathol. Res. 2019, 1, 31. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).