Thermostable and O2-Insensitive Pyruvate Decarboxylases from Thermoacidophilic Archaea Catalyzing the Production of Acetaldehyde
Simple Summary
Abstract
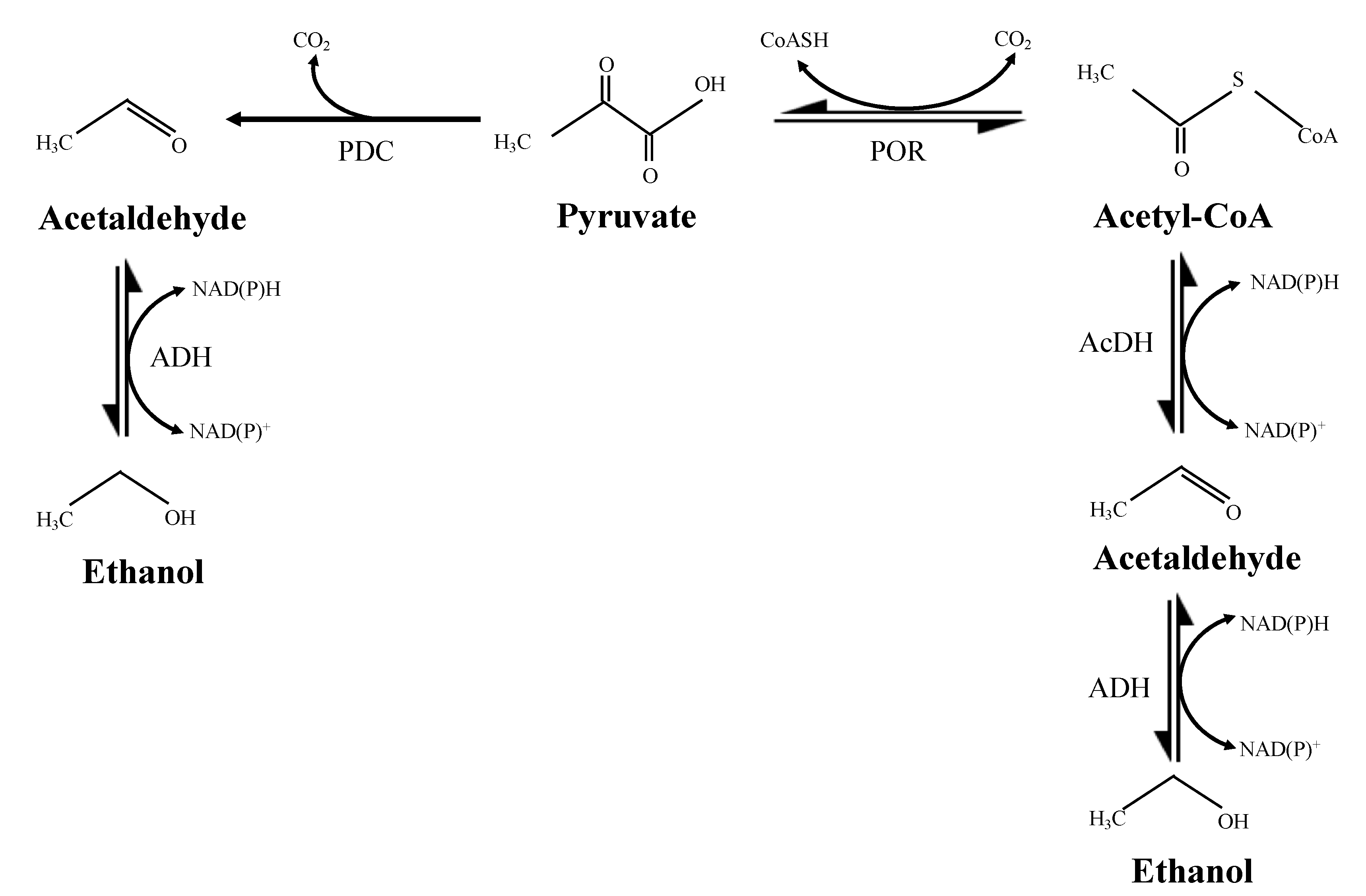
1. Introduction
2. Material and Methods
2.1. Microorganisms and Chemicals
2.2. Buffer Preparation
2.3. Enzyme Assay
2.4. Preparation of Cell-Free Extract
2.5. Purification of Enzymes
3. Results
3.1. Enzyme Purification
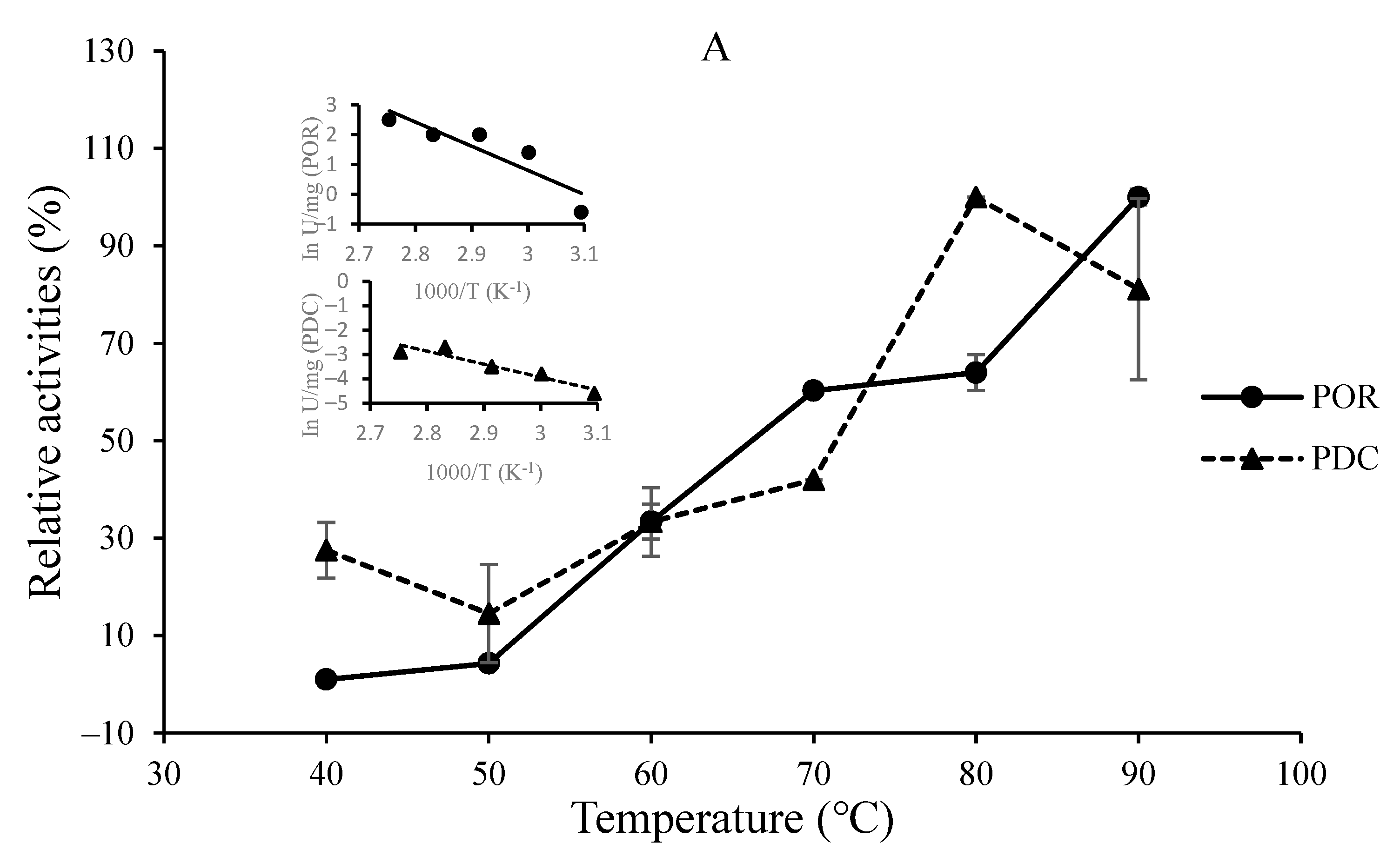
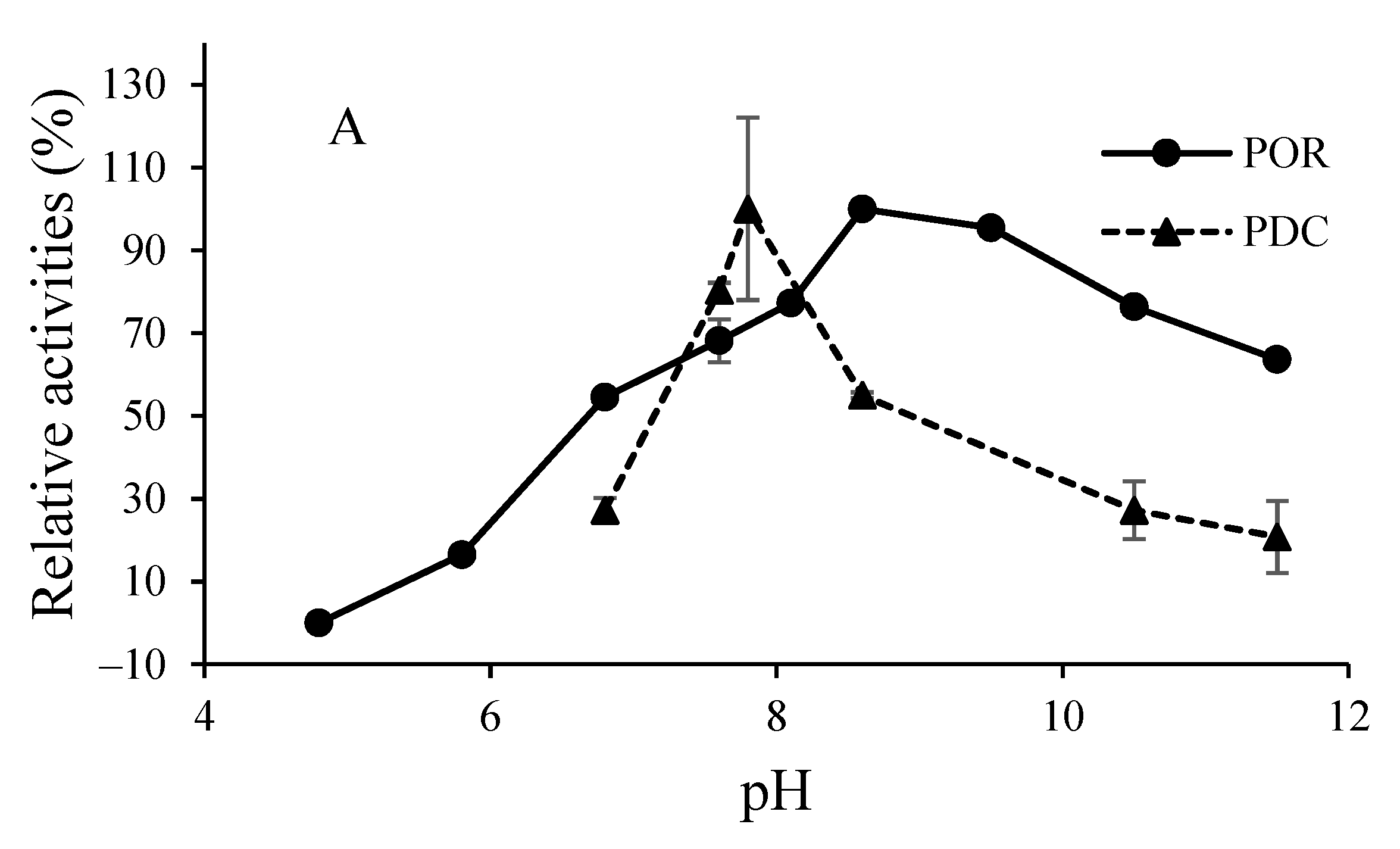
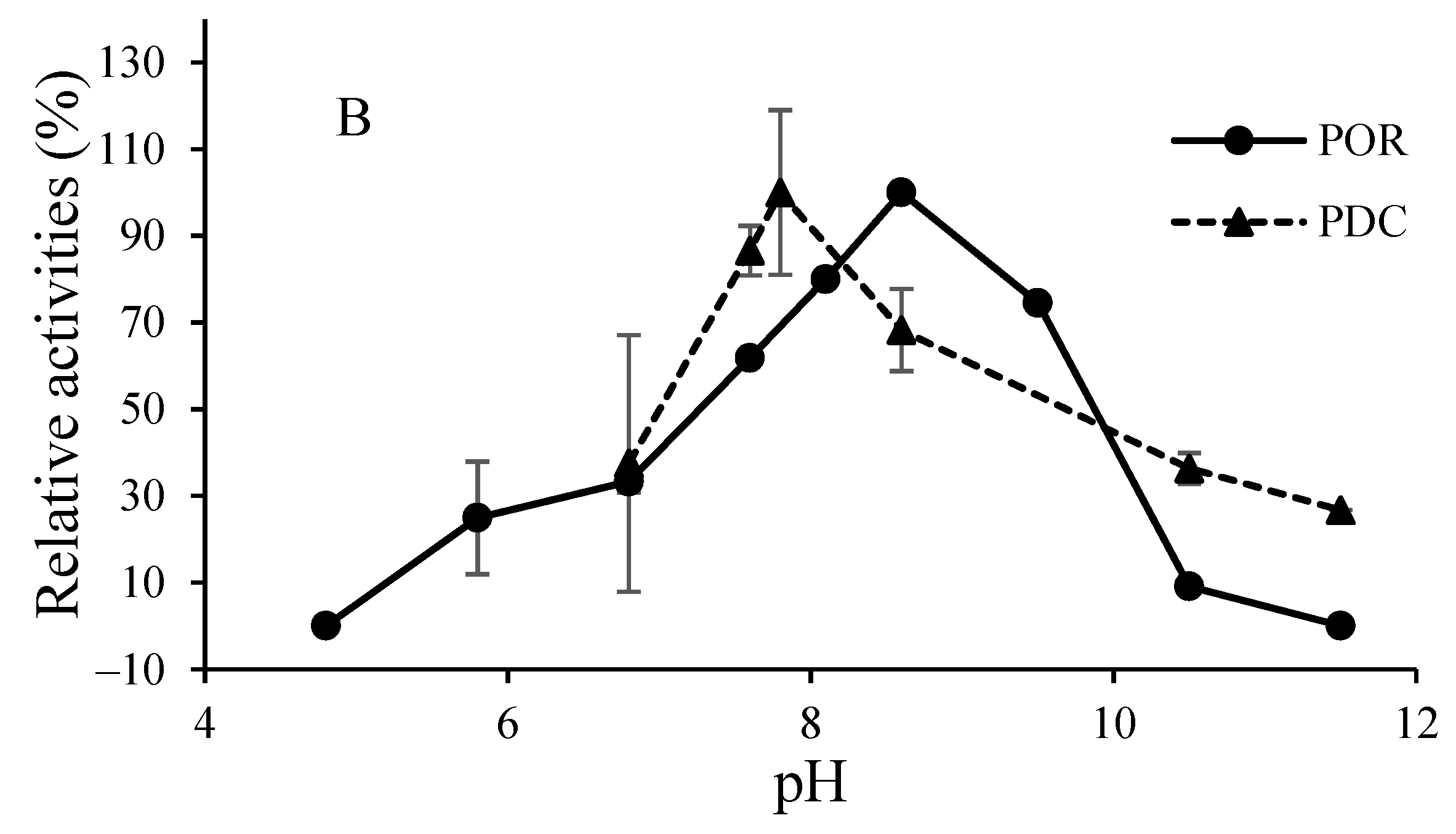
3.2. O2-Sensitivity and Thermostability of the Purified Enzymes
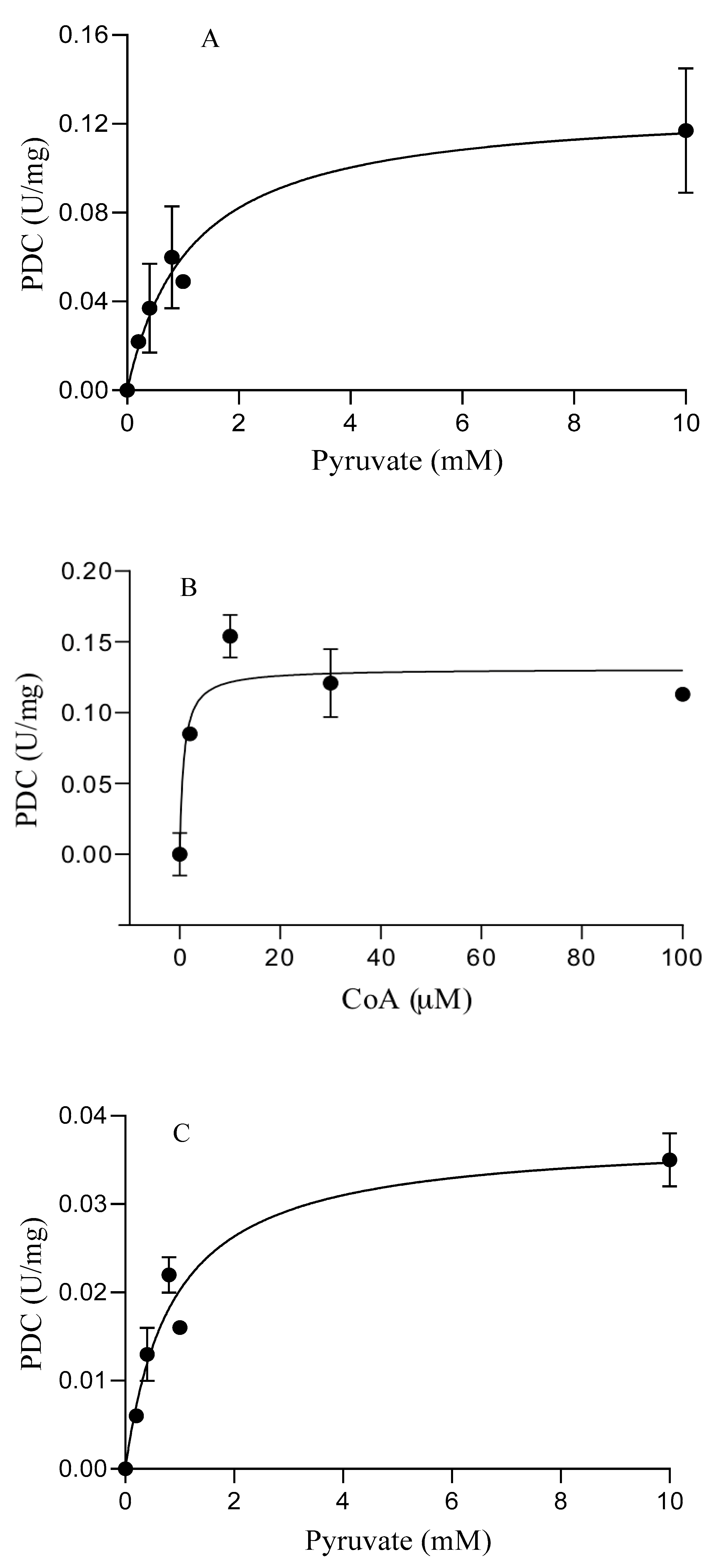
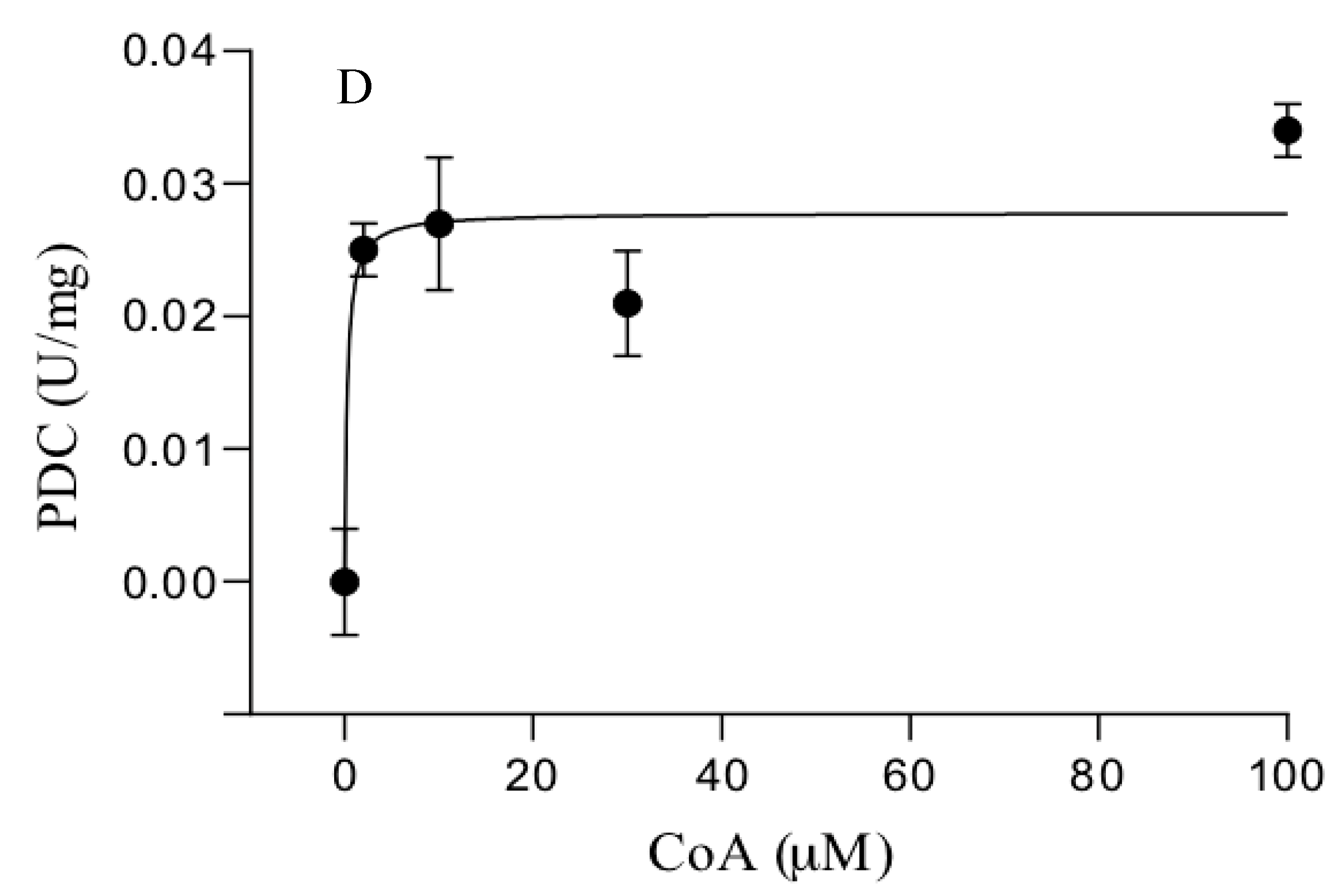
3.3. Catalytic Properties of the Purified Enzymes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, Y.; Tanaka, S. Ethanol fermentation from biomass resources: Current state and prospects. Appl. Microbiol. Biotechnol. 2006, 69, 627–642. [Google Scholar] [CrossRef]
- Tse, T.J.; Wiens, D.J.; Reaney, J.T. Production of bioethanol—A review of factors affecting ethanol yield. Fermentation 2021, 7, 268. [Google Scholar] [CrossRef]
- Eram, M.S.; Ma, K. Decarboxylation of Pyruvate to Acetaldehyde for Ethanol Production by Hyperthermophiles. Biomolecules 2013, 3, 578–596. [Google Scholar] [CrossRef]
- Bai, F.W.; Anderson, W.A.; Moo-Young, M. Ethanol fermentation technologies from sugar and starch feedstocks. Biotechnol. Adv. 2008, 26, 89–105. [Google Scholar] [CrossRef]
- Luan, G.; Qi, Y.; Wang, M.; Li, Z.; Duan, Y.; Tan, X.; Lu, X. Combinatory strategy for characterizing and understanding the ethanol synthesis pathway in cyanobacteria cell factories. Biotechnol. Biofuels 2015, 8, 184. [Google Scholar] [CrossRef]
- Straub, C.T.; Zeldes, B.M.; Schut, G.J.; Adams, M.W.; Kelly, R.M. Extremely thermophilic energy metabolisms: Biotechnological prospects. Cur. Opin. Biotechnol. 2017, 45, 104–112. [Google Scholar] [CrossRef]
- Ragsdale, S.W. Pyruvate ferredoxin oxidoreductase and its radical intermediate. Chem. Rev. 2003, 103, 2333–2346. [Google Scholar] [CrossRef]
- Eram, M.S.; Ma, K. The Bifunctional Pyruvate Decarboxylase/ Pyruvate Ferredoxin Oxidoreductase from Thermococcus guaymasensis. Archaea 2014, 2014, 349379. [Google Scholar] [CrossRef]
- Wang, Q.; Sha, C.; Wang, H.; Ma, K.; Wiegle, J.; Abomohra, A.E.-F.; Shao, W. A novel bifunctional aldehyde/alcohol dehydrogenase catalyzing reduction of acetyl-CoA to ethanol at temperatures up to 95 °C. Sci. Rep. 2021, 11, 1050. [Google Scholar] [CrossRef]
- Hoppner, T.C.; Doelle, H.W. Purification and kinetic characteristics of pyruvate decarboxylase and ethanol dehydrogenase from Zymomonas mobilis in relation to ethanol production. Eur. J. Appl. Microbiol. Biotechnol. 1983, 17, 152–157. [Google Scholar] [CrossRef]
- Talarico, L.A.; Ingram, L.O.; Maupin-Furlow, J.A. Production of the Gram-positive Sarcina ventriculi pyruvate decarboxylase in Escherichia coli. Microbiology 2001, 147, 2425–2435. [Google Scholar] [CrossRef]
- Fagernes, C.E.; Stensløkken, K.O.; Røhr, Å.K.; Berenbrink, M.; Ellefsen, S.; Nilsson, G.E. Extreme anoxia tolerance in crucian carp and goldfish through neofunctionalization of duplicated genes creating a new ethanol-producing pyruvate decarboxylase pathway. Sci. Rep. 2017, 7, 7884. [Google Scholar] [CrossRef]
- Berlowska, J.; Kregiel, D.; Ambroziak, W. Pyruvate decarboxylase activity assay in situ of different industrial yeast strains. Food Technol. Biotechnol. 2009, 47, 96–100. [Google Scholar]
- Eram, M.; Wong, A.; Oduaran, E.; Ma, K. Molecular and biochemical characterization of bifunctional pyruvate decarboxylases and pyruvate ferredoxin oxidoreductases from and. J. Biochem. 2015, 158, 459–466. [Google Scholar]
- Ma, K.; Hutchins, A.; Sung, S.S. Pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon, Pyrococcus furiosus, functions as a CoA-dependent pyruvate decarboxylase. Proc. Natl. Acad. Sci. USA 1997, 94, 9608–9613. [Google Scholar] [CrossRef]
- Yan, Z.; Fushinobu, S.; Wakagi, T. Four Cys residues in heterodimeric 2-oxoacid: Ferredoxin oxidoreductase are required for CoA-dependent oxidative decarboxylation but not for a non-oxidative decarboxylation. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2014, 1844, 736–743. [Google Scholar] [CrossRef]
- Eram, M.S.; Ma, K. Pyruvate decarboxylase activity of the acetohydroxyacid synthase of Thermotoga maritima. Biochem. Biophys. Rep. 2016, 7, 394–399. [Google Scholar] [CrossRef][Green Version]
- Duggleby, R.G.; McCourt, J.A.; Guddat, L.W. Structure and mechanism of inhibition of plant acetohydroxyacid synthase. Plant Physiol. Biochem. 2008, 46, 309–324. [Google Scholar] [CrossRef]
- Yan, Z.; Maruyama, A.; Arakawa, T.; Fushinobu, S.; Wakagi, T. Crystal structures of archaeal 2-oxoacid: Ferredoxin oxidoreductases from Sulfolobus tokodaii. Sci. Rep. 2016, 6, 33061. [Google Scholar] [CrossRef]
- Zhang, Q.; Iwasaki, T.; Wakagi, T.; Oshima, T. 2-Oxoacid: Ferredoxin Oxidoreductase from the Thermoacidophilic Archaeon, Sulfolobus sp. Strain 7. J. Biochem. 1996, 120, 587–599. [Google Scholar] [CrossRef]
- Pieulle, L.; Guigliarelli, B.; Asso, M.; Dole, F.; Bernadac, A.; Hatchikian, E.C. Isolation and characterization of the pyruvate-ferredoxin oxidoreductase from the sulfate-reducing bacterium Desulfovibrio africanus. BBA-Prot. Struc. Mol. Enzymol. 2017, 1250, 49–59. [Google Scholar] [CrossRef]
- Park, Y.J.; Yoo, C.B.; Choi, S.Y.; Lee, H.B. Purifications and characterizations of a ferredoxin and its related 2-oxoacid: Ferredoxin oxidoreductase from the hyperthermophilic archaeon, Sulfolobus solfataricus P1. BMB Rep. 2006, 39, 46–54. [Google Scholar] [CrossRef]
- Iwasaki, T.; Wakagi, T.; Oshima, T. Ferredoxin-dependent redox system of a thermoacidophilic archaeon, Sulfolobus sp. strain 7: Purification and characterization of a novel reduced ferredoxin-reoxidizing iron-sulfur flavoprotein. J. Biol. Chem. 1995, 270, 17878–17883. [Google Scholar] [CrossRef]
- Iwasaki, T.; Oshima, T. Ferredoxin and related enzymes from Sulfolobus. Meth. Enzymol. 2001, 334, 3–22. [Google Scholar]
- Nunn, C.E.M.; Johnsen, U.; Schönheit, P.; Fuhrer, T.; Sauer, U.; Hough, D.W.; Danson, M.J. Metabolism of pentose sugars in the hyperthermophilic archaea Sulfolobus solfataricus and Sulfolobus acidocaldarius. J. Biol. Chem. 2010, 285, 33701–33709. [Google Scholar] [CrossRef]
- Chen, L.M.; Brugger, K.; Skovgaard, M.; Redder, P.; She, Q.X.; Torarinsson, E.; Greve, B.; Awayez, M.; Zibat, A.; Klenk, H.-P.; et al. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 2005, 187, 4992–4999. [Google Scholar] [CrossRef]
- Grogan, D.W. Phenotypic characterization of the archaebacterial genus Sulfolobus: Comparison of five wild-type strains. J. Bacteriol. 1989, 171, 6710–6719. [Google Scholar] [CrossRef]
- Schocke, L.; Bräsen, C.; Siebers, B. Thermoacidophilic Sulfolobus species as source for extremozymes and as novel archaeal platform organisms. Curr. Opin. Biotechnol. 2019, 59, 71–77. [Google Scholar] [CrossRef]
- Lewis, A.M.; Recalde, A.; Bräsen, C.; Counts, J.A.; Nussbaum, P.; Bost, J.; Schocke, L.; Shen, L.; Willard, D.J.; Quax, T.E.F.; et al. The biology of thermoacidophilic archaea from the order Sulfolobales. FEMS Microbiol. Rev. 2021, 45, fuaa063. [Google Scholar] [CrossRef]
- Wagner, M.; Shen, L.; Albersmeier, A.; van der Kolk, N.; Kim, S.; Cha, J.; Bräsen, C.; Kalinowski, J.; Siebers, B.; Albers, S.-V. Sulfolobus acidocaldarius uptakes pentoses via a (CUT2)-type ABC transporter and metabolizes them through the aldolase-independent Weimberg pathway. Appl. Environ. Microbiol. 2018, 87, e01273-17. [Google Scholar] [CrossRef]
- Bräsen, C.; Esser, D.; Rauch, B.; Siebers, B. Carbohydrate metabolism in Archaea: Current insights into unusual enzymes and pathways and their regulation. Microbiol. Mol. Biol. Rev. 2014, 78, 176–197. [Google Scholar] [CrossRef]
- Quehenberger, J.; Shen, L.; Albers, S.V.; Siebers, B.; Spadiut, O. Sulfolobus—A potential key organism in future biotechnology. Front. Microbiol. 2017, 8, 2474. [Google Scholar] [CrossRef]
- Joshua, C.J.; Dahl, R.; Benke, P.I.; Keasling, J.D. Absence of diauxie during simultaneous utilization of glucose and xylose by Sulfolobus acidocaldarius. J. Bacteriol. 2011, 193, 1293–1301. [Google Scholar] [CrossRef]
- Fukuda, E.; Kino, H.; Matsuzawa, H.; Wakagi, T. Role of a highly conserved YPITP motif in 2-oxoacid: Ferredoxin oxidoreductase. Eur. J. Biochem. 2001, 268, 5639–5646. [Google Scholar] [CrossRef]
- Brock, T.; Brock, K.; Belly, R.; Weiss, R. Sulfolobus: A new genus of sulfur- oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 1972, 84, 54–68. [Google Scholar] [CrossRef]
- Mikoulinskaia, O.; Akimenko, V.; Galouchko, A.; Thauer, R.K.; Hedderich, R. Cytochrome c-dependent methacrylate reductase from Geobacter sulfurreducens AM-1. Eur. J. Biochem. 1999, 263, 346–352. [Google Scholar] [CrossRef]
- Yoon, K.S.; Ishii, M.; Kodama, T.; Igarashi, Y. Purification and characterization of pyruvate:ferredoxin oxidoreductase from Hydrogenobacter thermophilus TK-6. Arch. Microbiol. 1997, 167, 275–279. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Kunow, J.; Linder, D.; Thauer, R.K. Pyruvate: Ferredoxin oxidoreductase from the sulfate-reducing Archaeoglobus fulgidus: Molecular composition, catalytic properties, and sequence alignments. Arch. Microbiol. 1995, 163, 21–28. [Google Scholar]
- Kerscher, L.; Oesterhelt, D. Purification and properties of two 2-oxoacid: Ferredoxin oxidoreductases from Halobacterium halobium. Eur. J. Biochem. 1981, 116, 587–594. [Google Scholar] [CrossRef]
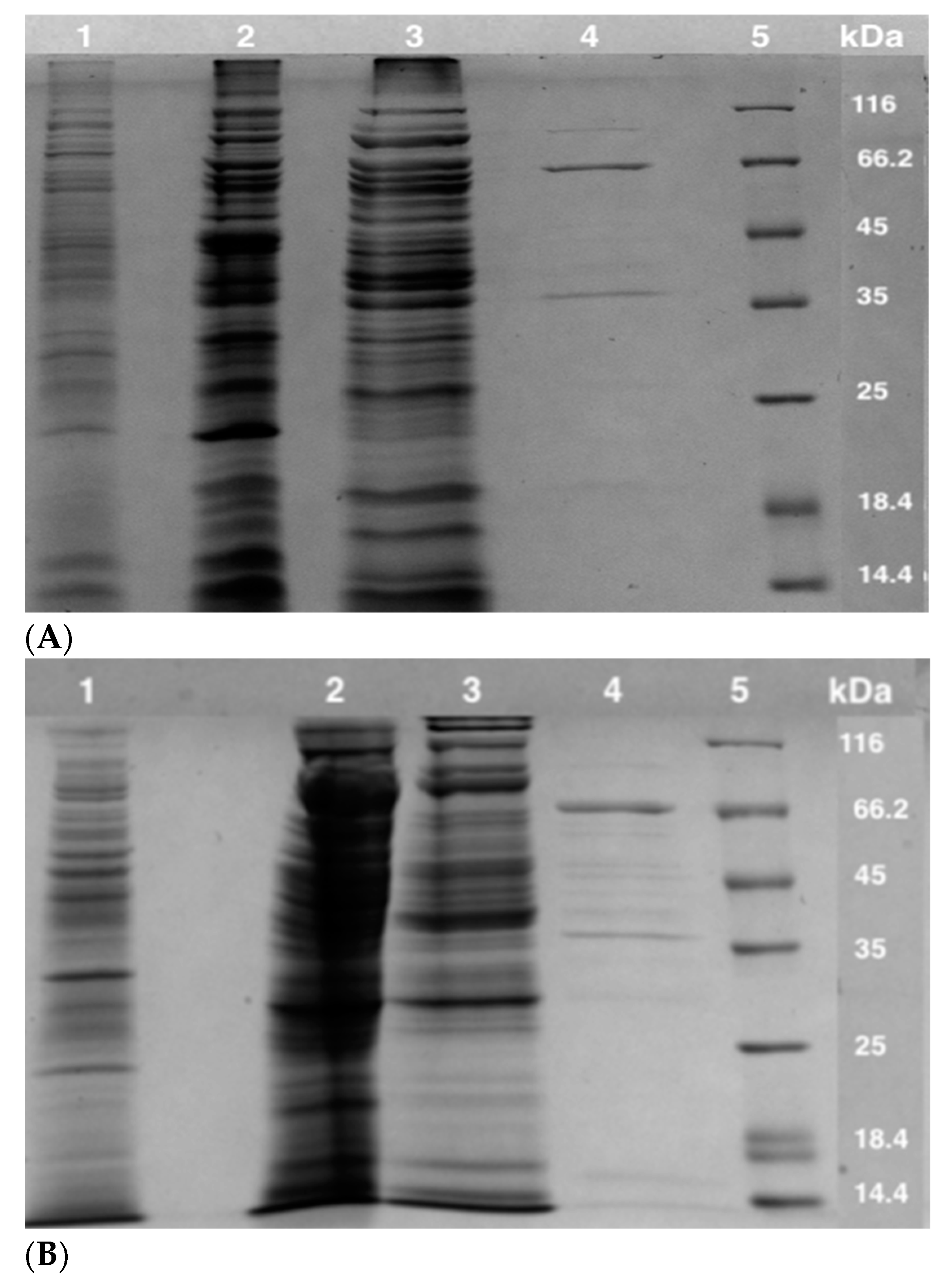

| Step | Enzyme | Protein (mg) | Specific Activity (U/mg) a,b | Total Activity (U) | Fold | Recovery (%) |
|---|---|---|---|---|---|---|
| CFE | POR | 354.2 ± 2.5 | 0.18 ± 0.01 | 63.8 ± 0.15 | 1 | 100 |
| PDC | 354.2 ± 2.5 | 0.0027 ± 0.0003 | 0.95 ± 0.05 | 1 | 100 | |
| DEAE | POR | 90 ± 1 | 0.43 ± 0.02 | 39 ± 0.1 | 2.4 | 61.4 |
| PDC | nd | nd | nd | nd | nd | |
| HAP | POR | 63.1 ± 0.5 | 0.6 ± 0.02 | 37.9 ± 0.05 | 3.3 | 59.3 |
| PDC | nd | nd | nd | nd | nd | |
| Phenyl-Sepharose | POR | 2.1 ± 0.1 | 7.5 ± 0.05 | 15.9 ± 0.01 | 41.6 | 25 |
| PDC | 2.1 ± 0.1 | 0.11 ± 0.004 | 0.23 ± 0.005 | 40.7 | 24 |
| Step | Enzyme | Protein (mg) | Specific Activity (U/mg) a,b | Total Activity (U) | Fold | Recovery (%) |
|---|---|---|---|---|---|---|
| CFE | POR | 231.8 ± 2 | 0.1 ± 0.01 | 23.2 ± 1 | 1 | 100 |
| PDC | 231.8 ± 2 | 0.0011 ± 0.0004 | 0.25 ± 0.04 | 1 | 100 | |
| DEAE | POR | 54.23 ± 1.5 | 0.28 ± 0.01 | 15.2 ± 0.3 | 2.38 | 65.4 |
| PDC | nd | nd | nd | nd | nd | |
| HAP | POR | 22.6 ± 0.4 | 0.45 ± 0.03 | 10 ± 0.2 | 4.5 | 39.4 |
| PDC | nd | nd | nd | nd | nd | |
| Phenyl-Sepharose | POR | 0.63 ± 0.03 | 7 ± 0.02 | 4.41 ± 0.01 | 70 | 19 |
| PDC | 0.63 ± 0.03 | 0.055 ± 0.003 | 0.035 ± 0.001 | 50 | 14 |
| Enzyme Sources | Enzyme Activity | a Pyruvate | b CoA | ||
|---|---|---|---|---|---|
| Km (mM) | Vmax (U/mg−1) | Km (µM) | Vmax (U/mg−1) | ||
| S. solfataricus | POR | 0.5 ± 0.1 | 6.3 ± 0.7 | 10.7 ± 0.4 | 7.7 ± 0.07 |
| PDC | 1.1 ± 0.2 | 0.12 ± 0.09 | 0.77 ± 0.27 c | 0.12 ± 0.08 c | |
| S. acidocaldarius | POR | 0.3 ± 0.05 | 1.9 ± 0.2 | 21.5 ± 3 | 1.7 ± 0.08 |
| PDC | 0.86 ± 0.2 | 0.04 ± 0.03 | 0.3 ± 0.06 c | 0.04 ± 0.03 c | |
| Organism (Growth Topt, °C) | Enzyme Activity (80 °C) | Pyruvate | CoA | References | |||
|---|---|---|---|---|---|---|---|
| Km (mM) | Vmax (U/mg) | Km (µM) | Vmax (U/mg) | ||||
| Bacteria | T. maritima (80) | POR | 0.4 ± 0.1 | 81 ± 6 | 63 ± 6 | 94 ± 2 | [14] |
| PDC | 0.92 ± 0.3 | 1.4 ± 0.04 | 3.1 ± 1.2 | 1.3 ± 0.03 | |||
| T. hypogea (70 a) | POR | 0.13 ± 0.03 | 99 ± 3 | 21 ± 2 | 73 ± 4 | [14] | |
| PDC | 1.4 ± 0.4 | 2.5 ± 0.18 | 1.4 ± 0.02 | 1.6 ± 0.13 | |||
| Archaea | T. guaymasensis (88) | POR | 0.53 ± 0.03 | 18 ± 0.23 | 70 ± 10 | 21.8 ± 0.8 | [8] |
| PDC | 0.25 ± 0.05 | 3.8 ± 0.14 | 20 ± 1 | 3.3 ± 0.09 | |||
| P. furiosus (100) | POR | 0.46 | 23.6 | 110 | 22 | [15] | |
| PDC b | 1.1 | 4.3 ± 0.3 | 110 | 4.3 ± 0.3 | |||
| S. solfataricus (80) | POR c | 0.5 ± 0.1 | 6.3 ± 0.7 | 10.7 ± 0.4 | 7.7 ± 0.07 | This study | |
| PDC d | 1.1 ± 0.2 | 0.12 ± 0.09 | 0.77 ± 0.27 | 0.12 ± 0.08 | |||
| S. acidocaldarius (80) | POR c | 0.3 ± 0.05 | 1.9 ± 0.2 | 21.5 ± 3 | 1.7 ± 0.08 | This study | |
| PDC d | 0.86 ± 0.2 | 0.04 ± 0.03 | 0.3 ± 0.06 | 0.04 ± 0.03 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alharbi, F.; Knura, T.; Siebers, B.; Ma, K. Thermostable and O2-Insensitive Pyruvate Decarboxylases from Thermoacidophilic Archaea Catalyzing the Production of Acetaldehyde. Biology 2022, 11, 1247. https://doi.org/10.3390/biology11081247
Alharbi F, Knura T, Siebers B, Ma K. Thermostable and O2-Insensitive Pyruvate Decarboxylases from Thermoacidophilic Archaea Catalyzing the Production of Acetaldehyde. Biology. 2022; 11(8):1247. https://doi.org/10.3390/biology11081247
Chicago/Turabian StyleAlharbi, Faisal, Thomas Knura, Bettina Siebers, and Kesen Ma. 2022. "Thermostable and O2-Insensitive Pyruvate Decarboxylases from Thermoacidophilic Archaea Catalyzing the Production of Acetaldehyde" Biology 11, no. 8: 1247. https://doi.org/10.3390/biology11081247
APA StyleAlharbi, F., Knura, T., Siebers, B., & Ma, K. (2022). Thermostable and O2-Insensitive Pyruvate Decarboxylases from Thermoacidophilic Archaea Catalyzing the Production of Acetaldehyde. Biology, 11(8), 1247. https://doi.org/10.3390/biology11081247






