Approaches in Gene Coexpression Analysis in Eukaryotes
Abstract
Simple Summary
Abstract
1. Introduction
- Protein–protein interaction (PPI) networks [6] describe the associations, either through physical contact or common pathway participation, between two or more proteins;
- Gene regulatory networks (GRNs) [7] depict the causal interactions between regulators and their target genes;
- Signal transduction networks [8] contain information on the interactions between biochemical signalling molecules and cell receptors;
- Metabolic and biochemical networks [9] display all metabolic reactions and molecules involved in biological pathways.
- Collection and integration of expression data
- Identification of modules using clustering techniques [32].
2. Collection and Processing of Transcriptomic Data and Construction of Gene Expression Matrices
- (A)
- ‘Condition independent’ approach uses a set of samples of a multitude of different conditions and source tissues. This method is suitable for studying the global coexpression landscape of an organism and demonstrates gene relationships regardless of experimental conditions [12].
- (B)
2.1. Microarray Data Analysis
- background correction
- normalisation
- probe summarisation
- log2 transformation (optional)
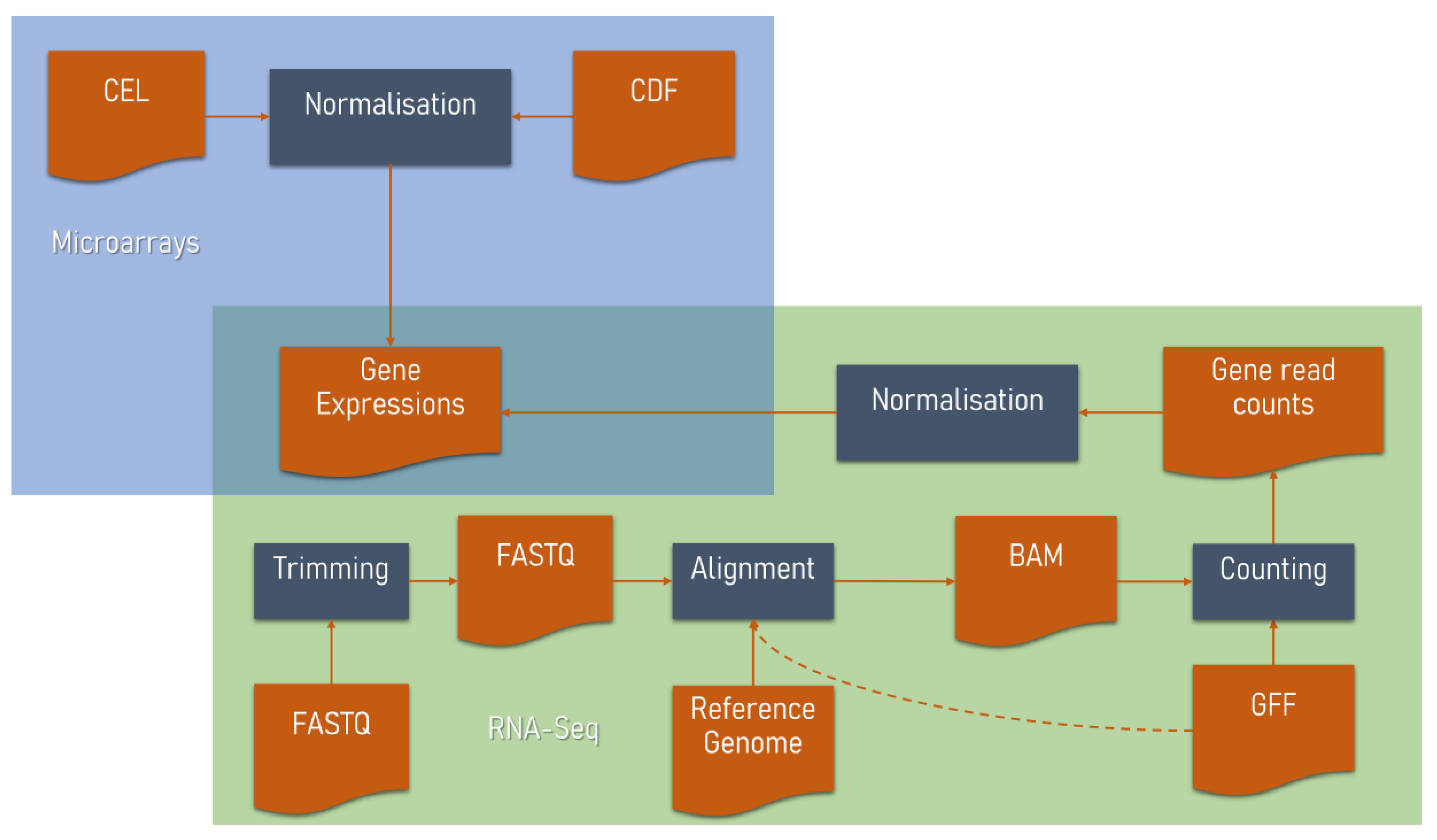
2.2. RNA-Seq Data Analysis
- quality control and trimming of sequence reads
- mapping reads to a reference genome or transcriptome
- producing gene read counts
- normalisation
2.3. Single-Cell RNA-Seq in Coexpression Analysis
2.4. Microarrays vs. RNA-Seq in Coexpression Analysis
2.5. Batch Correction
3. Selection of Coexpression Measure and Construction of Similarity Matrices
4. Selection of Significance Thresholds for Network Construction
5. Identification of Modules Using Clustering Techniques
6. Gene List Functional Enrichment Analysis
7. Coexpression Tools
7.1. Global Coexpression Web Tools
7.2. Condition-Specific Coexpression Web Tools
7.3. Stand-Alone Gene Coexpression Applications
8. Limitations and Perspectives in Coexpression Analysis
9. General Guidelines for Coexpression Tool Selection
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schneider, M.V.; Orchard, S. Omics Technologies, Data and Bioinformatics Principles. In Bioinformatics for Omics Data: Methods and Protocols; Mayer, B., Ed.; Humana Press: Totowa, NJ, USA, 2011; pp. 3–30. [Google Scholar]
- Barabasi, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Obayashi, T.; Mutwil, M.; Giorgi, F.M.; Bassel, G.W.; Tanimoto, M.; Chow, A.; Steinhauser, D.; Persson, S.; Provart, N.J. Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009, 32, 1633–1651. [Google Scholar] [CrossRef] [PubMed]
- Emamjomeh, A.; Saboori Robat, E.; Zahiri, J.; Solouki, M.; Khosravi, P. Gene co-expression network reconstruction: A review on computational methods for inferring functional information from plant-based expression data. Plant Biotechnol. Rep. 2017, 11, 71–86. [Google Scholar] [CrossRef]
- Pavlopoulos, G.A.; Secrier, M.; Moschopoulos, C.N.; Soldatos, T.G.; Kossida, S.; Aerts, J.; Schneider, R.; Bagos, P.G. Using graph theory to analyze biological networks. BioData Min. 2011, 4, 10. [Google Scholar] [CrossRef]
- Pellegrini, M.; Haynor, D.; Johnson, J.M. Protein interaction networks. Expert Rev. Proteom. 2004, 1, 239–249. [Google Scholar] [CrossRef]
- Emmert-Streib, F.; Dehmer, M.; Haibe-Kains, B. Gene regulatory networks and their applications: Understanding biological and medical problems in terms of networks. Front. Cell Dev. Biol. 2014, 2, 38. [Google Scholar] [CrossRef]
- Albert, R.; DasGupta, B.; Dondi, R.; Kachalo, S.; Sontag, E.; Zelikovsky, A.; Westbrooks, K. A novel method for signal transduction network inference from indirect experimental evidence. J. Comput. Biol. 2007, 14, 927–949. [Google Scholar] [CrossRef]
- Jeong, H.; Tombor, B.; Albert, R.; Oltvai, Z.N.; Barabasi, A.L. The large-scale organization of metabolic networks. Nature 2000, 407, 651–654. [Google Scholar] [CrossRef]
- Tieri, P.; Farina, L.; Petti, M.; Astolfi, L.; Paci, P.; Castiglione, F. Network Inference and Reconstruction in Bioinformatics. In Encyclopedia of Bioinformatics and Computational Biology; Ranganathan, S., Gribskov, M., Nakai, K., Schönbach, C., Eds.; Academic Press: Oxford, UK, 2019; pp. 805–813. [Google Scholar]
- Fionda, V. Networks in Biology. In Encyclopedia of Bioinformatics and Computational Biology; Ranganathan, S., Gribskov, M., Nakai, K., Schönbach, C., Eds.; Academic Press: Oxford, UK, 2019; pp. 915–921. [Google Scholar]
- Serin, E.A.R.; Nijveen, H.; Hilhorst, H.W.M.; Ligterink, W. Learning from Co-expression Networks: Possibilities and Challenges. Front. Plant Sci. 2016, 7, 444. [Google Scholar] [CrossRef]
- Michalopoulos, I.; Pavlopoulos, G.A.; Malatras, A.; Karelas, A.; Kostadima, M.A.; Schneider, R.; Kossida, S. Human gene correlation analysis (HGCA): A tool for the identification of transcriptionally co-expressed genes. BMC Res. Notes 2012, 5, 265. [Google Scholar] [CrossRef]
- Petereit, J.; Smith, S.; Harris, F.C., Jr.; Schlauch, K.A. Petal: Co-expression network modelling in R. BMC Syst. Biol. 2016, 10, 51. [Google Scholar] [CrossRef]
- He, F.; Maslov, S. Pan- and core- network analysis of co-expression genes in a model plant. Sci. Rep. 2016, 6, 38956. [Google Scholar] [CrossRef]
- Liseron-Monfils, C.; Ware, D. Revealing gene regulation and associations through biological networks. Curr. Plant Biol. 2015, 3–4, 30–39. [Google Scholar] [CrossRef]
- Obayashi, T.; Kagaya, Y.; Aoki, Y.; Tadaka, S.; Kinoshita, K. COXPRESdb v7: A gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res. 2019, 47, D55–D62. [Google Scholar] [CrossRef]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef]
- Jupiter, D.; Chen, H.; VanBuren, V. STARNET 2: A web-based tool for accelerating discovery of gene regulatory networks using microarray co-expression data. BMC Bioinform. 2009, 10, 332. [Google Scholar] [CrossRef]
- Yang, S.; Kim, C.Y.; Hwang, S.; Kim, E.; Kim, H.; Shim, H.; Lee, I. COEXPEDIA: Exploring biomedical hypotheses via co-expressions associated with medical subject headings (MeSH). Nucleic Acids Res. 2017, 45, D389–D396. [Google Scholar] [CrossRef]
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma’ayan, A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018, 9, 1366. [Google Scholar] [CrossRef]
- Obayashi, T.; Aoki, Y.; Tadaka, S.; Kagaya, Y.; Kinoshita, K. ATTED-II in 2018: A Plant Coexpression Database Based on Investigation of the Statistical Property of the Mutual Rank Index. Plant Cell Physiol. 2018, 59, e3. [Google Scholar] [CrossRef]
- Zogopoulos, V.L.; Saxami, G.; Malatras, A.; Angelopoulou, A.; Jen, C.H.; Duddy, W.J.; Daras, G.; Hatzopoulos, P.; Westhead, D.R.; Michalopoulos, I. Arabidopsis Coexpression Tool: A tool for gene coexpression analysis in Arabidopsis thaliana. iScience 2021, 24, 102848. [Google Scholar] [CrossRef]
- Leal, L.G.; Lopez, C.; Lopez-Kleine, L. Construction and comparison of gene co-expression networks shows complex plant immune responses. PeerJ 2014, 2, e610. [Google Scholar] [CrossRef] [PubMed]
- Narise, T.; Sakurai, N.; Obayashi, T.; Ohta, H.; Shibata, D. Co-expressed Pathways DataBase for Tomato: A database to predict pathways relevant to a query gene. BMC Genom. 2017, 18, 437. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; Oono, Y.; Wakimoto, H.; Ogata, J.; Kanamori, H.; Sasaki, H.; Mori, S.; Matsumoto, T.; Itoh, T. TENOR: Database for Comprehensive mRNA-Seq Experiments in Rice. Plant Cell Physiol. 2016, 57, e7. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Zou, D.; Sang, J.; Xu, X.; Yin, H.; Li, M.; Wu, S.; Hu, S.; Hao, L.; Zhang, Z. Rice Expression Database (RED): An integrated RNA-Seq-derived gene expression database for rice. J. Genet. Genom. 2017, 44, 235–241. [Google Scholar] [CrossRef]
- Yim, W.C.; Yu, Y.; Song, K.; Jang, C.S.; Lee, B.M. PLANEX: The plant co-expression database. BMC Plant Biol. 2013, 13, 83. [Google Scholar] [CrossRef]
- Proost, S.; Mutwil, M. PlaNet: Comparative Co-Expression Network Analyses for Plants. Methods Mol. Biol. 2017, 1533, 213–227. [Google Scholar] [CrossRef]
- Van Dam, S.; Craig, T.; de Magalhaes, J.P. GeneFriends: A human RNA-seq-based gene and transcript co-expression database. Nucleic Acids Res. 2015, 43, D1124–D1132. [Google Scholar] [CrossRef]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef]
- van Dam, S.; Vosa, U.; van der Graaf, A.; Franke, L.; de Magalhaes, J.P. Gene co-expression analysis for functional classification and gene-disease predictions. Brief. Bioinform. 2018, 19, 575–592. [Google Scholar] [CrossRef]
- Peng, J.; Wang, T.; Huc, J.; Wang, Y.; Chen, J. Constructing Networks of Organelle Functional Modules in Arabidopsis. Curr. Genom. 2016, 17, 427–438. [Google Scholar] [CrossRef]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Parkinson, H.; Kapushesky, M.; Shojatalab, M.; Abeygunawardena, N.; Coulson, R.; Farne, A.; Holloway, E.; Kolesnykov, N.; Lilja, P.; Lukk, M.; et al. ArrayExpress–A public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007, 35, D747–D750. [Google Scholar] [CrossRef]
- Papatheodorou, I.; Moreno, P.; Manning, J.; Fuentes, A.M.; George, N.; Fexova, S.; Fonseca, N.A.; Fullgrabe, A.; Green, M.; Huang, N.; et al. Expression Atlas update: From tissues to single cells. Nucleic Acids Res. 2020, 48, D77–D83. [Google Scholar] [CrossRef]
- Kodama, Y.; Shumway, M.; Leinonen, R.; International Nucleotide Sequence Database, C. The Sequence Read Archive: Explosive growth of sequencing data. Nucleic Acids Res. 2012, 40, D54–D56. [Google Scholar] [CrossRef]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Hutter, C.; Zenklusen, J.C. The Cancer Genome Atlas: Creating Lasting Value beyond Its Data. Cell 2018, 173, 283–285. [Google Scholar] [CrossRef]
- Amid, C.; Alako, B.T.F.; Balavenkataraman Kadhirvelu, V.; Burdett, T.; Burgin, J.; Fan, J.; Harrison, P.W.; Holt, S.; Hussein, A.; Ivanov, E.; et al. The European Nucleotide Archive in 2019. Nucleic Acids Res. 2020, 48, D70–D76. [Google Scholar] [CrossRef]
- Aoki, K.; Ogata, Y.; Shibata, D. Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 2007, 48, 381–390. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA Package FAQ. Available online: https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/faq.html (accessed on 5 June 2022).
- Lockhart, D.J.; Dong, H.; Byrne, M.C.; Follettie, M.T.; Gallo, M.V.; Chee, M.S.; Mittmann, M.; Wang, C.; Kobayashi, M.; Horton, H.; et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 1996, 14, 1675–1680. [Google Scholar] [CrossRef]
- Wolber, P.K.; Collins, P.J.; Lucas, A.B.; De Witte, A.; Shannon, K.W. The Agilent in situ-synthesized microarray platform. Methods Enzymol. 2006, 410, 28–57. [Google Scholar] [CrossRef]
- Kuhn, K.; Baker, S.C.; Chudin, E.; Lieu, M.H.; Oeser, S.; Bennett, H.; Rigault, P.; Barker, D.; McDaniel, T.K.; Chee, M.S. A novel, high-performance random array platform for quantitative gene expression profiling. Genome Res. 2004, 14, 2347–2356. [Google Scholar] [CrossRef]
- Hubbell, E.; Liu, W.M.; Mei, R. Robust estimators for expression analysis. Bioinformatics 2002, 18, 1585–1592. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Bolstad, B.M.; Collin, F.; Cope, L.M.; Hobbs, B.; Speed, T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003, 31, e15. [Google Scholar] [CrossRef]
- Wu, Z.; Irizarry, R.A.; Gentleman, R.; Martinez-Murillo, F.; Spencer, F. A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. J. Am. Stat. Assoc. 2004, 99, 909–917. [Google Scholar] [CrossRef]
- Hubbell, E. Affymetrix Technical Notes: Guide to Probe Logarithmic Intensity Error (PLIER) Estimation. Available online: http://tools.thermofisher.com/content/sfs/brochures/plier_technote.pdf (accessed on 5 June 2022).
- Piccolo, S.R.; Sun, Y.; Campbell, J.D.; Lenburg, M.E.; Bild, A.H.; Johnson, W.E. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics 2012, 100, 337–344. [Google Scholar] [CrossRef]
- Zogopoulos, V.L.; Malatras, A.; Michalopoulos, I. Gene coexpression analysis in Arabidopsis thaliana based on public microarray data. STAR Protoc. 2022, 3, 101208. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://cran.r-project.org/doc/manuals/r-release/fullrefman.pdf (accessed on 5 June 2022).
- Eijssen, L.M.; Jaillard, M.; Adriaens, M.E.; Gaj, S.; de Groot, P.J.; Muller, M.; Evelo, C.T. User-friendly solutions for microarray quality control and pre-processing on ArrayAnalysis.org. Nucleic Acids Res. 2013, 41, W71–W76. [Google Scholar] [CrossRef]
- Applied Biosystems. Applied Biosystems 3730 and 3730xl DNA Analyzers. Available online: http://tools.thermofisher.com/content/sfs/brochures/cms_042636.pdf (accessed on 5 June 2022).
- Jain, M.; Olsen, H.E.; Paten, B.; Akeson, M. The Oxford Nanopore MinION: Delivery of nanopore sequencing to the genomics community. Genome Biol. 2016, 17, 239. [Google Scholar] [CrossRef]
- Bentley, D.R.; Balasubramanian, S.; Swerdlow, H.P.; Smith, G.P.; Milton, J.; Brown, C.G.; Hall, K.P.; Evers, D.J.; Barnes, C.L.; Bignell, H.R.; et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008, 456, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.J.; Chen, Z.; et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005, 437, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Schadt, E.E.; Turner, S.; Kasarskis, A. A window into third-generation sequencing. Hum. Mol. Genet. 2010, 19, R227–R240. [Google Scholar] [CrossRef] [PubMed]
- Branton, D.; Deamer, D.W.; Marziali, A.; Bayley, H.; Benner, S.A.; Butler, T.; Di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.; et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [Google Scholar] [CrossRef]
- Cock, P.J.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.T.; Huang, X.; Huang, S.; Xie, S.J.; Xiao, Z.D.; Zhang, H. RNA sequencing: New technologies and applications in cancer research. J. Hematol. Oncol. 2020, 13, 166. [Google Scholar] [CrossRef]
- Macmanes, M.D. On the optimal trimming of high-throughput mRNA sequence data. Front. Genet. 2014, 5, 13. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 June 2022).
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Ermini, L.; Wang, H.; Carty, K.; Cheung, M.S. LongQC: A Quality Control Tool for Third Generation Sequencing Long Read Data. G3 Genes Genomes Genet. 2020, 10, 1193–1196. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 3. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; Busby, B.; Madden, T.L. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinform. 2019, 20, 405. [Google Scholar] [CrossRef]
- Marić, J.; Sović, I.; Križanović, K.; Nagarajan, N.; Šikić, M. Graphmap2—Splice-aware RNA-seq mapper for long reads. bioRxiv 2019. [Google Scholar] [CrossRef]
- Lin, H.N.; Hsu, W.L. DART: A fast and accurate RNA-seq mapper with a partitioning strategy. Bioinformatics 2018, 34, 190–197. [Google Scholar] [CrossRef]
- Liu, B.; Liu, Y.; Li, J.; Guo, H.; Zang, T.; Wang, Y. deSALT: Fast and accurate long transcriptomic read alignment with de Bruijn graph-based index. Genome Biol. 2019, 20, 274. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.D.; Reeder, J.; Lawrence, M.; Becker, G.; Brauer, M.J. GMAP and GSNAP for Genomic Sequence Alignment: Enhancements to Speed, Accuracy, and Functionality. Methods Mol. Biol. 2016, 1418, 283–334. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Stein, L. Generic Feature Format Version 3 (GFF3). Available online: https://github.com/The-Sequence-Ontology/Specifications/blob/master/gff3.md (accessed on 5 June 2022).
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Dillies, M.A.; Rau, A.; Aubert, J.; Hennequet-Antier, C.; Jeanmougin, M.; Servant, N.; Keime, C.; Marot, G.; Castel, D.; Estelle, J.; et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 2013, 14, 671–683. [Google Scholar] [CrossRef]
- Bolstad, B.M.; Irizarry, R.A.; Astrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef]
- Bullard, J.H.; Purdom, E.; Hansen, K.D.; Dudoit, S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinform. 2010, 11, 94. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Hicks, S.C.; Okrah, K.; Paulson, J.N.; Quackenbush, J.; Irizarry, R.A.; Bravo, H.C. Smooth quantile normalization. Biostatistics 2018, 19, 185–198. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Vandenbon, A. Evaluation of critical data processing steps for reliable prediction of gene co-expression from large collections of RNA-seq data. PLoS ONE 2022, 17, e0263344. [Google Scholar] [CrossRef]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef]
- Hwang, B.; Lee, J.H.; Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef]
- Chen, G.; Ning, B.; Shi, T. Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Front. Genet. 2019, 10, 317. [Google Scholar] [CrossRef] [PubMed]
- Li, W.V.; Li, J.J. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat. Commun. 2018, 9, 997. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Wang, J.; Torre, E.; Dueck, H.; Shaffer, S.; Bonasio, R.; Murray, J.I.; Raj, A.; Li, M.; Zhang, N.R. SAVER: Gene expression recovery for single-cell RNA sequencing. Nat. Methods 2018, 15, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, D.; Sharma, R.; Nainys, J.; Yim, K.; Kathail, P.; Carr, A.J.; Burdziak, C.; Moon, K.R.; Chaffer, C.L.; Pattabiraman, D.; et al. Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 2018, 174, 716–729.e727. [Google Scholar] [CrossRef]
- Dai, M.; Wang, P.; Boyd, A.D.; Kostov, G.; Athey, B.; Jones, E.G.; Bunney, W.E.; Myers, R.M.; Speed, T.P.; Akil, H.; et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005, 33, e175. [Google Scholar] [CrossRef]
- Chen, L.; Sun, F.; Yang, X.; Jin, Y.; Shi, M.; Wang, L.; Shi, Y.; Zhan, C.; Wang, Q. Correlation between RNA-Seq and microarrays results using TCGA data. Gene 2017, 628, 200–204. [Google Scholar] [CrossRef]
- Malatras, A.; Michalopoulos, I.; Duguez, S.; Butler-Browne, G.; Spuler, S.; Duddy, W.J. MyoMiner: Explore gene co-expression in normal and pathological muscle. BMC Med. Genom. 2020, 13, 67. [Google Scholar] [CrossRef]
- Leek, J.T.; Scharpf, R.B.; Bravo, H.C.; Simcha, D.; Langmead, B.; Johnson, W.E.; Geman, D.; Baggerly, K.; Irizarry, R.A. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010, 11, 733–739. [Google Scholar] [CrossRef]
- Pearson, K. LIII. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Sokal, R.R.; Michener, C.D. A statistical method for evaluating systematic relationships. Univ. Kansas Sci. Bull. 1958, 38, 1409–1438. [Google Scholar]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Leek, J.T.; Storey, J.D. A general framework for multiple testing dependence. Proc. Natl. Acad. Sci. USA 2008, 105, 18718–18723. [Google Scholar] [CrossRef]
- Buettner, F.; Pratanwanich, N.; McCarthy, D.J.; Marioni, J.C.; Stegle, O. f-scLVM: Scalable and versatile factor analysis for single-cell RNA-seq. Genome Biol. 2017, 18, 212. [Google Scholar] [CrossRef]
- Haghverdi, L.; Lun, A.T.L.; Morgan, M.D.; Marioni, J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018, 36, 421–427. [Google Scholar] [CrossRef]
- Buttner, M.; Miao, Z.; Wolf, F.A.; Teichmann, S.A.; Theis, F.J. A test metric for assessing single-cell RNA-seq batch correction. Nat. Methods 2019, 16, 43–49. [Google Scholar] [CrossRef]
- Minkowski, H. Geometrie Der Zahlen; Teubner: Leipzig, Germany, 1910. [Google Scholar]
- Pearson, K. VII. Note on regression and inheritance in the case of two parents. Proc. R. Soc. Lond. 1895, 58, 240–242. [Google Scholar] [CrossRef]
- Amaratunga, D.; Cabrera, J. Analysis of Data From Viral DNA Microchips. J. Am. Stat. Assoc. 2001, 96, 1161–1170. [Google Scholar] [CrossRef]
- Jaskowiak, P.A.; Campello, R.J.; Costa, I.G. On the selection of appropriate distances for gene expression data clustering. BMC Bioinform. 2014, 15, S2. [Google Scholar] [CrossRef]
- Spearman, C. ‘General intelligence’, objectively determined and measured. Am. J. Psychol. 1904, 15, 201–292. [Google Scholar] [CrossRef]
- Myers, J.L.; Well, A.D. Research Design and Statistical Analysis, 2nd ed.; Lawrence Erlbaum Associates: Mahwah, NJ, USA, 2003. [Google Scholar]
- Kendall, M.G. A new measure of rank correlation. Biometrika 1938, 30, 81–93. [Google Scholar] [CrossRef]
- Kumari, S.; Nie, J.; Chen, H.S.; Ma, H.; Stewart, R.; Li, X.; Lu, M.Z.; Taylor, W.M.; Wei, H. Evaluation of gene association methods for coexpression network construction and biological knowledge discovery. PLoS ONE 2012, 7, e50411. [Google Scholar] [CrossRef]
- Obayashi, T.; Hayashi, S.; Saeki, M.; Ohta, H.; Kinoshita, K. ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 2009, 37, D987–D991. [Google Scholar] [CrossRef]
- Obayashi, T.; Hibara, H.; Kagaya, Y.; Aoki, Y.; Kinoshita, K. ATTED-II v11: A Plant Gene Coexpression Database Using a Sample Balancing Technique by Subagging of Principal Components. Plant Cell Physiol. 2022, 63, 869–881. [Google Scholar] [CrossRef]
- Bansal, M.; Belcastro, V.; Ambesi-Impiombato, A.; di Bernardo, D. How to infer gene networks from expression profiles. Mol. Syst. Biol. 2007, 3, 78. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423+623–656. [Google Scholar] [CrossRef]
- Steuer, R.; Kurths, J.; Daub, C.O.; Weise, J.; Selbig, J. The mutual information: Detecting and evaluating dependencies between variables. Bioinformatics 2002, 18, S231–S240. [Google Scholar] [CrossRef]
- Mousavi, A.; Baraniuk, R.G. An information-theoretic measure of dependency among variables in large datasets. In Proceedings of the 2015 53rd Annual Allerton Conference on Communication Control, and Computing (Allerton), Monticello, IL, USA, 29 September–2 October 2015; pp. 650–657. [Google Scholar]
- Obayashi, T.; Kinoshita, K. Coexpression landscape in ATTED-II: Usage of gene list and gene network for various types of pathways. J. Plant Res. 2010, 123, 311–319. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Borate, B.R.; Chesler, E.J.; Langston, M.A.; Saxton, A.M.; Voy, B.H. Comparison of threshold selection methods for microarray gene co-expression matrices. BMC Res. Notes 2009, 2, 240. [Google Scholar] [CrossRef]
- Ala, U.; Piro, R.M.; Grassi, E.; Damasco, C.; Silengo, L.; Oti, M.; Provero, P.; Di Cunto, F. Prediction of human disease genes by human-mouse conserved coexpression analysis. PLoS Comput. Biol. 2008, 4, e1000043. [Google Scholar] [CrossRef]
- De la Fuente, A. From ‘differential expression’ to ‘differential networking’—Identification of dysfunctional regulatory networks in diseases. Trends Genet. 2010, 26, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Christensen, C.; Thakar, J.; Albert, R. Systems-level insights into cellular regulation: Inferring, analysing, and modelling intracellular networks. IET Syst. Biol. 2007, 1, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Franz, M.; Lopes, C.T.; Huck, G.; Dong, Y.; Sumer, O.; Bader, G.D. Cytoscape.js: A graph theory library for visualisation and analysis. Bioinformatics 2016, 32, 309–311. [Google Scholar] [CrossRef]
- D’Haeseleer, P. How does gene expression clustering work? Nat. Biotechnol. 2005, 23, 1499. [Google Scholar] [CrossRef]
- Perkins, A.D.; Langston, M.A. Threshold selection in gene co-expression networks using spectral graph theory techniques. BMC Bioinform. 2009, 10, S4. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Jen, C.H.; Manfield, I.W.; Michalopoulos, I.; Pinney, J.W.; Willats, W.G.; Gilmartin, P.M.; Westhead, D.R. The Arabidopsis co-expression tool (ACT): A WWW-based tool and database for microarray-based gene expression analysis. Plant J. 2006, 46, 336–348. [Google Scholar] [CrossRef]
- Manfield, I.W.; Jen, C.H.; Pinney, J.W.; Michalopoulos, I.; Bradford, J.R.; Gilmartin, P.M.; Westhead, D.R. Arabidopsis Co-expression Tool (ACT): Web server tools for microarray-based gene expression analysis. Nucleic Acids Res. 2006, 34, W504–W509. [Google Scholar] [CrossRef]
- Chen, P.; Wang, F.; Feng, J.; Zhou, R.; Chang, Y.; Liu, J.; Zhao, Q. Co-expression network analysis identified six hub genes in association with metastasis risk and prognosis in hepatocellular carcinoma. Oncotarget 2017, 8, 48948–48958. [Google Scholar] [CrossRef]
- Yuan, L.; Chen, L.; Qian, K.; Qian, G.; Wu, C.L.; Wang, X.; Xiao, Y. Co-expression network analysis identified six hub genes in association with progression and prognosis in human clear cell renal cell carcinoma (ccRCC). Genom. Data 2017, 14, 132–140. [Google Scholar] [CrossRef]
- Olsen, G. The ”Newick’s 8:45” Tree Format Standard. Available online: https://evolution.genetics.washington.edu/phylip/newick_doc.html (accessed on 5 June 2022).
- Hartigan, J.A. Direct Clustering of a Data Matrix. J. Am. Stat. Assoc. 1972, 67, 123–129. [Google Scholar] [CrossRef]
- Padilha, V.A.; Campello, R.J.G.B. A systematic comparative evaluation of biclustering techniques. BMC Bioinform. 2017, 18, 55. [Google Scholar] [CrossRef]
- Eren, K.; Deveci, M.; Kucuktunc, O.; Catalyurek, U.V. A comparative analysis of biclustering algorithms for gene expression data. Brief. Bioinform. 2012, 14, 279–292. [Google Scholar] [CrossRef]
- Hartigan, J. Clustering Algorithms; John Wiley & Sons: New York, NY, USA, 1975. [Google Scholar]
- Heyer, L.J.; Kruglyak, S.; Yooseph, S. Exploring expression data: Identification and analysis of coexpressed genes. Genome Res. 1999, 9, 1106–1115. [Google Scholar] [CrossRef]
- Tamayo, P.; Slonim, D.; Mesirov, J.; Zhu, Q.; Kitareewan, S.; Dmitrovsky, E.; Lander, E.S.; Golub, T.R. Interpreting patterns of gene expression with self-organizing maps: Methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA 1999, 96, 2907–2912. [Google Scholar] [CrossRef]
- Farris, J.S. On the Cophenetic Correlation Coefficient. Syst. Biol. 1969, 18, 279–285. [Google Scholar] [CrossRef]
- Saraçli, S.; Doğan, N.; Doğan, İ. Comparison of hierarchical cluster analysis methods by cophenetic correlation. J. Inequal. Appl. 2013, 2013, 203. [Google Scholar] [CrossRef]
- Fisher, R.A. On the Interpretation of χ2 from Contingency Tables, and the Calculation of P. J. R. Stat. Soc. 1922, 85, 87–94. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Royal Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Pinero, J.; Ramirez-Anguita, J.M.; Sauch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Berhanu Lemma, R.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Manosalva Perez, N.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef]
- Encode Project Consortium. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Thanati, F.; Karatzas, E.; Baltoumas, F.A.; Stravopodis, D.J.; Eliopoulos, A.G.; Pavlopoulos, G.A. FLAME: A Web Tool for Functional and Literature Enrichment Analysis of Multiple Gene Lists. Biology 2021, 10, 665. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pomaznoy, M.; Ha, B.; Peters, B. GOnet: A tool for interactive Gene Ontology analysis. BMC Bioinform. 2018, 19, 470. [Google Scholar] [CrossRef] [PubMed]
- Okamura, Y.; Aoki, Y.; Obayashi, T.; Tadaka, S.; Ito, S.; Narise, T.; Kinoshita, K. COXPRESdb in 2015: Coexpression database for animal species by DNA-microarray and RNAseq-based expression data with multiple quality assessment systems. Nucleic Acids Res. 2015, 43, D82–D86. [Google Scholar] [CrossRef]
- Okamura, Y.; Kinoshita, K. Matataki: An ultrafast mRNA quantification method for large-scale reanalysis of RNA-Seq data. BMC Bioinform. 2018, 19, 266. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tseng, K.C.; Li, G.Z.; Hung, Y.C.; Chow, C.N.; Wu, N.Y.; Chien, Y.Y.; Zheng, H.Q.; Lee, T.Y.; Kuo, P.L.; Chang, S.B.; et al. EXPath 2.0: An Updated Database for Integrating High-Throughput Gene Expression Data with Biological Pathways. Plant Cell Physiol. 2020, 61, 1818–1827. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ogata, Y.; Suzuki, H.; Sakurai, N.; Shibata, D. CoP: A database for characterizing co-expressed gene modules with biological information in plants. Bioinformatics 2010, 26, 1267–1268. [Google Scholar] [CrossRef][Green Version]
- Ogata, Y.; Sakurai, N.; Suzuki, H.; Aoki, K.; Saito, K.; Shibata, D. The prediction of local modular structures in a co-expression network based on gene expression datasets. Genome Inform. 2009, 23, 117–127. [Google Scholar]
- De Bodt, S.; Hollunder, J.; Nelissen, H.; Meulemeester, N.; Inze, D. CORNET 2.0: Integrating plant coexpression, protein-protein interactions, regulatory interactions, gene associations and functional annotations. New Phytol. 2012, 195, 707–720. [Google Scholar] [CrossRef]
- Zhang, W.; Morris, Q.D.; Chang, R.; Shai, O.; Bakowski, M.A.; Mitsakakis, N.; Mohammad, N.; Robinson, M.D.; Zirngibl, R.; Somogyi, E.; et al. The functional landscape of mouse gene expression. J. Biol. 2004, 3, 21. [Google Scholar] [CrossRef]
- Zhu, Q.; Wong, A.K.; Krishnan, A.; Aure, M.R.; Tadych, A.; Zhang, R.; Corney, D.C.; Greene, C.S.; Bongo, L.A.; Kristensen, V.N.; et al. Targeted exploration and analysis of large cross-platform human transcriptomic compendia. Nat. Methods 2015, 12, 211–214. [Google Scholar] [CrossRef]
- Adler, P.; Kolde, R.; Kull, M.; Tkachenko, A.; Peterson, H.; Reimand, J.; Vilo, J. Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol. 2009, 10, R139. [Google Scholar] [CrossRef]
- Zoubarev, A.; Hamer, K.M.; Keshav, K.D.; McCarthy, E.L.; Santos, J.R.; Van Rossum, T.; McDonald, C.; Hall, A.; Wan, X.; Lim, R.; et al. Gemma: A resource for the reuse, sharing and meta-analysis of expression profiling data. Bioinformatics 2012, 28, 2272–2273. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Zuberi, K.; Franz, M.; Rodriguez, H.; Montojo, J.; Lopes, C.T.; Bader, G.D.; Morris, Q. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013, 41, W115–W122. [Google Scholar] [CrossRef]
- Raina, P.; Lopes, I.; Chatsirisupachai, K.; Farooq, Z.; de Magalhães, J.P. GeneFriends 2021: Updated co-expression databases and tools for human and mouse genes and transcripts. bioRxiv 2021. [Google Scholar] [CrossRef]
- Miller, H.E.; Bishop, A.J.R. Correlation AnalyzeR: Functional predictions from gene co-expression correlations. BMC Bioinform. 2021, 22, 206. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Wang, P.; Qi, H.; Song, S.; Li, S.; Huang, N.; Han, W.; Ma, D. ImmuCo: A database of gene co-expression in immune cells. Nucleic Acids Res. 2015, 43, D1133–D1139. [Google Scholar] [CrossRef]
- Vandenbon, A.; Dinh, V.H.; Mikami, N.; Kitagawa, Y.; Teraguchi, S.; Ohkura, N.; Sakaguchi, S. Immuno-Navigator, a batch-corrected coexpression database, reveals cell type-specific gene networks in the immune system. Proc. Natl. Acad. Sci. USA 2016, 113, E2393–E2402. [Google Scholar] [CrossRef]
- Farber, C.R.; Mesner, L.D. A Systems-Level Understanding of Cardiovascular Disease through Networks. In Translational Cardiometabolic Genomic Medicine; Rodriguez-Oquendo, A., Ed.; Academic Press: Boston, MA, USA, 2016; pp. 59–81. [Google Scholar]
- Langfelder, P.; Zhang, B.; Horvath, S. Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinformatics 2008, 24, 719–720. [Google Scholar] [CrossRef]
- Xu, X.; Lu, X.; Tang, Z.; Zhang, X.; Lei, F.; Hou, L.; Li, M. Combined analysis of carotenoid metabolites and the transcriptome to reveal the molecular mechanism underlying fruit colouration in zucchini (Cucurbita pepo L.). Food Chem. Mol. Sci. 2021, 2, 100021. [Google Scholar] [CrossRef]
- Xie, J.; Ma, A.; Zhang, Y.; Liu, B.; Cao, S.; Wang, C.; Xu, J.; Zhang, C.; Ma, Q. QUBIC2: A novel and robust biclustering algorithm for analyses and interpretation of large-scale RNA-Seq data. Bioinformatics 2019, 36, 1143–1149. [Google Scholar] [CrossRef]
- Hochreiter, S.; Bodenhofer, U.; Heusel, M.; Mayr, A.; Mitterecker, A.; Kasim, A.; Khamiakova, T.; Van Sanden, S.; Lin, D.; Talloen, W.; et al. FABIA: Factor analysis for bicluster acquisition. Bioinformatics 2010, 26, 1520–1527. [Google Scholar] [CrossRef]
- Bergmann, S.; Ihmels, J.; Barkai, N. Iterative signature algorithm for the analysis of large-scale gene expression data. Phys. Rev. E 2003, 67, 031902. [Google Scholar] [CrossRef]
- Pontes, B.; Giráldez, R.; Aguilar-Ruiz, J.S. Biclustering on expression data: A review. J. Biomed. Inform. 2015, 57, 163–180. [Google Scholar] [CrossRef]
- Barrett, T.; Suzek, T.O.; Troup, D.B.; Wilhite, S.E.; Ngau, W.C.; Ledoux, P.; Rudnev, D.; Lash, A.E.; Fujibuchi, W.; Edgar, R. NCBI GEO: Mining millions of expression profiles—Database and tools. Nucleic Acids Res. 2005, 33, D562–D566. [Google Scholar] [CrossRef]
- Russo, P.S.T.; Ferreira, G.R.; Cardozo, L.E.; Burger, M.C.; Arias-Carrasco, R.; Maruyama, S.R.; Hirata, T.D.C.; Lima, D.S.; Passos, F.M.; Fukutani, K.F.; et al. CEMiTool: A Bioconductor package for performing comprehensive modular co-expression analyses. BMC Bioinform. 2018, 19, 56. [Google Scholar] [CrossRef] [PubMed]
- Cardozo, L.E.; Russo, P.S.T.; Gomes-Correia, B.; Araujo-Pereira, M.; Sepulveda-Hermosilla, G.; Maracaja-Coutinho, V.; Nakaya, H.I. webCEMiTool: Co-expression Modular Analysis Made Easy. Front. Genet. 2019, 10, 146. [Google Scholar] [CrossRef] [PubMed]
- Vivian Li, W.; Li, Y. scLink: Inferring Sparse Gene Co-expression Networks from Single-cell Expression Data. Genom. Proteom. Bioinform. 2021, 19, 475–492. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.H.; Davis, F.G.; et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef]
- Haque, A.; Engel, J.; Teichmann, S.A.; Lonnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017, 9, 75. [Google Scholar] [CrossRef]
- Moll, P.; Ante, M.; Seitz, A.; Reda, T. QuantSeq 3′ mRNA sequencing for RNA quantification. Nat. Methods 2014, 11, i–iii. [Google Scholar] [CrossRef]
- Corley, S.M.; Troy, N.M.; Bosco, A.; Wilkins, M.R. QuantSeq. 3′ Sequencing combined with Salmon provides a fast, reliable approach for high throughput RNA expression analysis. Sci. Rep. 2019, 9, 18895. [Google Scholar] [CrossRef]
- Logotheti, S.; Pavlopoulou, A.; Galtsidis, S.; Vojtesek, B.; Zoumpourlis, V. Functions, divergence and clinical value of TAp73 isoforms in cancer. Cancer Metastasis Rev. 2013, 32, 511–534. [Google Scholar] [CrossRef]
- Policastro, R.A.; Zentner, G.E. Global approaches for profiling transcription initiation. Cell Rep. Methods 2021, 1, 100081. [Google Scholar] [CrossRef]
- Policastro, R.A.; Zentner, G.E. Genome-Wide Profiling of Transcription Initiation with STRIPE-seq. Methods Mol. Biol. 2022, 2477, 21–34. [Google Scholar] [CrossRef]
- Cole, C.; Byrne, A.; Beaudin, A.E.; Forsberg, E.C.; Vollmers, C. Tn5Prime, a Tn5 based 5′ capture method for single cell RNA-seq. Nucleic Acids Res. 2018, 46, e62. [Google Scholar] [CrossRef]
- Picelli, S.; Faridani, O.R.; Bjorklund, A.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171–181. [Google Scholar] [CrossRef]
- Ostlund, G.; Sonnhammer, E.L. Avoiding pitfalls in gene (co)expression meta-analysis. Genomics 2014, 103, 21–30. [Google Scholar] [CrossRef]
- Michiels, S.; Koscielny, S.; Hill, C. Prediction of cancer outcome with microarrays: A multiple random validation strategy. Lancet 2005, 365, 488–492. [Google Scholar] [CrossRef]
- Huang, J.; Vendramin, S.; Shi, L.; McGinnis, K.M. Construction and Optimization of a Large Gene Coexpression Network in Maize Using RNA-Seq Data. Plant. Physiol. 2017, 175, 568–583. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zogopoulos, V.L.; Saxami, G.; Malatras, A.; Papadopoulos, K.; Tsotra, I.; Iconomidou, V.A.; Michalopoulos, I. Approaches in Gene Coexpression Analysis in Eukaryotes. Biology 2022, 11, 1019. https://doi.org/10.3390/biology11071019
Zogopoulos VL, Saxami G, Malatras A, Papadopoulos K, Tsotra I, Iconomidou VA, Michalopoulos I. Approaches in Gene Coexpression Analysis in Eukaryotes. Biology. 2022; 11(7):1019. https://doi.org/10.3390/biology11071019
Chicago/Turabian StyleZogopoulos, Vasileios L., Georgia Saxami, Apostolos Malatras, Konstantinos Papadopoulos, Ioanna Tsotra, Vassiliki A. Iconomidou, and Ioannis Michalopoulos. 2022. "Approaches in Gene Coexpression Analysis in Eukaryotes" Biology 11, no. 7: 1019. https://doi.org/10.3390/biology11071019
APA StyleZogopoulos, V. L., Saxami, G., Malatras, A., Papadopoulos, K., Tsotra, I., Iconomidou, V. A., & Michalopoulos, I. (2022). Approaches in Gene Coexpression Analysis in Eukaryotes. Biology, 11(7), 1019. https://doi.org/10.3390/biology11071019








