Incipient Sympatric Speciation and Evolution of Soil Bacteria Revealed by Metagenomic and Structured Non-Coding RNAs Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, DNA Isolation, and Sequencing
2.2. Quality Control, Reads Assembly, Binning, Refinement, Assessment, and Annotation
2.3. Taxonomic Compositions of Basalt and Chalk Sample
2.4. Function Compositions of Basalt and Chalk Sample
- Step 1: Calculation of the copy number of each gene: bi = xi/Li;
- Step 2: Calculation of the relative abundance of gene i: ai = bi/∑bi:
- ai: the relative abundance of gene I;
- bi: the copy number of gene i from sample N;
- Li: the length of gene i;
- xi: the number of mapped reads.
2.5. Analysis of Structured Noncoding RNAs in the Metagenome-Assembled Genomic Reads (MAGs) Sequences
2.6. Metagenomic Single Nucleotide Variants (SNVs) Analysis
2.7. Single Nucleotide Polymorphisms (SNPs) Analysis
3. Results
3.1. Metagenome Assembly
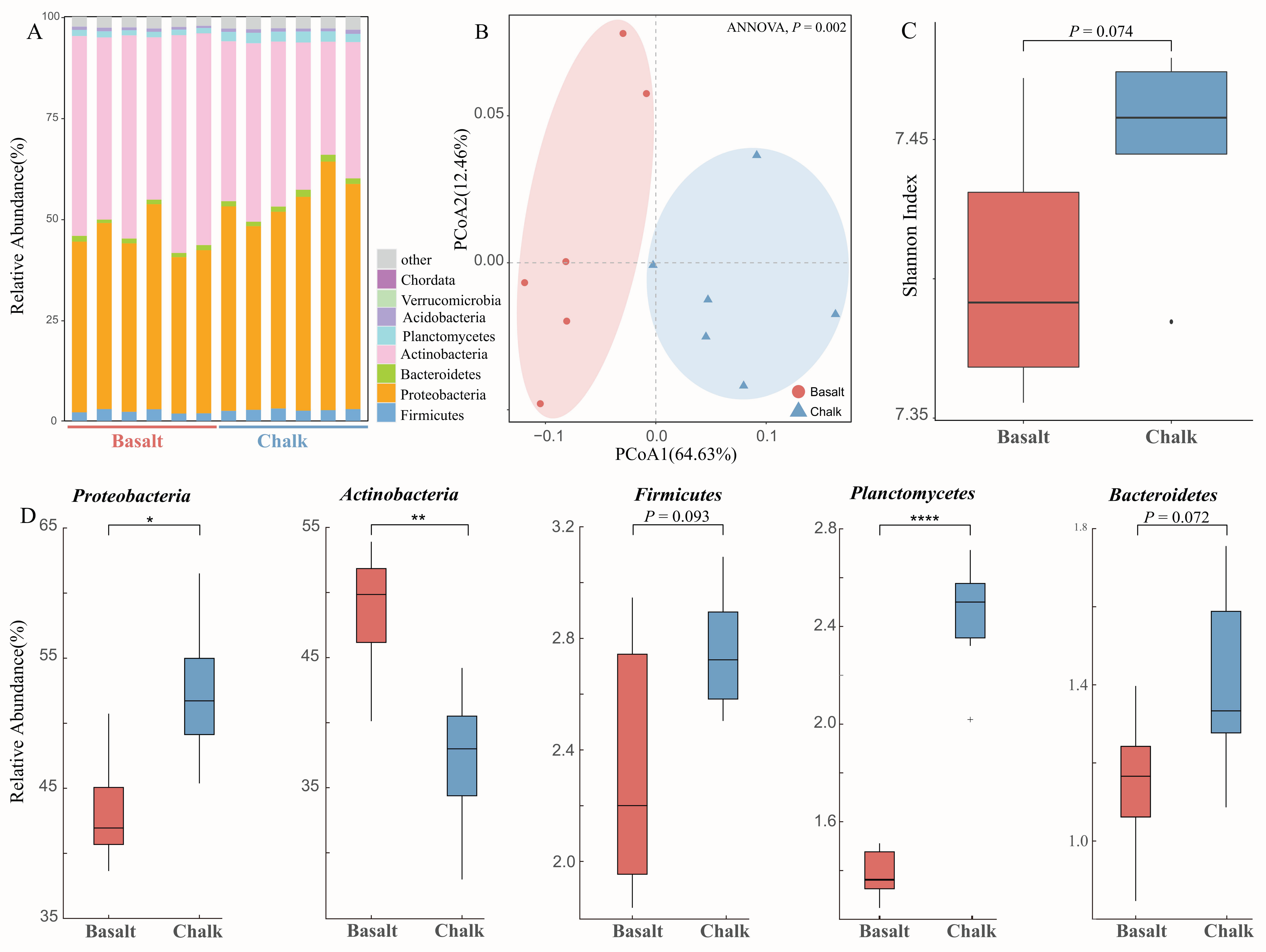
3.2. Bacterial Community Composition
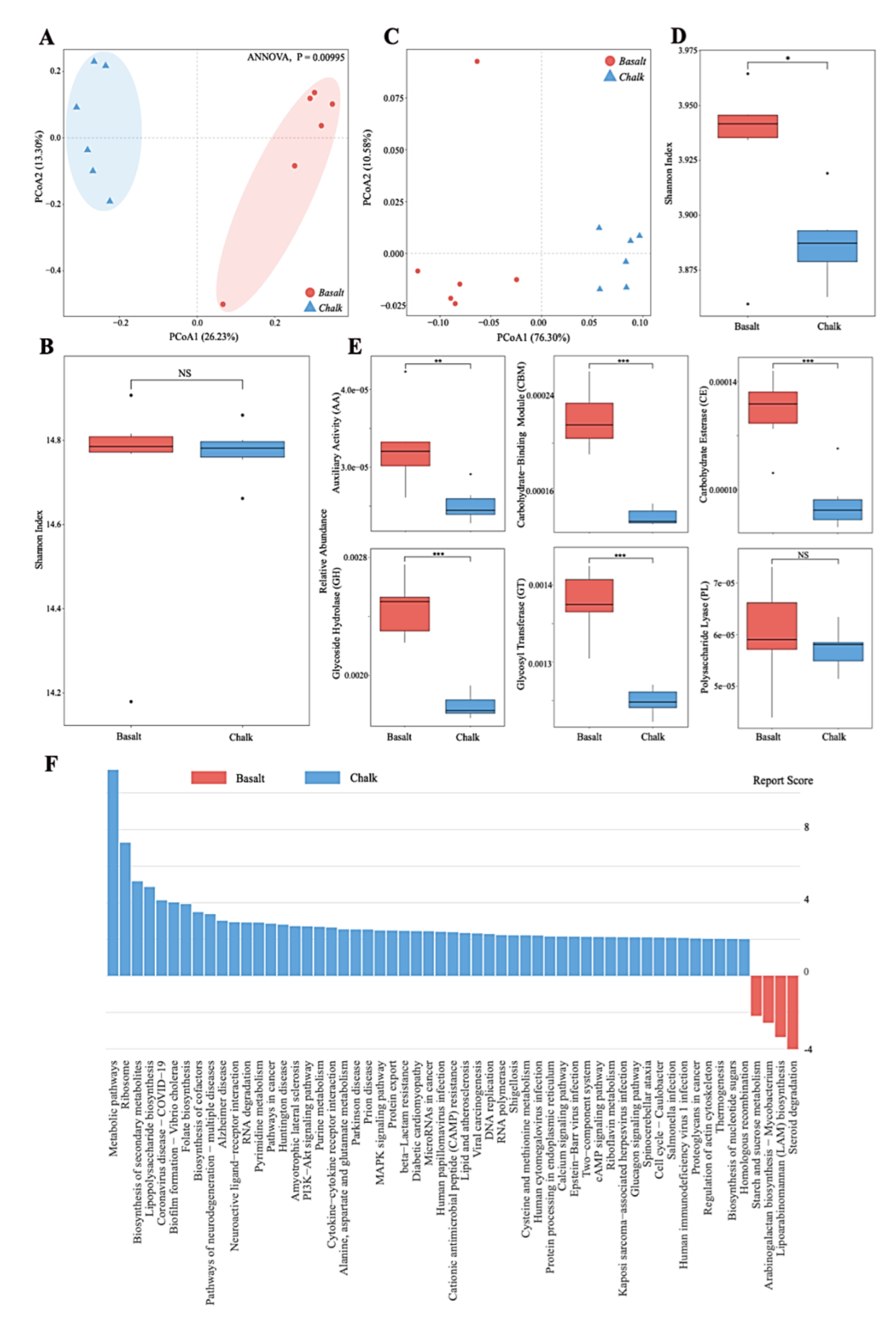
3.3. Functional Analysis of Metagenomic Data
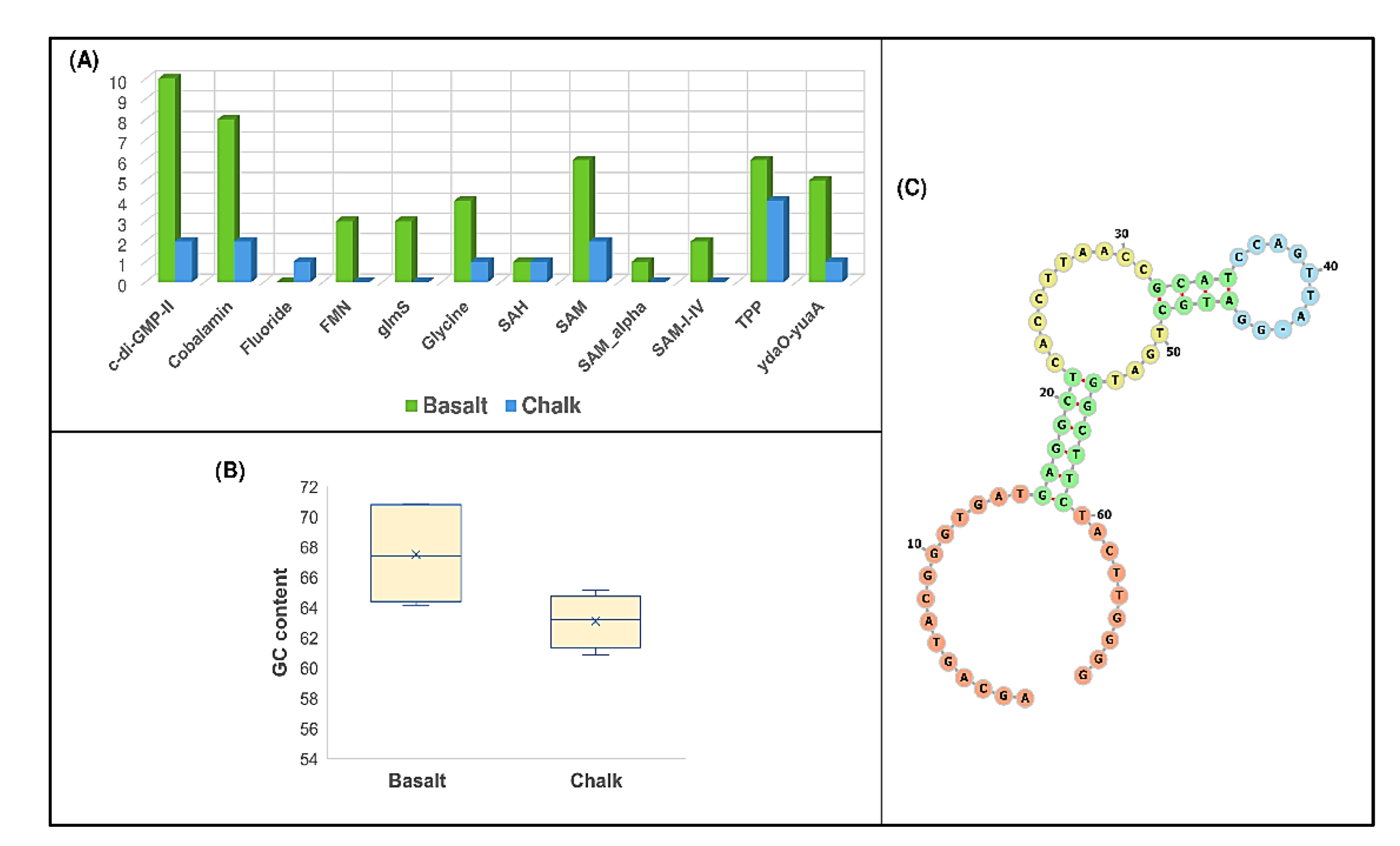
3.4. Analysis of Structured Noncoding RNAs in Metagenome-Assembled Genomic Reads (MAGs) Sequences
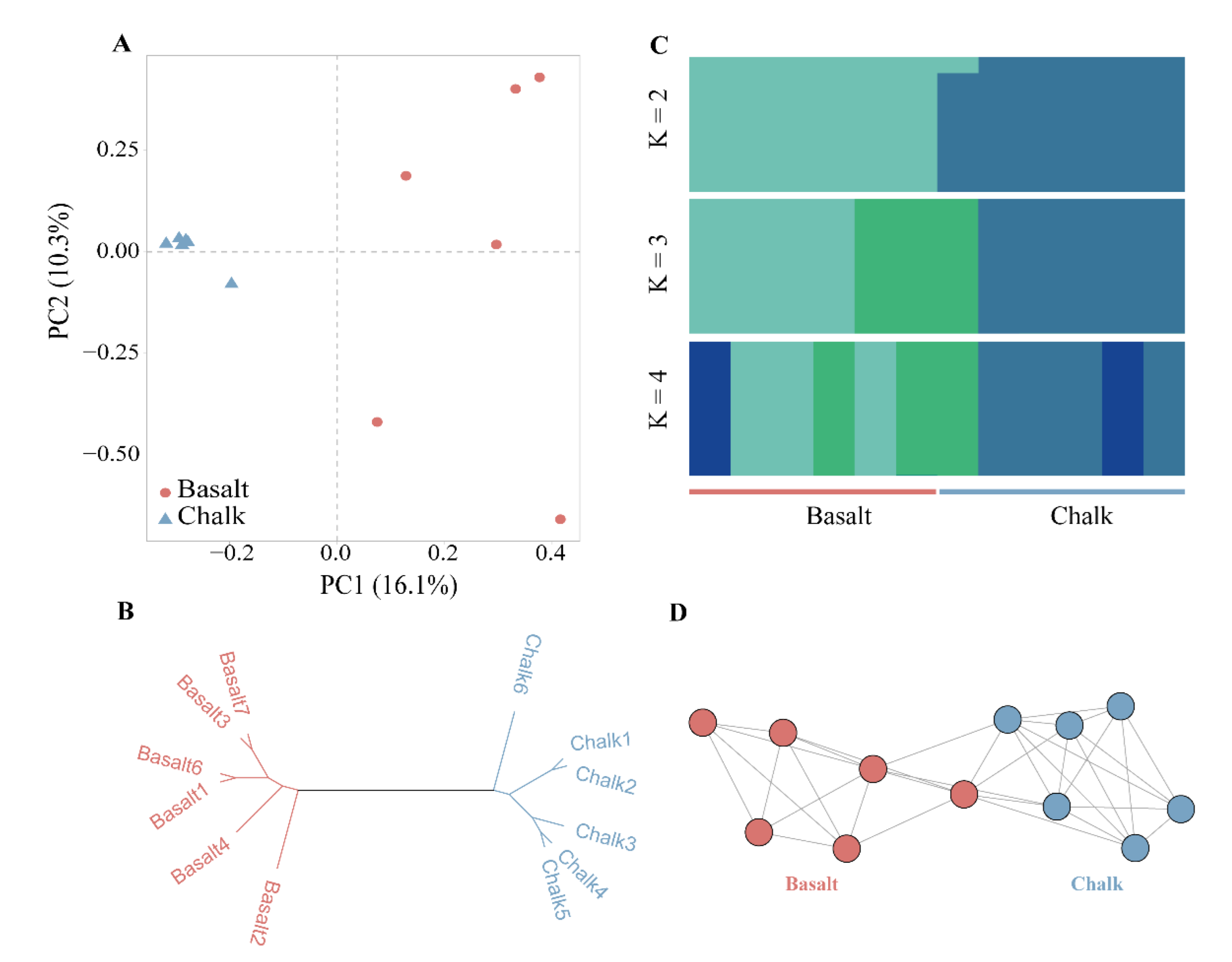
3.5. Genetic Divergence between the Two Microsites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharpton, T.J. An introduction to the analysis of shotgun metagenomic data. Front. Plant Sci. 2014, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- Dunivin, T.K.; Choi, J.; Howe, A.; Shade, A. RefSoil+: A Reference Database for Genes and Traits of Soil Plasmids. mSystems 2019, 4, e00349-18. [Google Scholar] [CrossRef] [PubMed]
- Altieri, M.A. The ecological role of biodiversity in agroecosystems. Agric. Ecosyst. Environ. 1999, 74, 19–31. [Google Scholar] [CrossRef]
- Choi, J.; Yang, F.; Stepanauskas, R.; Cardenas, E.; Garoutte, A.; Williams, R.; Flater, J.; Tiedje, J.M.; Hofmockel, K.S.; Gelder, B.; et al. Strategies to improve reference databases for soil microbiomes. ISME J. 2017, 11, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.D.; Martiny, J.B.H. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef] [PubMed]
- Greenblum, S.; Carr, R.; Borenstein, E. Extensive strain-level copy-number variation across human gut microbiome species. Cell 2015, 160, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Sunagawa, S.; Mende, D.R.; Bork, P. Inter-individual differences in the gene content of human gut bacterial species. Genome Biol. 2015, 16, 82. [Google Scholar] [CrossRef]
- Shapiro, B.J.; Friedman, J.; Cordero, O.X.; Preheim, S.P.; Timberlake, S.C.; Szabó, G.; Polz, M.F.; Alm, E.J. Population genomics of early events in the ecological differentiation of bacteria. Science 2012, 335, 48–51. [Google Scholar] [CrossRef]
- Kashtan, N.; Roggensack, S.E.; Rodrigue, S.; Thompson, J.W.; Biller, S.J.; Coe, A.; Ding, H.; Marttinen, P.; Malmstrom, R.R.; Stocker, R.; et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 2014, 344, 416–420. [Google Scholar] [CrossRef]
- Nguyen, J.; Lara-Gutiérrez, J.; Stocker, R. Environmental fluctuations and their effects on microbial communities, populations and individuals. FEMS Microbiol. Rev. 2021, 45, fuaa068. [Google Scholar] [CrossRef]
- Vicuña, R.; González, B. The microbial world in a changing environment. Rev. Chil. Hist. Nat. 2021, 94, 2. [Google Scholar] [CrossRef]
- Collins, S.; Boyd, P.W.; Doblin, M.A. Evolution, Microbes, and Changing Ocean Conditions. Ann. Rev. Mar. Sci. 2020, 12, 181–208. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, P.D. Microbial evolution and ecological opportunity in the gut environment. Proc. R. Soc. B Boil. Sci. 2019, 286, 20191964. [Google Scholar] [CrossRef]
- Chase, A.B.; Weihe, C.; Martiny, J.B.H. Adaptive differentiation and rapid evolution of a soil bacterium along a climate gradient. Proc. Natl. Acad. Sci. USA 2021, 118, e2101254118. [Google Scholar] [CrossRef]
- Massey, R.C.; Rainey, P.B.; Sheehan, B.J.; Keane, O.M.; Dorman, C.J. Environmentally constrained mutation and adaptive evolution in Salmonella. Curr. Biol. 1999, 9, 1477–1481. [Google Scholar] [CrossRef]
- Foster, P.L.; Lee, H.; Popodi, E.; Townes, J.P.; Tang, H. Determinants of spontaneous mutation in the bacterium Escherichia coli as revealed by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 2015, 112, E5990–E5999. [Google Scholar] [CrossRef] [PubMed]
- Barash, D.; Sikorski, J.; Perry, E.B.; Nevo, E.; Nudler, E. Adaptive mutations in RNA-based regulatory mechanisms: Computational and experimental investigations. Isr. J. Ecol. Evol. 2006, 52, 263–269. [Google Scholar] [CrossRef]
- Graf, M.; Haas, T.; Müller, F.; Buchmann, A.; Harm-Bekbenbetova, J.; Freund, A.; Nieß, A.; Persicke, M.; Kalinowski, J.; Blombach, B.; et al. Continuous Adaptive Evolution of a Fast-Growing Corynebacterium glutamicum Strain Independent of Protocatechuate. Front. Microbiol. 2019, 10, 1648. [Google Scholar] [CrossRef]
- Espejo, R.T.; García, K.; Plaza, N. Insight into the origin and evolution of the Vibrio parahaemolyticus pandemic strain. Front. Microbiol. 2017, 8, 1397. [Google Scholar] [CrossRef]
- Darwin, C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life; John Murray: London, UK, 1859; Available online: https://books.google.co.il/books/about/The_Origin_of_Species_by_Means_of_Natura.html?id=dOQEAAAAYAAJ&printsec=frontcover&source=kp_read_button&hl=en&redir_esc=y#v=onepage&q&f=false (accessed on 13 May 2022).
- Coyne, J.A. Speciation in a small space. Proc. Natl. Acad. Sci. USA 2011, 108, 12975–12976. [Google Scholar] [CrossRef]
- Wang, Y.; Qiao, Z.; Mao, L.; Li, F.; Liang, X.; An, X.; Zhang, S.; Liu, X.; Kuang, Z.; Wan, N.; et al. Sympatric speciation of the spiny mouse from Evolution Canyon in Israel substantiated genomically and methylomically. Proc. Natl. Acad. Sci. USA 2022, 119, e2121822119. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wang, H.; Cai, Z.; Wang, L.; Xu, Q.; Lövy, M.; Wang, Z.; Nevo, E. Sympatric speciation of spiny mice, Acomys, unfolded transcriptomically at Evolution Canyon, Israel. Proc. Natl. Acad. Sci. USA 2016, 113, 8254–8259. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wang, L.; Knisbacher, B.A.; Xu, Q.; Levanon, E.Y.; Wang, H.; Frenkel-Morgenstern, M.; Tagore, S.; Fang, X.; Bazak, L.; et al. Transcriptome, genetic editing, and microRNA divergence substantiate sympatric speciation of blind mole rat, Spalax. Proc. Natl. Acad. Sci. USA 2016, 113, 7584–7589. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Hong, W.; Jiao, H.; Wang, G.D.; Rodriguez, K.A.; Buffenstein, R.; Zhao, Y.; Nevo, E.; Zhao, H. Sympatric speciation revealed by genome-wide divergence in the blind mole rat Spalax. Proc. Natl. Acad. Sci. USA 2015, 112, 11905–11910. [Google Scholar] [CrossRef]
- Koeppel, A.; Perry, E.B.; Sikorski, J.; Krizanc, D.; Warner, A.; Ward, D.M.; Rooney, A.P.; Brambilla, E.; Connor, N.; Ratcliff, R.M.; et al. Identifying the fundamental units of bacterial diversity: A paradigm shift to incorporate ecology into bacterial systematics. Proc. Natl. Acad. Sci. USA 2008, 105, 2504–2509. [Google Scholar] [CrossRef]
- Nevo, E. Asian, African and European biota meet at “Evolution Canyon” Israel: Local tests of global biodiversity and genetic diversity patterns. Proc. R. Soc. B Biol. Sci. 1995, 262, 149–155. [Google Scholar] [CrossRef]
- Nevo, E. “Evolution Canyon,” a potential microscale monitor of global warming across life. Proc. Natl. Acad. Sci. USA 2012, 109, 2960–2965. [Google Scholar] [CrossRef]
- Nevo, E. Evolution in action across life at “evolution canyons”, Israel. Trends Evol. Biol. 2009, 1, e3. [Google Scholar] [CrossRef]
- Nevo, E. Evolution of wild emmer wheat and crop improvement. J. Syst. Evol. 2014, 52, 673–696. [Google Scholar] [CrossRef]
- Hadid, Y.; Tzur, S.; Pavlíček, T.; Šumbera, R.; Šklíba, J.; Lövy, M.; Fragman-Sapir, O.; Beiles, A.; Arieli, R.; Raz, S.; et al. Possible incipient sympatric ecological speciation in blind mole rats (Spalax). Proc. Natl. Acad. Sci. USA 2013, 110, 2587–2592. [Google Scholar] [CrossRef]
- Sikorski, J.; Nevo, E. Adaptation and incipient sympatric speciation of Bacillus simplex under microclimatic contrast at “Evolution Canyons” I and II, Israel. Proc. Natl. Acad. Sci. USA 2005, 102, 15924–15929. [Google Scholar] [CrossRef] [PubMed]
- Nayfach, S.; Rodriguez-Mueller, B.; Garud, N.; Pollard, K.S. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 2016, 26, 1612–1625. [Google Scholar] [CrossRef]
- Costea, P.I.; Munch, R.; Coelho, L.P.; Paoli, L.; Sunagawa, S.; Bork, P. metaSNV: A tool for metagenomic strain level analysis. PLoS ONE 2017, 12, e0182392. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Cui, L.; Wang, X.; Yang, G.; Huo, F.; Ling, H.; Chen, L.; She, K.; Du, X.; Levi, B.; et al. Genomic and Phenotypic Divergence in Wild Barley Driven by Microgeographic Adaptation. Adv. Sci. 2020, 7, 2000709. [Google Scholar] [CrossRef] [PubMed]
- Polyakov, A.; Beharav, A.; Avivi, A.; Nevo, E. Mammalian microevolution in action: Adaptive edaphic genomic divergence in blind subterranean mole-rats. Proc. R. Soc. B Biol. Sci. 2004, 271, S156–S159. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Ren, X.; Song, X.; Li, X.; Zhou, Y.; Harlev, E.; Sun, D.; Nevo, E. Incipient sympatric speciation in wild barley caused by geological-edaphic divergence. Life Sci. Alliance 2020, 3, e202000827. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zhang, S.; Song, X.; Weyrich, A.; Wang, Y.; Liu, X.; Wan, N.; Liu, J.; Lovy, M.; Cui, H.; et al. Genome evolution of blind subterranean mole rats: Adaptive peripatric versus sympatric speciation. Proc. Natl. Acad. Sci. USA 2020, 117, 32499–32508. [Google Scholar] [CrossRef]
- Lövy, M.; Šklíba, J.; Hrouzková, E.; Dvoráková, V.; Nevo, E.; Šumbera, R. Habitat and burrow system characteristics of the blind mole rat spalax galili in an area of supposed sympatric speciation. PLoS ONE 2015, 10, e0133157. [Google Scholar] [CrossRef]
- Lövy, M.; Šklíba, J.; Šumbera, R.; Nevo, E. Soil preference in blind mole rats in an area of supposed sympatric speciation: Do they choose the fertile or the familiar? J. Zool. 2017, 303, 291–300. [Google Scholar] [CrossRef]
- Lövy, M.; Šumbera, R.; Heth, G.; Nevo, E. Presumed ecological speciation in blind mole rats: Does soil type influence mate preferences? Ethol. Ecol. Evol. 2020, 32, 46–59. [Google Scholar] [CrossRef]
- Nevo, E. Evolution under environmental stress at macro- and microscales. Genome Biol. Evol. 2011, 3, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A quality control tool for high throughput sequence data. Babraham Bioinforma. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 19 May 2022).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating species abundance in metagenomics data. PeerJ Comput. Sci. 2017, 3, e104. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. DbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- McCown, P.J.; Corbino, K.A.; Stav, S.; Sherlock, M.E.; Breaker, R.R. Riboswitch diversity and distribution. RNA 2017, 23, 995–1011. [Google Scholar] [CrossRef]
- Barrick, J.E.; Breaker, R.R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007, 8, R239. [Google Scholar] [CrossRef]
- Mukherjee, S.; Das Mandal, S.; Gupta, N.; Drory-Retwitzer, M.; Barash, D.; Sengupta, S. RiboD: A comprehensive database for prokaryotic riboswitches. Bioinformatics 2019, 35, 3541–3543. [Google Scholar] [CrossRef] [PubMed]
- Nudler, E.; Mironov, A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004, 29, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Barash, D.; Sengupta, S. Comparative genomics and phylogenomic analyses of lysine riboswitch distributions in bacteria. PLoS ONE 2017, 12, e0184314. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Burge, S.W.; Bateman, A.; Daub, J.; Eberhardt, R.Y.; Eddy, S.R.; Floden, E.W.; Gardner, P.P.; Jones, T.A.; Tate, J.; et al. Rfam 12.0: Updates to the RNA families database. Nucleic Acids Res. 2015, 43, D130–D137. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Sengupta, S. Riboswitch Scanner: An efficient pHMM-based web-server to detect riboswitches in genomic sequences. Bioinformatics 2016, 32, 776–778. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Höner Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Retwitzer, M.D.; Barash, D.; Sengupta, S. Phylogenomic and comparative analysis of the distribution and regulatory patterns of TPP riboswitches in fungi. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Li, H.; Chen, Z.; Coghlan, A.; Coin, L.J.M.; Guo, Y.; Hériché, J.K.; Hu, Y.; Kristiansen, K.; Li, R.; et al. TreeFam: 2008 Update. Nucleic Acids Res. 2008, 36, D735–D740. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Steinig, E.J.; Neuditschko, M.; Khatkar, M.S.; Raadsma, H.W.; Zenger, K.R. Netview p: A network visualization tool to unravel complex population structure using genome-wide SNPs. Mol. Ecol. Resour. 2016, 16, 216–227. [Google Scholar] [CrossRef]
- Bastet, L.; Dub, A.; Mass, E.; Lafontaine, D.A. New insights into riboswitch regulation mechanisms. Mol. Microbiol. 2011, 80, 1148–1154. [Google Scholar] [CrossRef]
- Breaker, R.R. Prospects for riboswitch discovery and analysis. Mol. Cell 2011, 43, 867–879. [Google Scholar] [CrossRef]
- Serganov, A.; Nudler, E. A Decade of Riboswitches. Cell 2013, 152, 17–24. [Google Scholar] [CrossRef]
- Barash, D.; Gabdank, I. Energy minimization methods applied to riboswitches: A perspective and challenges. RNA Biol. 2010, 7, 90–97. [Google Scholar] [CrossRef]
- Barash, D.; Churkin, A. Mutational analysis in RNAs: Comparing programs for RNA deleterious mutation prediction. Brief. Bioinform. 2011, 12, 104–114. [Google Scholar] [CrossRef][Green Version]
- Markham, N.R.; Zuker, M. UNAFold: Software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008, 453, 3–31. [Google Scholar] [CrossRef]
- Ivry, T.; Michal, S.; Avihoo, A.; Sapiro, G.; Barash, D. An image processing approach to computing distances between RNA secondary structures dot plots. Algorithms Mol. Biol. 2009, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.L.; Sudarsan, N.; Weinberg, Z.; Roth, A.; Stockbridge, R.B.; Breaker, R.R. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 2012, 335, 233–235. [Google Scholar] [CrossRef]
- Savolainen, V.; Anstett, M.C.; Lexer, C.; Hutton, I.; Clarkson, J.J.; Norup, M.V.; Powell, M.P.; Springate, D.; Salamin, N.; Baker, W.J. Sympatric speciation in palms on an oceanic island. Nature 2006, 441, 210–213. [Google Scholar] [CrossRef] [PubMed]
- Papadopulos, A.S.T.; Igea, J.; Dunning, L.T.; Osborne, O.G.; Quan, X.; Pellicer, J.; Turnbull, C.; Hutton, I.; Baker, W.J.; Butlin, R.K.; et al. Ecological speciation in sympatric palms: Genetic map reveals genomic islands underlying species divergence in Howea. Evolution 2019, 73, 1986–1995. [Google Scholar] [CrossRef] [PubMed]
- Papadopulos, A.S.T.; Igea, J.; Smith, T.P.; Hutton, I.; Baker, W.J.; Butlin, R.K.; Savolainen, V. Ecological speciation in sympatric palms: Demographic analyses support speciation of Howea in the face of high gene flow. Evolution 2019, 73, 1996–2002. [Google Scholar] [CrossRef]
- Nevo, E.; Korol, A.; Beiles, A.; Fahima, T. Evolution of Wild Emmer and Wheat Improvement: Population Genetics, Genetic Resources, and Genome Organization of Wheat’s Progenitor, Triticum Dicoccoides; Springer: Berlin, Germany, 2002. [Google Scholar]
- Wang, H.; Yin, H.; Jiao, C.; Fang, X.; Wang, G.; Li, G.; Ni, F.; Li, P.; Su, P.; Ge, W.; et al. Sympatric speciation of wild emmer wheat driven by ecology and chromosomal rearrangements. Proc. Natl. Acad. Sci. USA 2020, 117, 5955–5963. [Google Scholar] [CrossRef]
- Qian, C.; Yan, X.; Yin, H.; Fan, X.; Yin, X.; Sun, P.; Li, Z.; Nevo, E.; Ma, X.F. Transcriptomes Divergence of Ricotia lunaria Between the Two Micro-Climatic Divergent Slopes at “Evolution Canyon” I, Israel. Front. Genet. 2018, 9, 506. [Google Scholar] [CrossRef]
- Hong, W.; Li, K.; Sharaf, K.; Song, X.; Pavlìcek, T.; Zhao, H.; Nevo, E. Genome-wide analysis revisits incipient sympatric and allopatric speciation in a beetle. Isr. J. Ecol. Evol. 2021, 67, 69–80. [Google Scholar] [CrossRef]
- Nevo, E. Evolution Canyons model: Biodiversity, adaptation, and incipient sympatric ecological speciation across life: A revisit. In New Horizons in Evolution; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Perry, E.B.; Krizanc, D.; Rooney, A.P.; Sikorski, J.; Nevo, E.; Cohan, F.M. Identifying the fundamental units of diversity among Bacillus isolates from “Evolution Canyons” III. Isr. J. Ecol. Evol. 2006, 52, 543–552. [Google Scholar] [CrossRef]
- Takebayashi, S.; Narihiro, T.; Fujii, Y.; Hiraishi, A. Water Availability is a Critical Determinant of a Population Shift from Proteobacteria to Actinobacteria during Start-Up Operation of Mesophilic Fed-Batch Composting. Microbes Environ. 2007, 22, 279–289. [Google Scholar] [CrossRef]
- Zhang, R.; Chen, L.; Niu, Z.; Song, S.; Zhao, Y. Water stress affects the frequency of Firmicutes, Clostridiales and Lysobacter in rhizosphere soils of greenhouse grape. Agric. Water Manag. 2019, 226, 105776. [Google Scholar] [CrossRef]
- Gupta, A.; Dutta, A.; Sarkar, J.; Panigrahi, M.K.; Sar, P. Low-abundance members of the firmicutes facilitate bioremediation of soil impacted by highly acidic mine drainage from the Malanjkhand copper project, India. Front. Microbiol. 2018, 9, 2882. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukherjee, S.; Kuang, Z.; Ghosh, S.; Detroja, R.; Carmi, G.; Tripathy, S.; Barash, D.; Frenkel-Morgenstern, M.; Nevo, E.; Li, K. Incipient Sympatric Speciation and Evolution of Soil Bacteria Revealed by Metagenomic and Structured Non-Coding RNAs Analysis. Biology 2022, 11, 1110. https://doi.org/10.3390/biology11081110
Mukherjee S, Kuang Z, Ghosh S, Detroja R, Carmi G, Tripathy S, Barash D, Frenkel-Morgenstern M, Nevo E, Li K. Incipient Sympatric Speciation and Evolution of Soil Bacteria Revealed by Metagenomic and Structured Non-Coding RNAs Analysis. Biology. 2022; 11(8):1110. https://doi.org/10.3390/biology11081110
Chicago/Turabian StyleMukherjee, Sumit, Zhuoran Kuang, Samrat Ghosh, Rajesh Detroja, Gon Carmi, Sucheta Tripathy, Danny Barash, Milana Frenkel-Morgenstern, Eviatar Nevo, and Kexin Li. 2022. "Incipient Sympatric Speciation and Evolution of Soil Bacteria Revealed by Metagenomic and Structured Non-Coding RNAs Analysis" Biology 11, no. 8: 1110. https://doi.org/10.3390/biology11081110
APA StyleMukherjee, S., Kuang, Z., Ghosh, S., Detroja, R., Carmi, G., Tripathy, S., Barash, D., Frenkel-Morgenstern, M., Nevo, E., & Li, K. (2022). Incipient Sympatric Speciation and Evolution of Soil Bacteria Revealed by Metagenomic and Structured Non-Coding RNAs Analysis. Biology, 11(8), 1110. https://doi.org/10.3390/biology11081110









