Histochemical Evidence for Nitrogen-Transfer Endosymbiosis in Non-Photosynthetic Cells of Leaves and Inflorescence Bracts of Angiosperms
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Histochemical Staining of Intracellular Bacteria
2.2.1. Reducing Sugar Staining
2.2.2. Ethylene Staining
2.2.3. Superoxide Staining
2.2.4. Hydrogen Peroxide Staining
2.2.5. Nitric Oxide Staining
2.2.6. Nitrate Staining
2.3. Differential Nitrogen Assimilation Experiments
2.4. Bacterial Isolation, Identification, and Characterization
2.5. Experiments to Assess Effects on Trichomes by Reduction in Seedling Bacteria
2.5.1. Rhus glabra Seed Sterilization Experiment
2.5.2. Perilla frutescens Seed Sterilization Experiment
2.5.3. Bacterial Replication Repression in Ailanthus altissima Seedlings Using Elevated Carbon Dioxide
2.6. Fluorescent Protein mCherry Transformation of Klebsiella oxytoca
2.7. Inoculation Experiments
2.8. Confocal Microscopy
2.9. Statistical Analyses
3. Results
3.1. Histochemical Staining of Intracellular Bacteria
3.2. Summary of Types of Endosymbiosis in Cells of Leaves and Bracts
- (1)
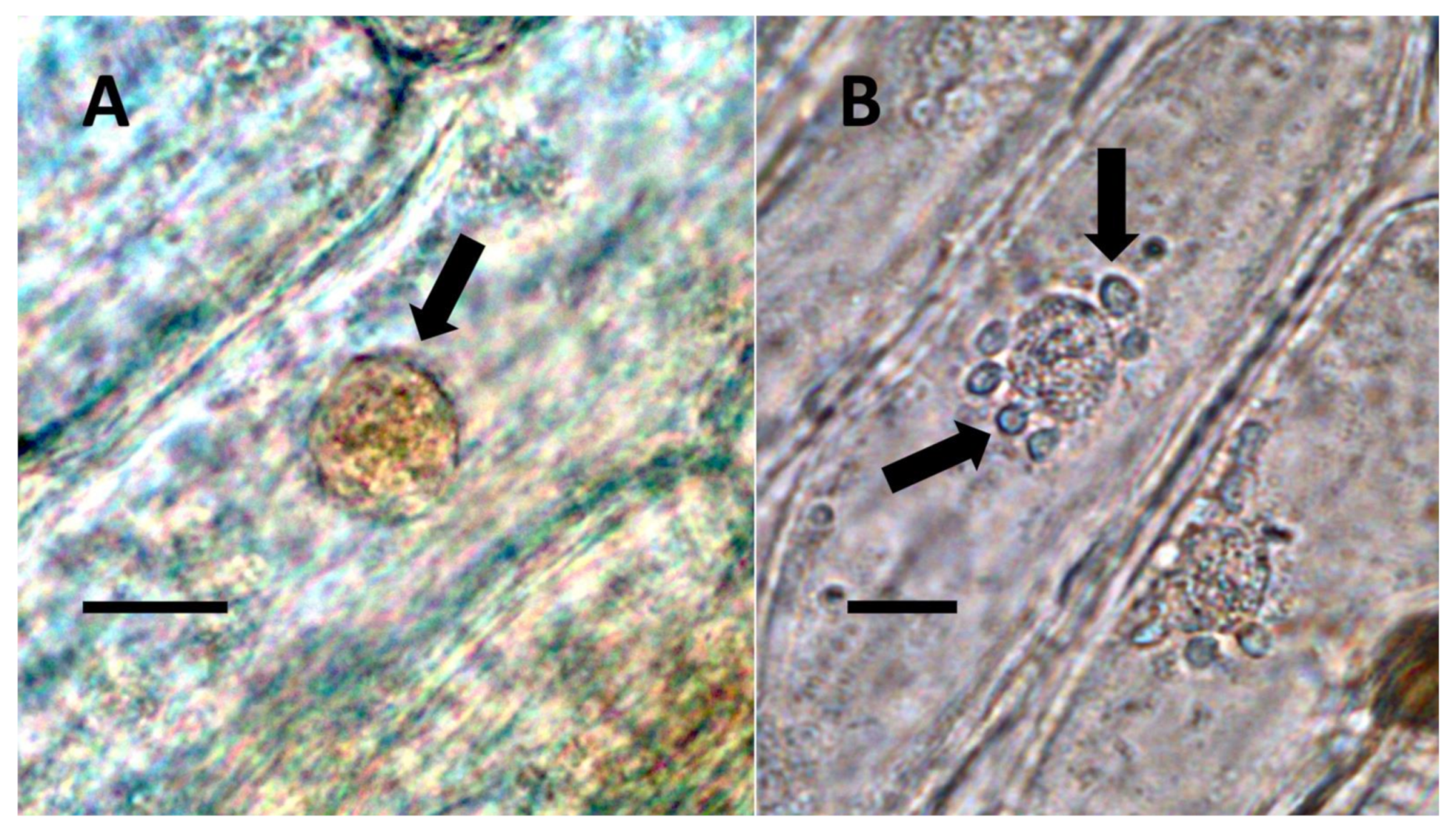
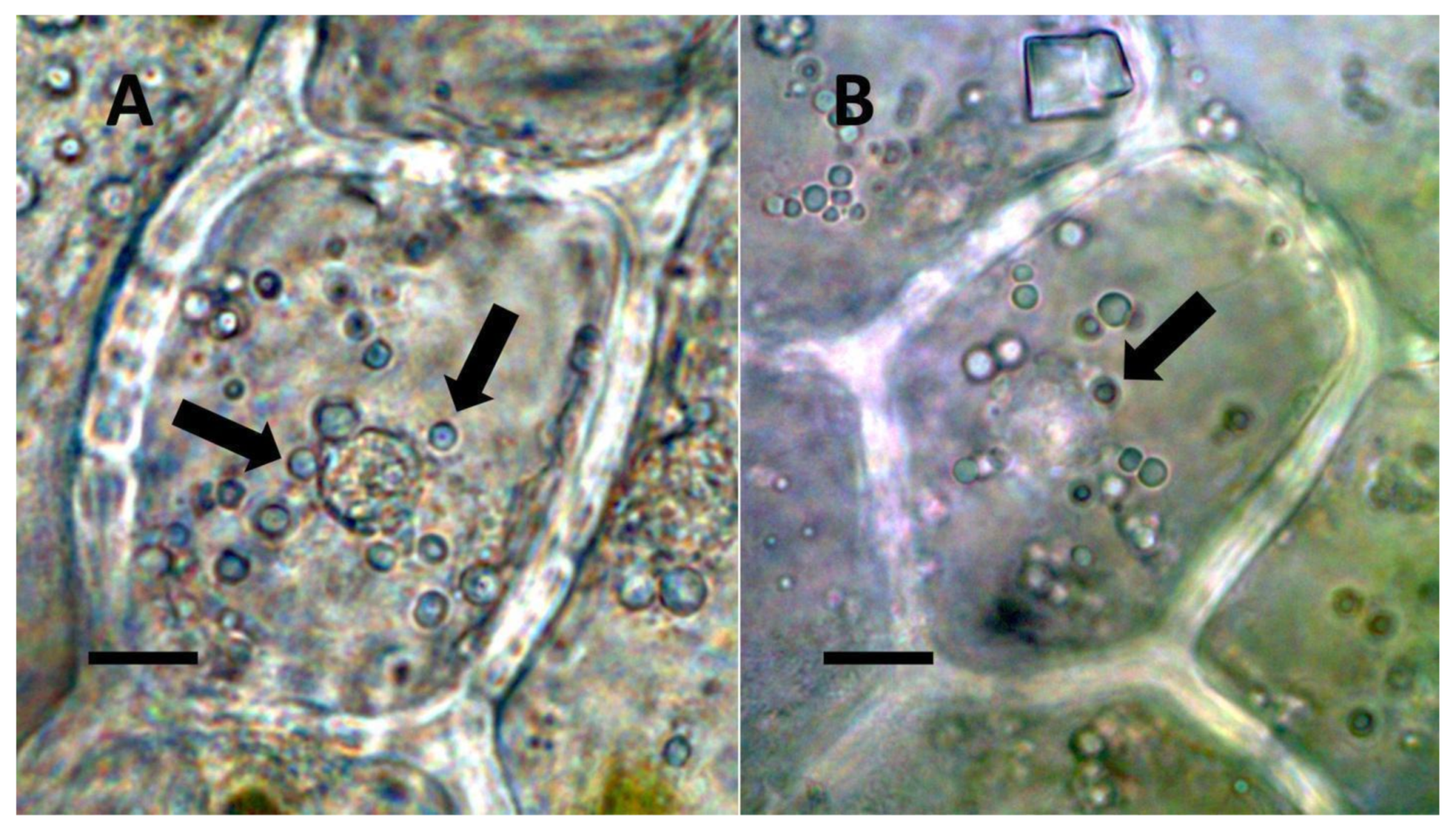
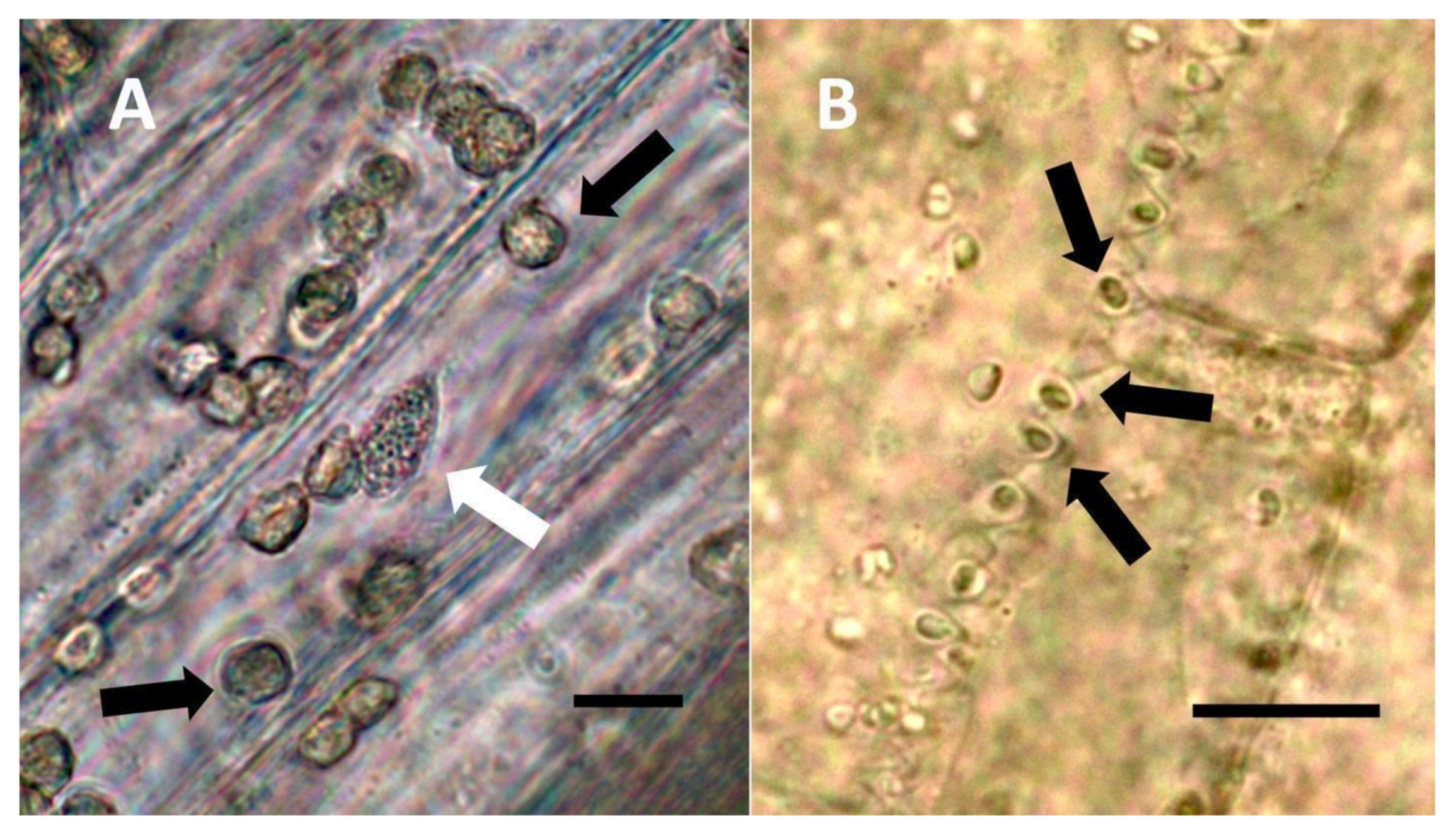
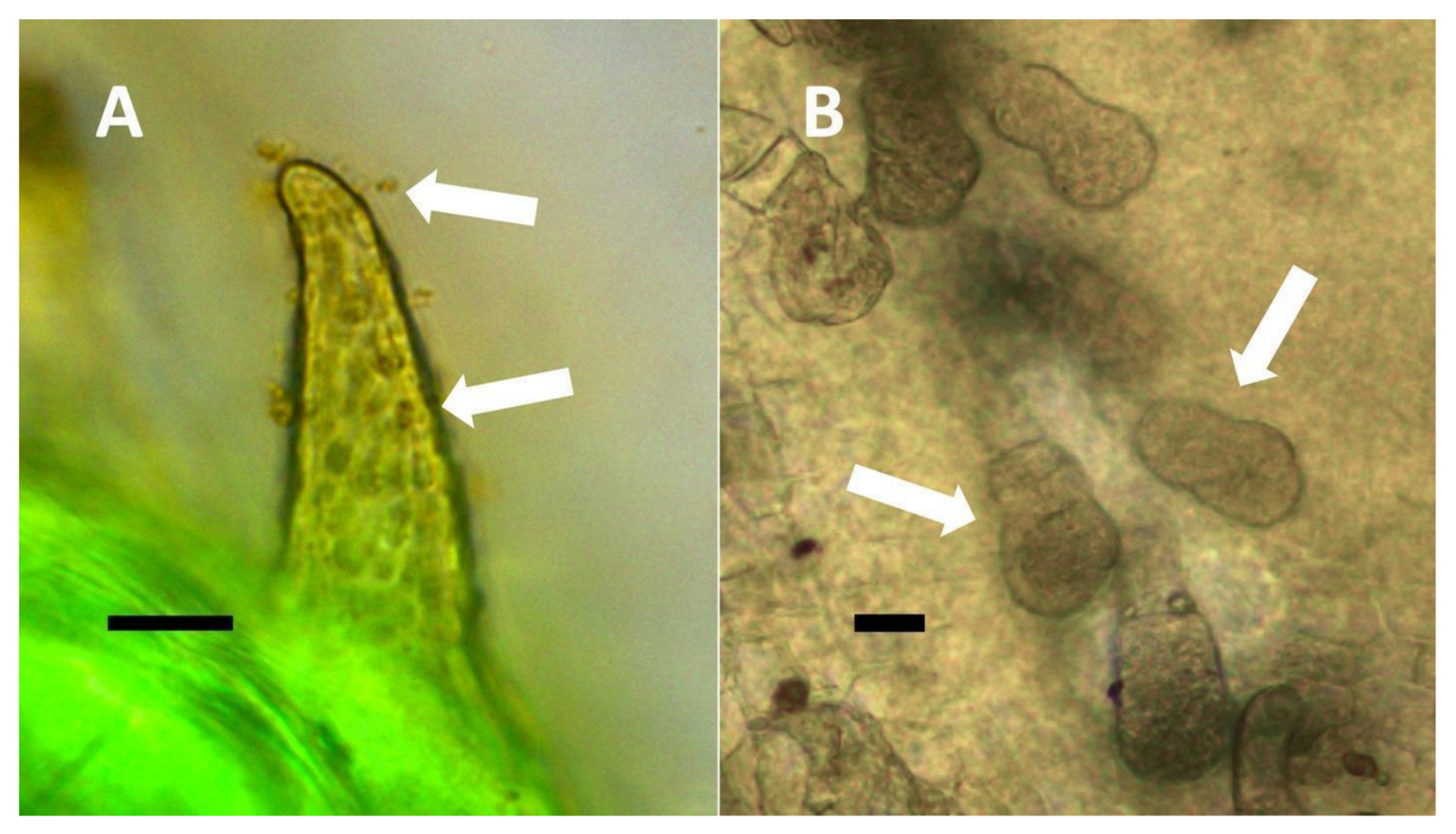
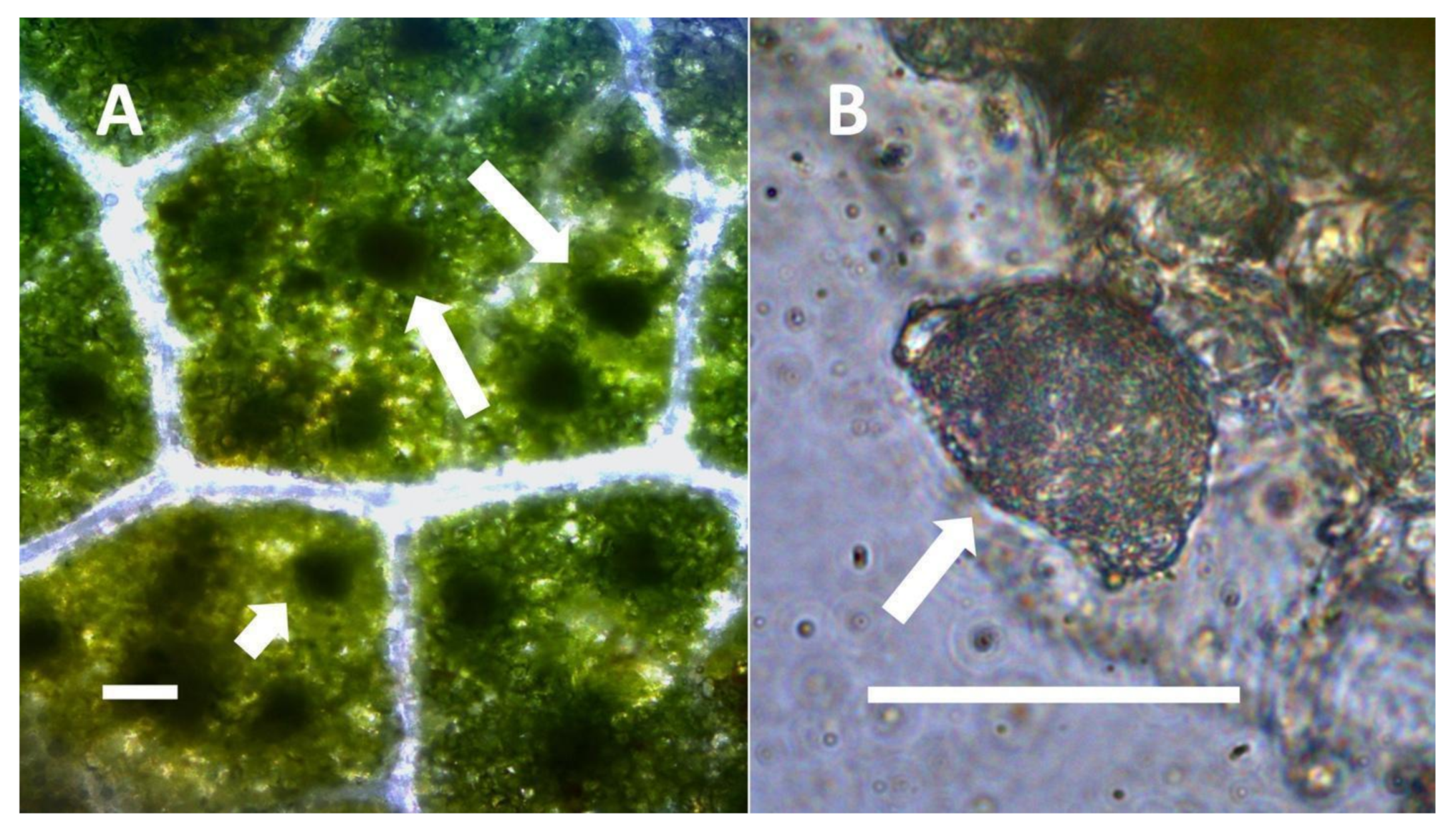
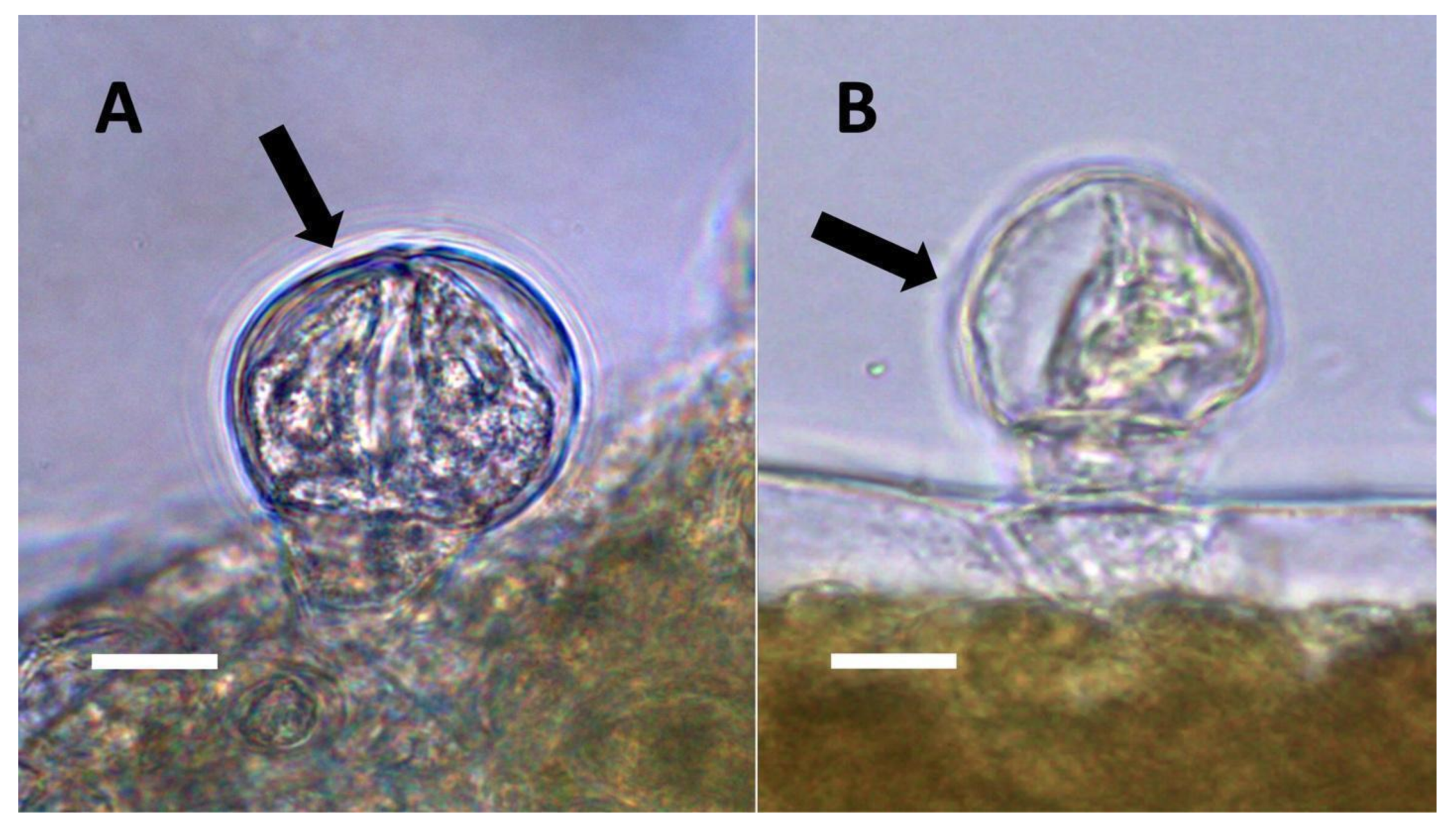
- Nuclear symbiosis (Figure 1, Figure 2 and Figure 3): In this symbiosis, bacteria are cultivated within nuclei, where sugars may fuel bacterial replication and metabolic activities. The bacteria are released into the cytoplasm of the cell in vesicles. Bacteria, once released from nuclei, begin to secrete ethylene. Bacteria in the cytoplasm are exposed to host-produced superoxide. Bacteria were also seen to stain for nitric oxide and nitrate, perhaps as an antioxidant in response to host-cell-produced superoxide. Nuclear symbioses were seen in plants without trichomes, including Agave, Hosta, and Vanilla. Nuclear symbioses were also seen in the grasses Phragmites australis, Digitaria sanguinalis, and Festuca rubra along with simple non-pitted trichomes. These trichomes are filaments, typically unicellular, and frequently contain bacteria. Grasses show additional epidermal cell modifications, where the lateral walls of cells develop serrations or convolutions. Bacteria in developing epidermal cells accumulate in the wall serrations (Figure 3). Some trichomes produced in the Asteraceae (e.g., Eupatorium, Helianthus, and Solidago) on bracts show evidence of nuclear symbiosis in cells of the trichome. Typically, the trichomes bearing nuclear symbiosis are thick-walled with striations but do not have lateral pits. Previous experiments with vanilla orchids [57] that possess nuclear symbiosis suggest that the epidermal cells become colonized by bacteria in the shoot meristem, where biofilms of bacteria are cultivated, or in the recently differentiated leaves.
- (2)
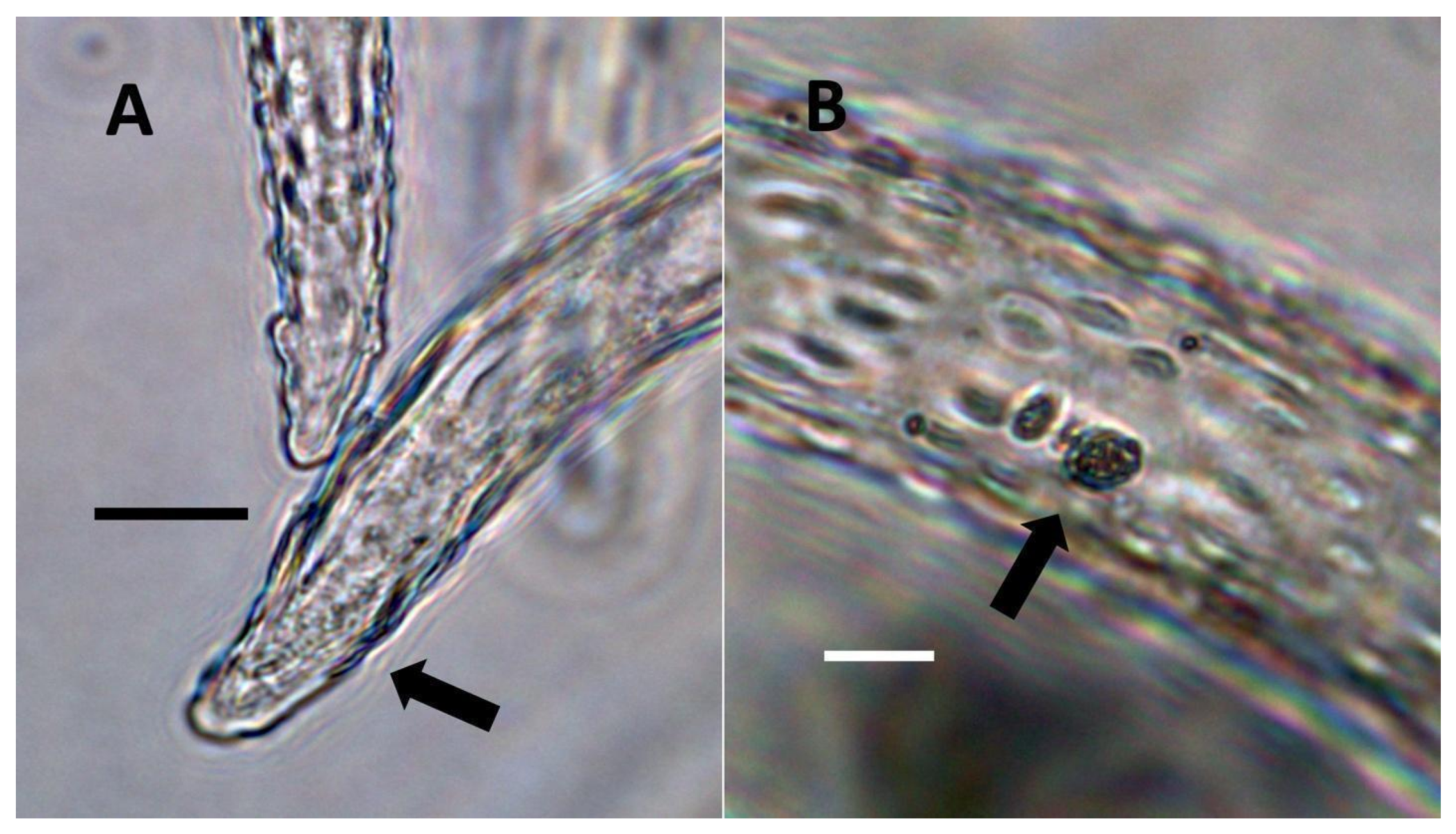
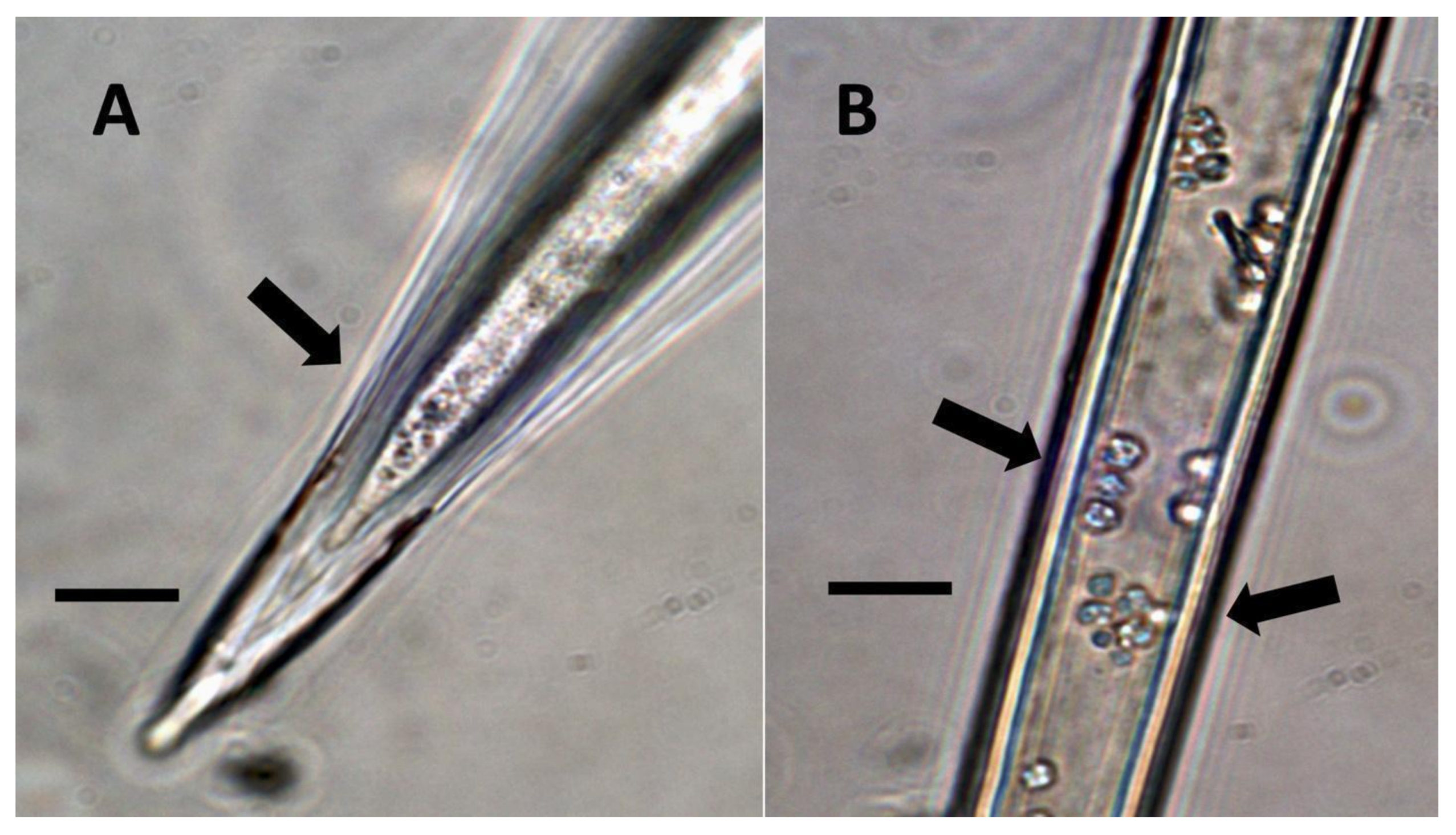
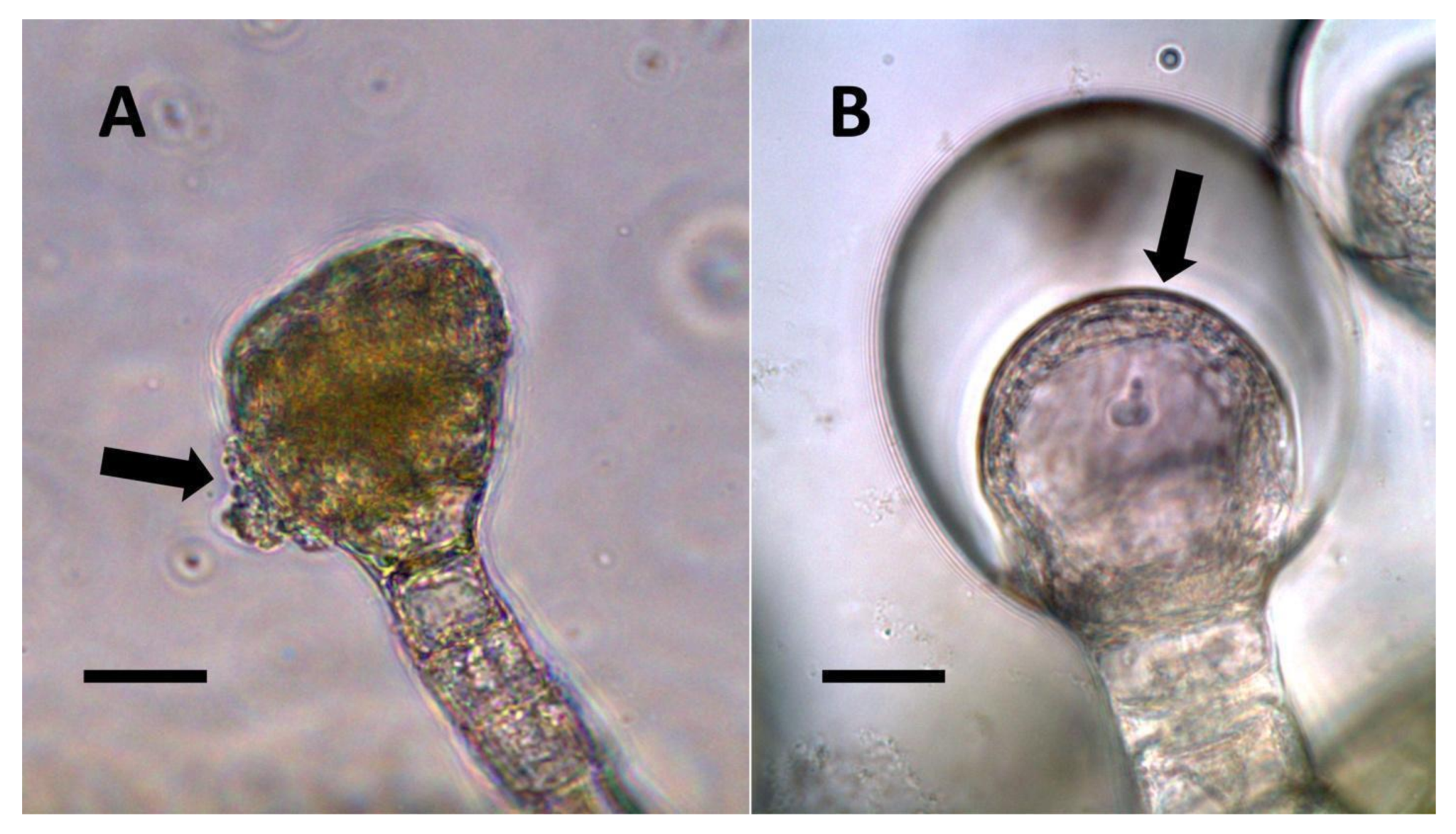
- Pitted filamentous trichome symbiosis (Figure 4): In this symbiosis, bacteria are seen to be replicated within trichomes, where they are moved through periplasmic streaming, or cyclosis, within hairs to accumulate in equidistantly spaced depressions on the surface of the trichome plasma membrane. The pores develop in the lateral trichome walls just over the bacterial clusters in the trichome plasma membrane depressions. These trichomes often show reducing sugars around bacteria throughout the trichome. Ethylene, nitrogenous compounds, and superoxide can be seen around bacteria, especially associated with lateral wall pits. These bacteria are often seen to spill from hairs through the pits in the trichome walls; this is especially evident in the basal parts of the trichome. These pitted trichomes may also function to populate the plant surface (phyllosphere) with bacteria. Pitted trichomes were observed in many different dicotyledonous plants (Table 1), including, for example, Celtis occidentalis and Eutrochium maculatum, but were predominant in Ailanthus altissima, where a Bacillus sp. was isolated from leaf washings of young plants. In addition, endospores could be observed on and within trichomes.
- (3)
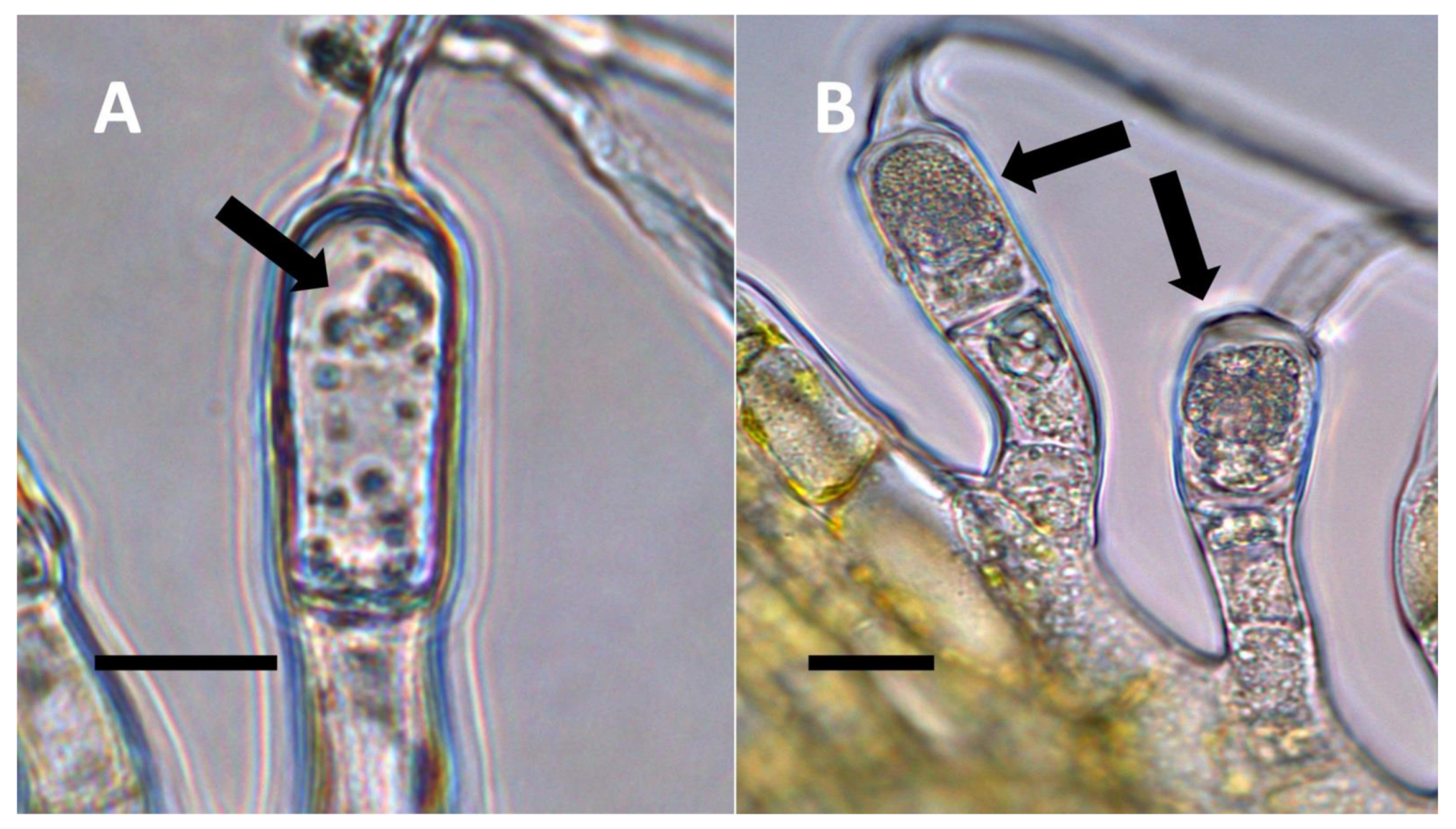
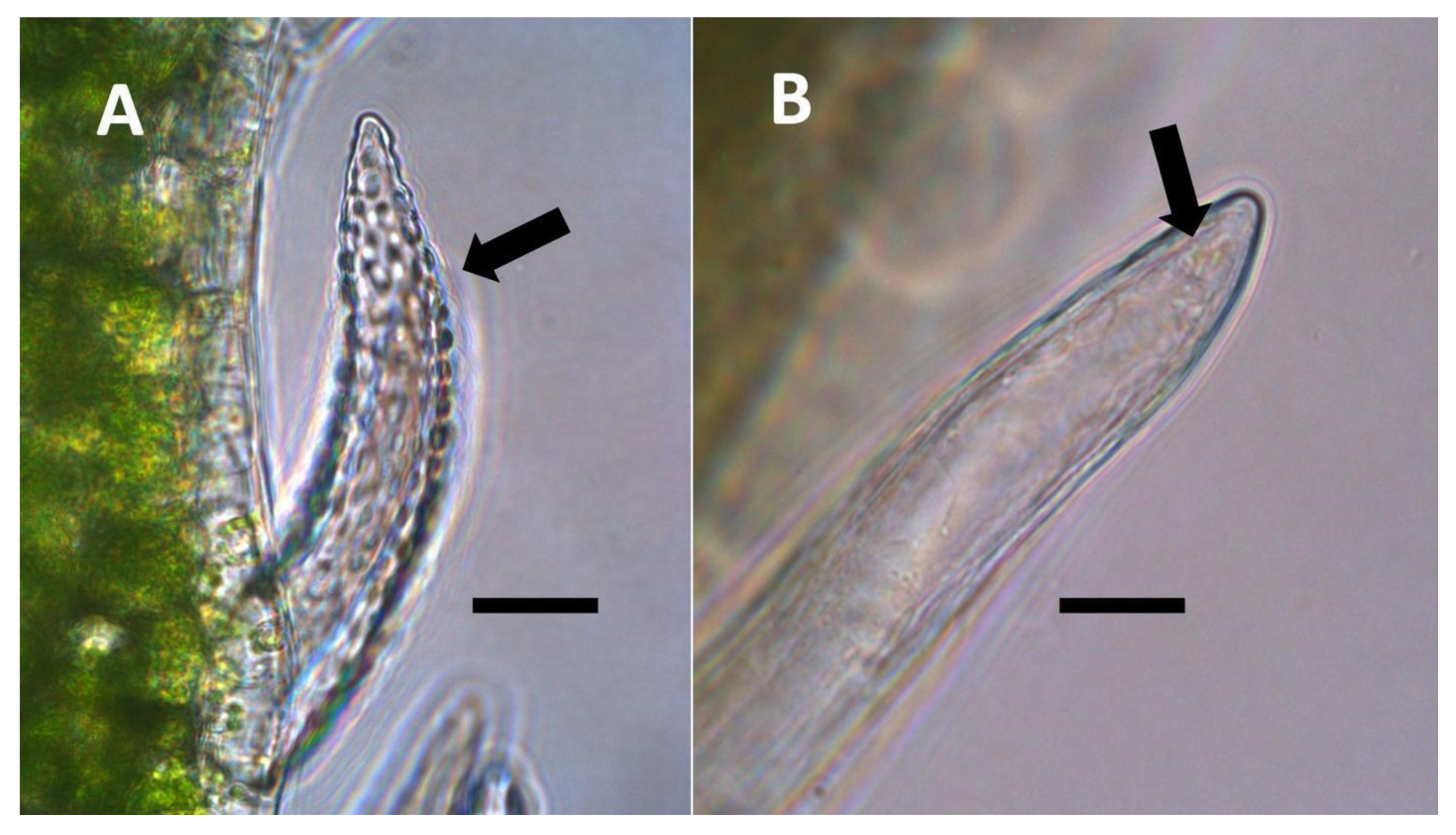
- Non-pitted filamentous trichome symbiosis (Figure 5 and Figure 6): These trichomes often contain bacteria that are evident in the tips of hairs. Nitrogenous chemicals were evident around bacteria in histochemical experiments. Ligule trichomes observed in Phragmites australis and leaf sheath trichomes from the grass Digitaria sanguinalis appear to be this type. Another example is the highly branched trichomes observed covering heavily tomentose leaves of Verbascum thapsus. Bacteria do not appear to exit trichomes in this endosymbiosis. Peltate trichomes observed in Thespesia populnea and other species are a special case of this type, where multiple filaments fuse to form a circular sheet, with bacteria present in the tips of each cell of the trichome. Some trichomes of this non-pitted filamentous type do not show evidence of bacteria within them. An example here is Stachys byzantina, where leaves are covered with very long filamentous trichomes that do not contain bacteria (Table 1).
- (4)
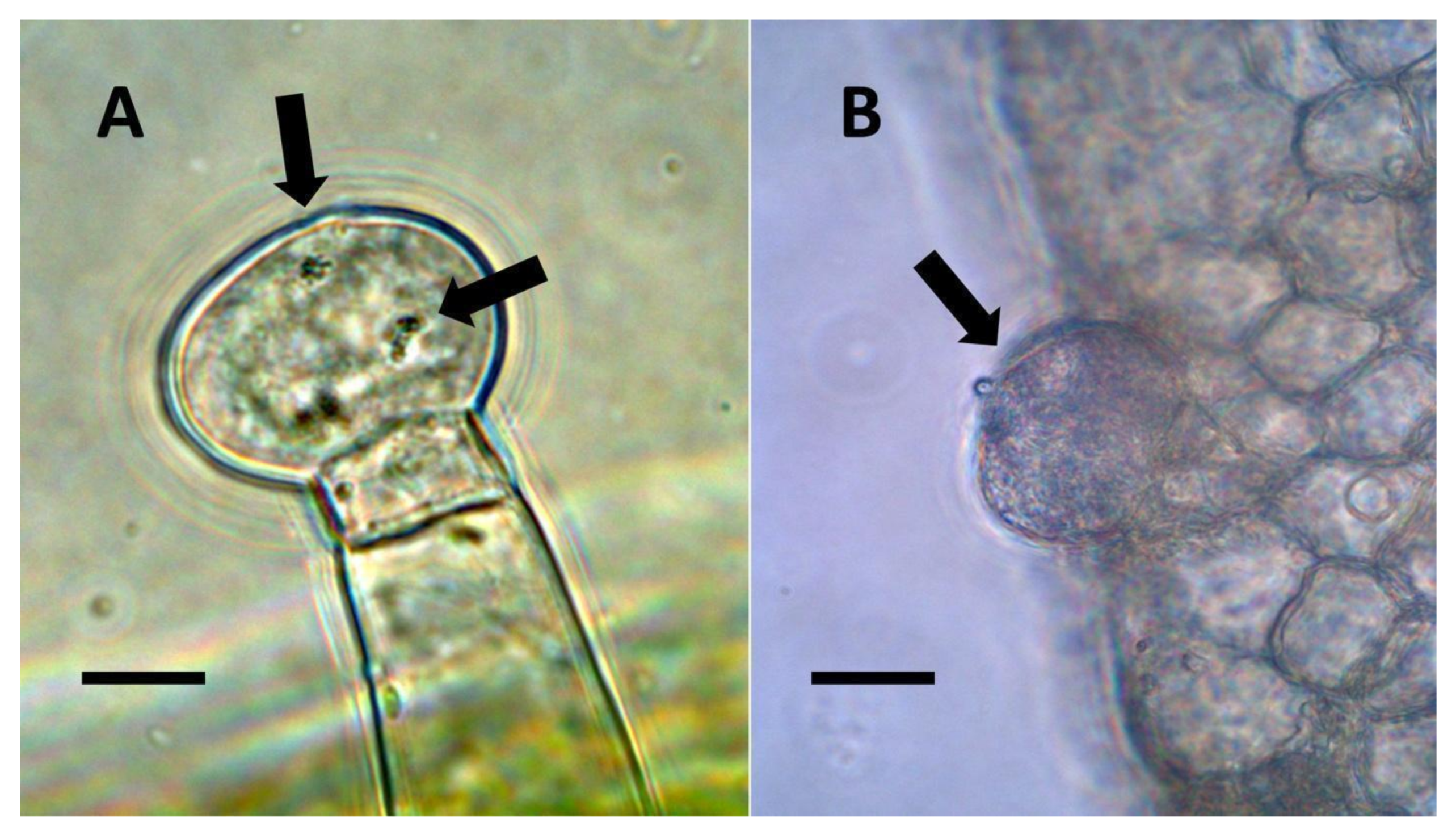
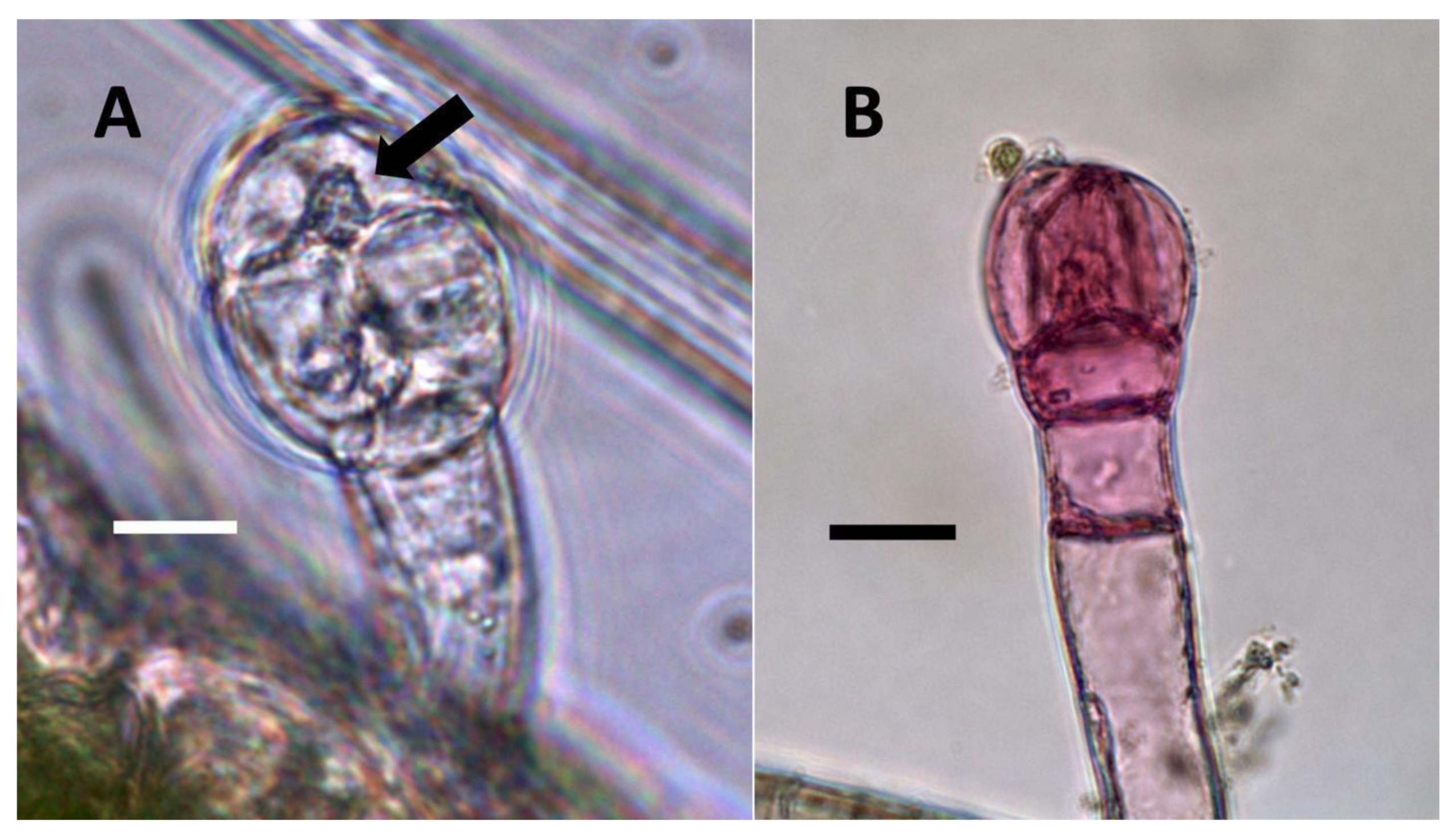
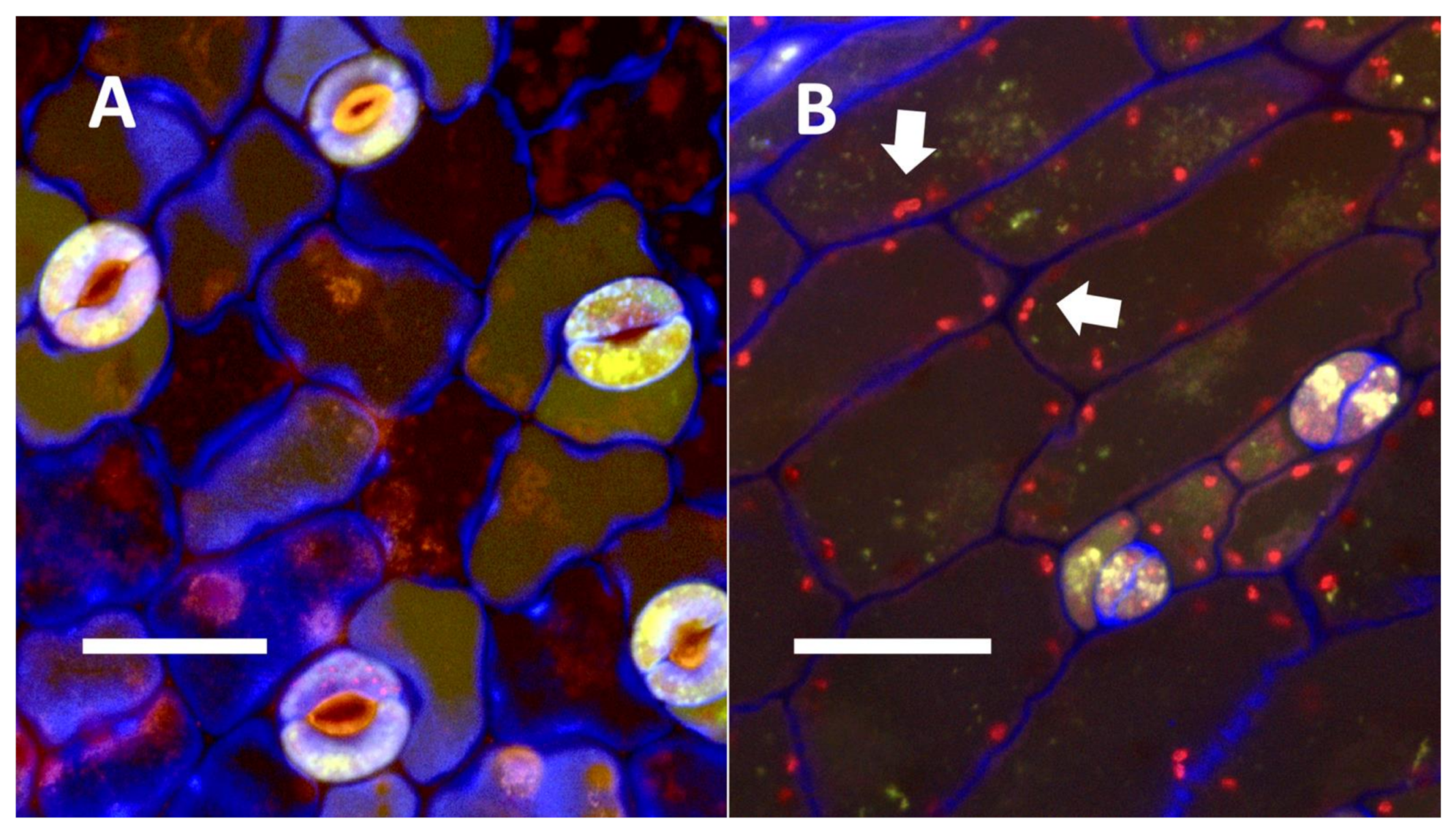
- Glandular trichome symbiosis (Figure 7, Figure 8, Figure 9 and Figure 10): Some of the dicotyledonous plants examined possessed glandular trichomes that contained bacteria (Table 1). Typically, the tip or head of the glandular trichomes contained several non-photosynthetic plant cells in addition to bacteria. Glandular trichomes tended to stain densely for nitrate compared to other trichome types, suggesting that they are more efficient than other trichomes in nitrogen acquisition. Glandular trichomes with bacteria were notable in Cannabis sativa, Citronella mucronata, Humulus lupulus, Perilla frutescens, Rhus glabra, Solanum dulcamara, Apocynum cannabinum, and Solanum lycopersicum.
- (5)
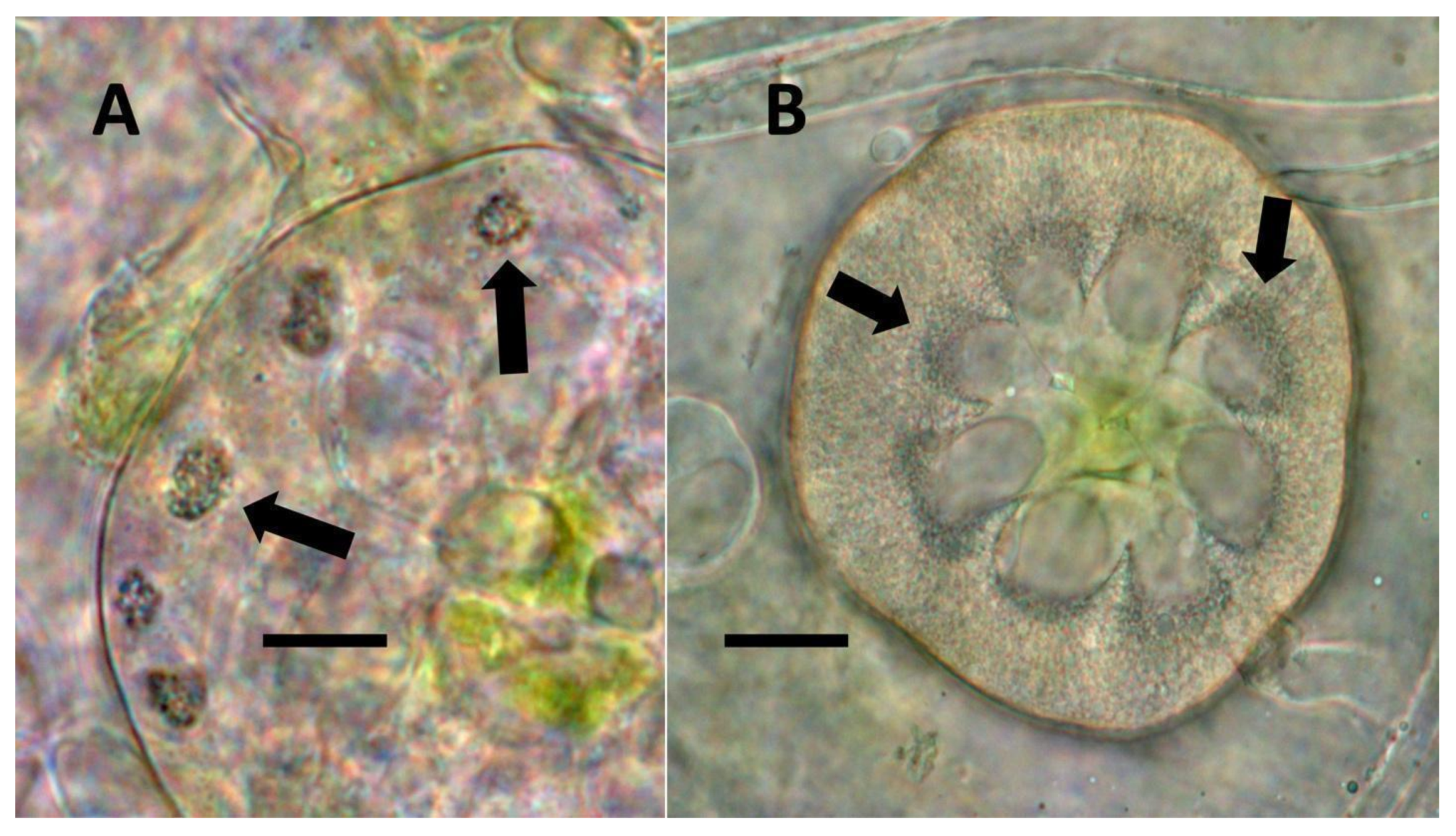
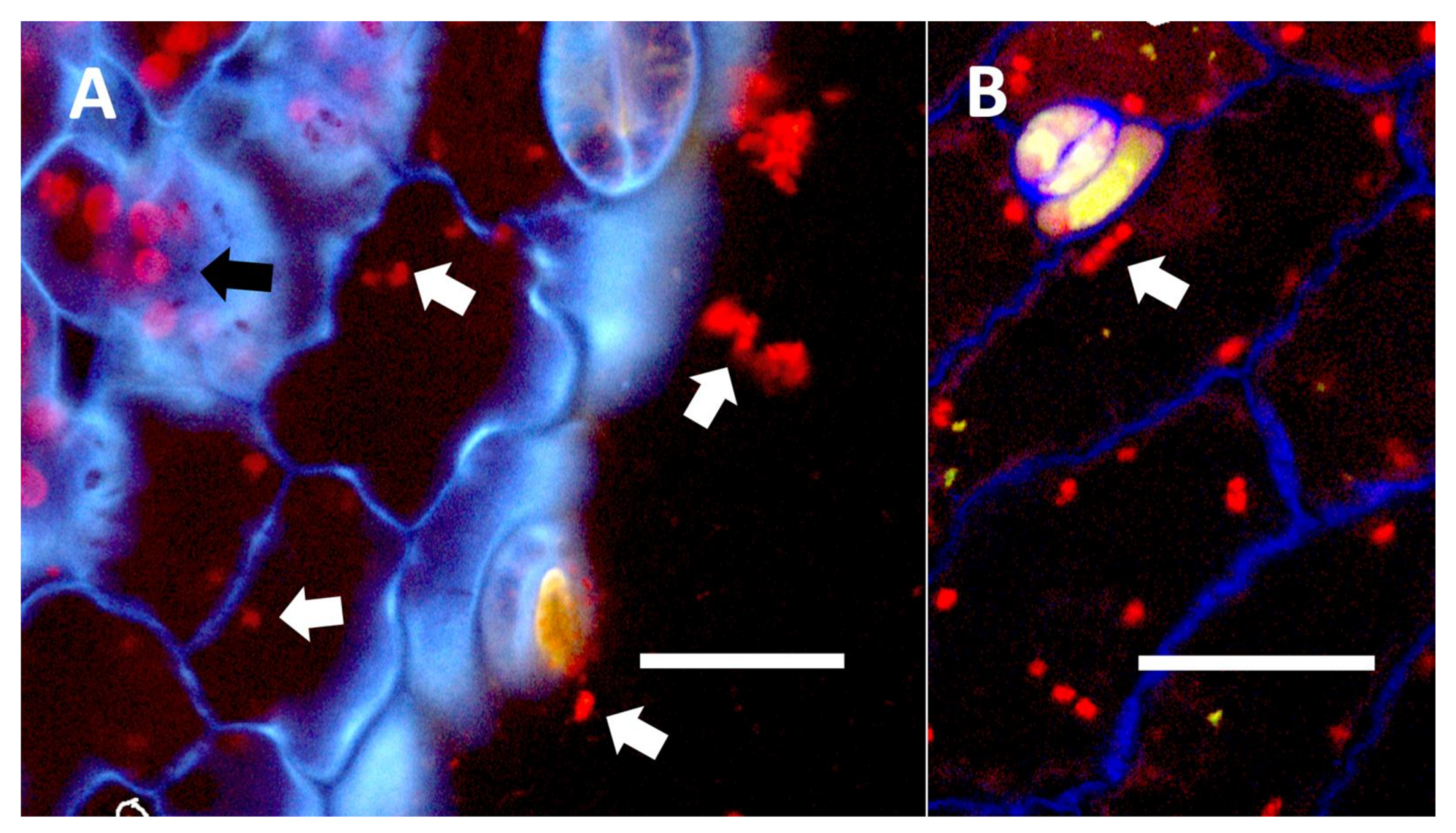
- Leaf nodule symbiosis (Figure 11): We observed large masses of regularly spaced bacteria in leaves of Thespesia populnea (family Malvaceae). These masses were found to stain densely for nitrate using acidified diphenylamine, suggesting that they were active in producing nitrogen. These structures in leaves correspond to previously described structures called ‘leaf nodules’ [80]. We often observed abundant trichomes with bacteria in developing leaves of Thespesia populnea. This emphasizes that plants often show multiple types of structures that may produce nitrogen in their tissues.
3.3. Isotopic Nitrogen Assimilation Experiments
3.4. Summary Data on Leaf Bacteria
| Host | Organs | Bacterium | GenBank Accession | Growth on N-Free Media | Acetylene Reduction Assay | Nif Genes Assessed | Article |
|---|---|---|---|---|---|---|---|
| Agave palmeri | Seeds, leaves, roots | Klebsiella oxytoca | KJ667735.1 | +1 | + | N/A | [69] |
| Ailanthus altissima | Leaves | Bacillus sp. | OM223869 | + | N/A | N/A | This article |
| Digitaria ischaemum | Seeds, leaves, roots | Pantoea sp. | MK733357 | + | N/A | N/A | [81] |
| Staphylococcus sp. | MT275650.1 | + | N/A | N/A | [81] | ||
| Glycine max | Leaves | Bacillus megaterium | OL870610 | + | N/A | N/A | This article |
| Hedera helix | Seeds, leaves, roots | Bacillus amyloliquefaciens | KM822602 | + | + | + | [82] |
| Hosta plantaginea | Seeds, leaves, | Bacillus amyloliquefaciens | KM454171 | + | + | + | [83] |
| Seeds, leaves | Curtobacterium sp. | - | + | N/A | N/A | [83] | |
| Humulus lupulus | Inflorescence bracts | Pseudomonas fluorescens | GCA004794015 | N/A | N/A | +2 | [84] |
| Inflorescence bracts | Pseudomonas stutzeri | GCA_004793985 | N/A | N/A | +2 | [84] | |
| Inflorescence bracts | Massilia sp. | OM223867 | + | N/A | N/A | This article | |
| Inflorescence bracts | Pantoea sp. | OM223868 | + | N/A | N/A | This article | |
| Phragmites australis | Tillers, leaves | Bacillus amyloliquefaciens | KP860304.1 | + | + | + | [44] |
| Tillers, leaves | Microbacterium oxydans | KP860310.1 | + | + | N/A | [44] | |
| Tillers, leaves | Achromobacter spanius | KP860309.1 | + | + | N/A | [44] | |
| Thespesia populnea | Seeds, leaves | Bacillus amyloliquefaciens | KX622564 | + | N/A | N/A | [61] |
| Seeds, leaves, roots | Pseudomonas oryzihabitans | KY471285 | + | N/A | N/A | [61] | |
| Vanilla phaeantha | Leaves, roots | Bacillus amyloliquefaciens | KF765481 | + | N/A | N/A | [57] |
3.5. Experiments to Assess Effects on Trichomes by Reduction in Seedling Bacteria
3.5.1. Rhus glabra Seed Sterilization Experiment
3.5.2. Perilla frutescens Seed Sterilization Experiment
3.5.3. Bacterial Replication Repression in Ailanthus altissima Seedlings Using Elevated Carbon Dioxide
3.5.4. Inoculation Experiments
4. Discussion
4.1. Patterns of Nitrogen Assimilation into Leaves and Bracts
4.2. Symbiosis Stacking
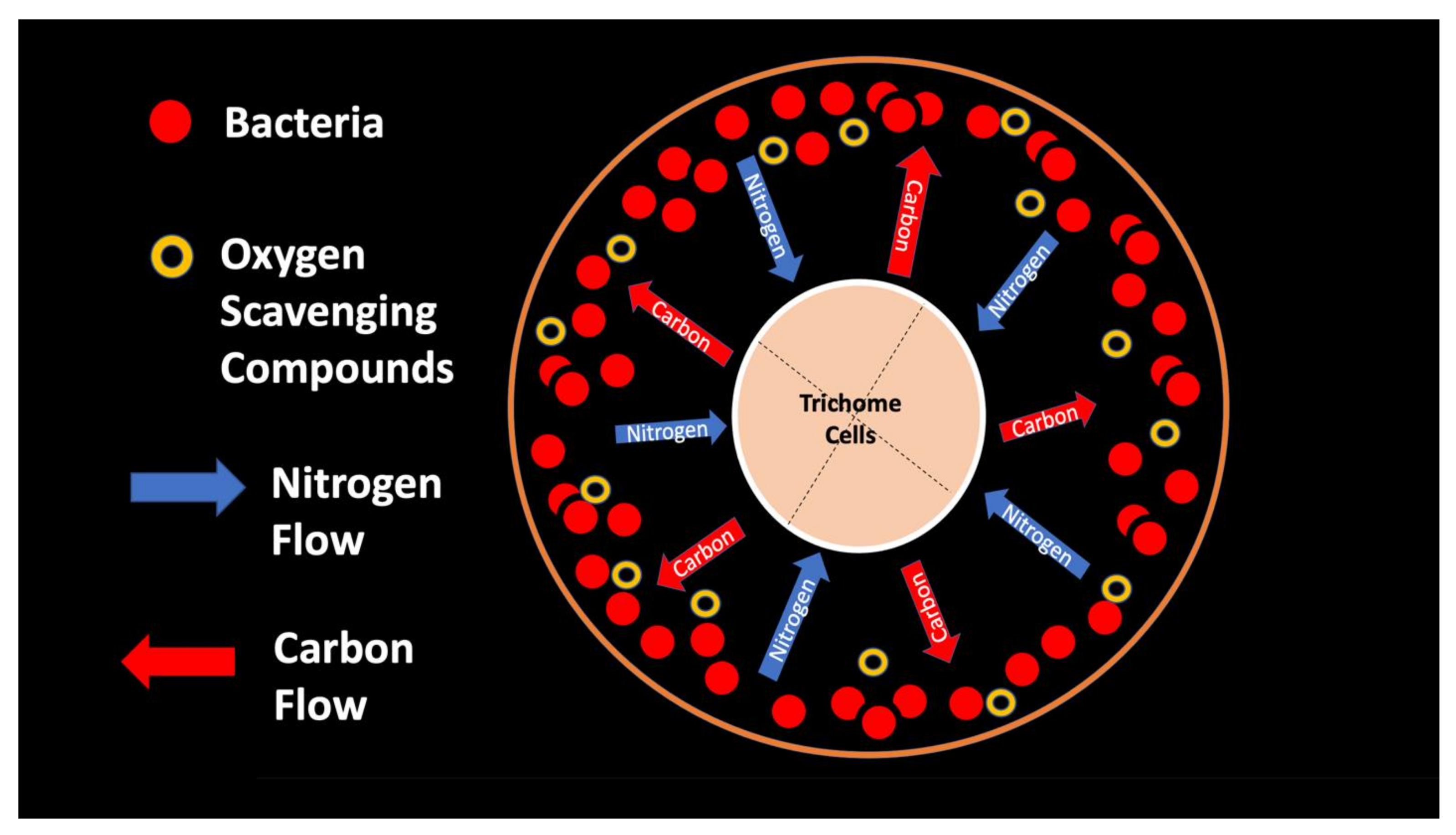
4.3. Trichome Endosymbiosis
4.4. Factors That May Affect Nitrogen Accumulation in Plant Cells
4.4.1. Photosynthate
4.4.2. Oxygen Levels
4.4.3. Variability in Nitrogen Absorption in Plants
4.5. Bacteria in Phyllospheres of Plants
4.6. Evolutionary Considerations
5. Conclusions
- (1)
- Can nitrogen fixation within plant cells be confirmed?
- (2)
- How are these endosymbioses regulated by the plant?
- (3)
- What bacteria are involved in these endosymbioses?
- (4)
- How robust are these endosymbioses?
- (5)
- Are these endosymbioses lost in plants under cultivation?
- (6)
- How might these endosymbioses function when plants are treated with nitrogen or other agrochemicals?
- (7)
- Are there ways to treat plants to support or enhance these native endosymbioses in plant leaves?
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Remy, W.; Taylor, T.N.; Hass, H.; Kerp, H. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. USA 1994, 91, 11841–11843. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Castillo-Avila, D.K.; Raizada, M.N.; Becerra Lopez-Lavalle, L.A. Paying the rent: How endophytic microorganisms help plant hosts obtain nutrients. In Comprehensive Biotechnology, 3rd ed.; Moo-Young, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 770–788. [Google Scholar] [CrossRef]
- Chanway, C.P.; Anand, R.; Yang, H. Nitrogen fixation outside and inside plant tissues. In Advances in Biology and Ecology of Nitrogen Fixation; InTechOpen: London, UK, 2014. [Google Scholar] [CrossRef]
- Adamczyk, B.; Sietiö, O.M.; Straková, P.; Prommer, J.; Wild, B.; Hagner, M.; Pihlatie, M.; Fritze, H.; Richter, A.; Heinonsalo, J. Plant roots increase both decomposition and stable organic matter formation in boreal forest soil. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Martinez-Rodriguez, A.; Macedo-Raygoza, G.; Huerta-Robles, A.X.; Reyes-Sepulveda, I.; Lozano-Lopez, J.; García-Ochoa, E.Y.; Fierro-Kong, L.; Medeiros, M.H.G.; Di Mascio, P.; White, J.F.; et al. Agave seed endophytes: Ecology and impacts on root architecture, nutrient acquisition, and cold stress tolerance. In Seed Endophytes; Springer: Cham, Switzerland, 2019; pp. 139–170. [Google Scholar] [CrossRef]
- Adamczyk, B. How do terrestrial plants access high molecular mass organic nitrogen, and why does it matter for soil organic matter stabilization? Plant Soil 2021, 465, 583–592. [Google Scholar] [CrossRef]
- Simon, J.; Dannenmann, M.; Pena, R.; Gessler, A.; Rennenberg, H. Nitrogen nutrition of beech forests in a changing climate: Importance of plant-soil-microbe water, carbon, and nitrogen interactions. Plant Soil 2017, 418, 89–114. [Google Scholar] [CrossRef]
- Puente, M.E.; Bashan, Y. The desert epiphyte Tillandsia recurvata harbours the nitrogen-fixing bacterium Pseudomonas stutzeri. Can. J. Bot. 2011, 72, 406–408. [Google Scholar] [CrossRef]
- Hestrin, R.; Hammer, E.C.; Mueller, C.W.; Lehmann, J. Synergies between mycorrhizal fungi and soil microbial communities increase plant nitrogen acquisition. Commun. Biol. 2019, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- De La Peña, T.C.; Fedorova, E.; Pueyo, J.J.; Mercedes Lucas, M. The symbiosome: Legume and rhizobia co-evolution toward a nitrogen-fixing organelle? Front. Plant Sci. 2018, 8, 2229. [Google Scholar] [CrossRef]
- Maity, P.J.; Pawlowski, K. Anthropogenic influences on the distribution of the Casuarina-Frankia symbiosis. Symbiosis 2021, 84, 353–367. [Google Scholar] [CrossRef]
- Fujita, H.; Aoki, S.; Kawaguchi, M. Evolutionary dynamics of nitrogen fixation in the legume–rhizobia Symbiosis. PLoS ONE 2014, 9, e93670. [Google Scholar] [CrossRef]
- Paungfoo-Lonhienne, C.; Rentsch, D.; Robatzek, S.; Webb, R.I.; Sagulenko, E.; Näsholm, T.; Schmidt, S.; Lonhienne, T.G.A. Turning the table: Plants consume microbes as a source of nutrients. PLoS ONE 2010, 5, e11915. [Google Scholar] [CrossRef]
- Hill, P.W.; Marsden, K.A.; Jones, D.L. How significant to plant N nutrition is the direct consumption of soil microbes by roots? New Phytol. 2013, 199, 948–955. [Google Scholar] [CrossRef]
- White, J.; Kingsley, K.; Verma, S.; Kowalski, K. Rhizophagy cycle: An oxidative process in plants for nutrient extraction from symbiotic microbes. Microorganisms 2018, 6, 95. [Google Scholar] [CrossRef]
- White, J.F.; Torres, M.S.; Verma, S.K.; Elmore, M.T.; Kowalski, K.P.; Kingsley, K.L. Evidence for widespread microbivory of endophytic bacteria in roots of vascular plants through oxidative degradation in root cell periplasmic spaces. In PGPR Amelioration in Sustainable Agriculture; Woodhead Publishing: Thorston, UK, 2019; pp. 167–193. [Google Scholar] [CrossRef]
- Morris, C.E.; Kinkel, L.L. Fifty years of phylosphere microbiology: Significant contributions to research in related fields. In Phyllosphere Microbiology; Lindow, S.E., Hecht-Poinar, E.I., Elliott, V., Eds.; APS Press: St. Paul, MN, USA, 2002; pp. 365–375. [Google Scholar]
- Lindow, S.E.; Brandl, M.T. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef]
- Redford, A.J.; Bowers, R.M.; Knight, R.; Linhart, Y.; Fierer, N. The ecology of the phyllosphere: Geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 2010, 12, 2885–2893. [Google Scholar] [CrossRef]
- Fürnkranz, M.; Wanek, W.; Richter, A.; Abell, G.; Rasche, F.; Sessitsch, A. Nitrogen fixation by phyllosphere bacteria associated with higher plants and their colonizing epiphytes of a tropical lowland rainforest of Costa Rica. ISME J. 2008, 2, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Freiberg, E. Microclimatic parameters influencing nitrogen fixation in the phyllosphere in a Costa Rican premontane rain forest. Oecologia 1998, 117, 9–18. [Google Scholar] [CrossRef]
- Stanton, D.E.; Batterman, S.A.; Von Fischer, J.C.; Hedin, L.O.; Stanton, D.E.; Batterman, S.A.; Von Fischer, J.C.; Hedin, L.O. Rapid nitrogen fixation by canopy microbiome in tropical forest determined by both phosphorus and molybdenum. Ecology 2019, 100, e02795. [Google Scholar] [CrossRef]
- Gargallo-Garriga, A.; Sardans, J.; Pérez-Trujillo, M.; Guenther, A.; Llusià, J.; Rico, L.; Terradas, J.; Farré-Armengol, G.; Filella, I.; Parella, T.; et al. Shifts in plant foliar and floral metabolomes in response to the suppression of the associated microbiota. BMC Plant Biol. 2016, 16, 78. [Google Scholar] [CrossRef]
- Santi, C.; Bogusz, D.; Franche, C. Biological nitrogen fixation in non-legume plants. Ann Bot. 2013, 111, 743–767. [Google Scholar] [CrossRef]
- Beattie, G.A.; Lindow, S.E. The secret life of foliar bacterial pathogens on leaves. Annu. Rev. Phytopathol. 2003, 33, 145–172. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.W. Plant trichomes as microbial habitats and infection sites. Eur. J. Plant Pathol. 2019, 154, 157–169. [Google Scholar] [CrossRef]
- Papen, H.; Geßler, A.; Zumbusch, E.; Rennenberg, H. Chemolithoautotrophic nitrifiers in the phyllosphere of a spruce ecosystem receiving high atmospheric nitrogen input. Curr. Microbiol. 2002, 44, 56–60. [Google Scholar] [CrossRef]
- Abadi, V.A.J.M.; Sepehri, M.; Rahmani, H.A.; Zarei, M.; Ronaghi, A.; Taghavi, S.M.; Shamshiripour, M. Role of dominant phyllosphere bacteria with plant growth–promoting characteristics on growth and nutrition of maize (Zea mays L.). J. Soil Sci. Plant Nutr. 2020, 20, 2348–2363. [Google Scholar] [CrossRef]
- Bashir, I.; Assad, R.; War, A.F.; Rafiq, I.; Sofi, I.A.; Reshi, Z.A.; Rashid, I. Application of phyllosphere microbiota as biofertilizers. Microbiota Biofertil. 2021, 2, 311–327. [Google Scholar] [CrossRef]
- Wani, S.P.; Dart, P.J.; Upadhyaya, M.N. Factors affecting nitrogenase activity (C2H2 reduction) associated with sorghum and millet estimated using the soil core assay. Can. J. Microbiol. 2011, 29, 1063–1069. [Google Scholar] [CrossRef][Green Version]
- Johnston-Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2011, 6, e20396. [Google Scholar] [CrossRef]
- Beltran-Garcia, M.J.; White, J.F.; Prado, F.M.; Prieto, K.R.; Yamaguchi, L.F.; Torres, M.S.; Kato, M.J.; Medeiros, M.H.G.; Di Mascio, P. Nitrogen acquisition in Agave tequilana from degradation of endophytic bacteria. Sci. Rep. 2014, 4, 1–7. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Thomas, P.; Agrawal, M.; Bharathkumar, C.B. Diverse cellular colonizing endophytic bacteria in field shoots and in vitro cultured papaya with physiological and functional implications. Physiol. Plant. 2019, 166, 729–747. [Google Scholar] [CrossRef]
- Macedo-Raygoza, G.M.; Valdez-Salas, B.; Prado, F.M.; Prieto, K.R.; Yamaguchi, L.F.; Kato, M.J.; Canto-Canché, B.B.; Carrillo-Beltrán, M.; Di Mascio, P.; White, J.F.; et al. Enterobacter cloacae, an endophyte that establishes a nutrient-transfer symbiosis with banana plants and protects against the black sigatoka pathogen. Front. Microbiol. 2019, 10, 804. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Sahu, P.K.; Kumar, K.; Pal, G.; Gond, S.K.; Kharwar, R.N.; White, J.F. Endophyte roles in nutrient acquisition, root system architecture development and oxidative stress tolerance. J. Appl. Microbiol. 2021, 131, 2161–2177. [Google Scholar] [CrossRef] [PubMed]
- Johnston-Monje, D.; Gutiérrez, J.P.; Lopez-Lavalle, L.A.B. Seed-transmitted bacteria and fungi dominate juvenile plant microbiomes. Front Microbiol. 2021, 12, 737616. [Google Scholar] [CrossRef] [PubMed]
- Van Deynze, A.; Zamora, P.; Delaux, P.M.; Heitmann, C.; Jayaraman, D.; Rajasekar, S.; Graham, D.; Maeda, J.; Gibson, D.; Schwartz, K.D.; et al. Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLoS Biol. 2018, 16, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Rosenblueth, M.; Ormeño-Orrillo, E.; López-López, A.; Rogel, M.A.; Reyes-Hernández, B.J.; Martínez-Romero, J.C.; Reddy, P.M.; Martínez-Romero, E. Nitrogen fixation in cereals. Front. Microbiol. 2018, 9, 1794. [Google Scholar] [CrossRef]
- James, E.K. Nitrogen fixation in endophytic and associative symbiosis. F. Crop. Res. 2000, 65, 197–209. [Google Scholar] [CrossRef]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef]
- White, J.F.; Torres, M.S.; Johnson, H.; Irizarry, I.; Tadych, M. A Functional view of plant microbiomes: Endosymbiotic systems that enhance plant growth and survival. In Advances in Endophytic Research; Verma, V., Gange, A., Eds.; Springer: New Delhi, India, 2014. [Google Scholar] [CrossRef]
- Larrea-Álvarez, M.; Purton, S. The chloroplast of Chlamydomonas reinhardtii as a testbed for engineering nitrogen fixation into plants. Int. J. Mol. Sci. 2021, 22, 8806. [Google Scholar] [CrossRef]
- Dent, D.; Cocking, E. Establishing symbiotic nitrogen fixation in cereals and other non-legume crops: The Greener Nitrogen Revolution. Agric. Food Secur. 2017, 6, 1–9. [Google Scholar] [CrossRef]
- Dobereiner, J. History and new perspectives of diazotrophs in association with non-leguminous plants. Symbiosis 1992, 13, 1–13. [Google Scholar]
- Goyal, R.K.; Schmidt, M.A.; Hynes, M.F. Molecular biology in the improvement of biological nitrogen fixation by rhizobia and extending the scope to cereals. Microorganisms 2021, 9, 125. [Google Scholar] [CrossRef]
- Pauly, N.; Pucciariello, C.; Mandon, K.; Innocenti, G.; Jamet, A.; Baudouin, E.; Hérouart, D.; Frendo, P.; Puppo, A. Reactive oxygen and nitrogen species and glutathione: Key players in the legume–Rhizobium symbiosis. J. Exp. Bot. 2006, 57, 1769–1776. [Google Scholar] [CrossRef]
- Kraiser, T.; Gras, D.E.; Gutiérrez, A.G.; González, B.; Gutiérrez, R.A. A holistic view of nitrogen acquisition in plants. J. Exp. Bot. 2011, 62, 1455–1466. [Google Scholar] [CrossRef] [PubMed]
- Waters, J.K.; Mawhinney, T.P.; Emerich, D.W. Nitrogen assimilation and transport by ex planta nitrogen-fixing Bradyrhizobium diazoefficiens bacteroids is modulated by oxygen, bacteroid density and l-malate. Int. J. Mol. Sci. 2020, 21, 7542. [Google Scholar] [CrossRef]
- Glyan’ko, A.K.; Vasil’eva, G.G. Reactive oxygen and nitrogen species in legume-rhizobial symbiosis: A review. Appl. Biochem. Microbiol. 2010, 46, 15–22. [Google Scholar] [CrossRef]
- Chang, X.; Kingsley, K.L.; White, J.F.; Egamberdieva, D. Chemical interactions at the interface of plant root hair cells and intracellular bacteria. Microorganisms 2021, 9, 1041. [Google Scholar] [CrossRef] [PubMed]
- Leveau, J.H.J.; Lindow, S.E. Appetite of an epiphyte: Quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. USA 2001, 98, 3446–3453. [Google Scholar] [CrossRef]
- Remus-Emsermann, M.N.P.; de Oliveira, S.; Schreiber, L.; Leveau, J.H.J. Quantification of lateral heterogeneity in carbohydrate permeability of isolated plant leaf cuticles. Front. Microbiol. 2011, 2, 197. [Google Scholar] [CrossRef]
- Monier, J.M.; Lindow, S.E. Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl. Environ. Microbiol. 2004, 70, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Baldotto, L.E.B.; Olivares, F.L. Phylloepiphytic interaction between bacteria and different plant species in a tropical agricultural system. Can. J. Microbiol. 2008, 54, 918–931. [Google Scholar] [CrossRef] [PubMed]
- White, J.F.; Torres, M.S.; Sullivan, R.F.; Jabbour, R.E.; Chen, Q.; Tadych, M.; Irizarry, I.; Bergen, M.S.; Havkin-Frenkel, D.; Belanger, F.C. Occurrence of Bacillus amyloliquefaciens as a systemic endophyte of vanilla orchids. Microsc. Res. Tech. 2014, 77, 874–885. [Google Scholar] [CrossRef]
- Zenkteler, M.; Stefaniak, B. Ailanthus altissima Mill. Swingle (Tree of Heaven). In Trees IV; Springer: Berlin, Heidelberg, 1996; pp. 18–30. [Google Scholar] [CrossRef]
- Wang, G.; Tian, L.; Aziz, N.; Broun, P.; Dai, X.; He, J.; King, A.; Zhao, P.X.; Dixon, R.A. Terpene biosynthesis in glandular trichomes of hop. Plant Physiol. 2008, 148, 1254–1266. [Google Scholar] [CrossRef] [PubMed]
- Rodziewicz, P.; Loroch, S.; Marczak, Ł.; Sickmann, A.; Kayser, O. Cannabinoid synthases and osmoprotective metabolites accumulate in the exudates of Cannabis sativa L. glandular trichomes. Plant Sci. 2019, 284, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, I.; White, J.F. Application of bacteria from non-cultivated plants to promote growth, alter root architecture and alleviate salt stress of cotton. J. Appl. Microbiol. 2017, 122, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Holtzclaw, H.; Robinson, W. College Chemistry, with Qualitative Analysis, 8th ed.; Raytheon: Waltham, MA, USA, 1988; pp. 1006–1007. [Google Scholar] [CrossRef]
- Lang, C.; Hübert, T. A Colour ripeness indicator for apples. Food Bioprocess Technol. 2012, 5, 3244–3249. [Google Scholar] [CrossRef]
- Álvarez-Hernández, M.H.; Martínez-Hernández, G.B.; Avalos-Belmontes, F.; Castillo-Campohermoso, M.A.; Contreras-Esquivel, J.C.; Artés-Hernández, F. Potassium permanganate-based ethylene scavengers for fresh horticultural produce as an active packaging. Food Eng. Rev. 2019, 11, 159–183. [Google Scholar] [CrossRef]
- Wei, H.; Seidi, F.; Zhang, T.; Jin, Y.; Xiao, H. Ethylene scavengers for the preservation of fruits and vegetables: A review. Food Chem. 2021, 337, 127750. [Google Scholar] [CrossRef]
- Conant, J.B.; Hartshorn, E.B.; Richardson, G.O. The mechnism of the reaction between ethylene and sulfur chloride. J. Am. Chem. Soc. 2002, 42, 585–595. [Google Scholar] [CrossRef][Green Version]
- Chae, H.S.; Lee, W.S. Ethylene- and enzyme-mediated superoxide production and cell death in carrot cells grown under carbon starvation. Plant Cell Rep. 2001, 20, 256–261. [Google Scholar] [CrossRef]
- Javvaji, P.K.; Dhali, A.; Francis, J.R.; Kolte, A.P.; Mech, A.; Roy, S.C.; Mishra, A.; Bhatta, R. An efficient nitroblue tetrazolium staining and bright-field microscopy based method for detecting and quantifying intracellular reactive oxygen species in oocytes, cumulus cells and embryos. Front. Cell Dev. Biol. 2020, 8, 764. [Google Scholar] [CrossRef] [PubMed]
- White, J.F.; Crawford, H.; Torres, M.S.; Mattera, R.; Irizarry, I.; Bergen, M. A proposed mechanism for nitrogen acquisition by grass seedlings through oxidation of symbiotic bacteria. Symbiosis 2012, 57, 161–171. [Google Scholar] [CrossRef] [PubMed]
- White, J.F.; Torres, M.S.; Somu, M.P.; Johnson, H.; Irizarry, I.; Chen, Q.; Zhang, N.; Walsh, E.; Tadych, M.; Bergen, M. Hydrogen peroxide staining to visualize intracellular bacterial infections of seedling root cells. Microsc. Res. Tech. 2014, 77, 566–573. [Google Scholar] [CrossRef]
- Coldwell, B.B.; McLean, S.R. The reaction between diphenylamine and nitrates in ultraviolet light. Can. J. Chem. 1959, 37, 1637–1643. [Google Scholar] [CrossRef]
- Kahn, M.L.; Parra-Colmenares, A.; Ford, C.L.; Kaser, F.; McCaskill, D.; Ketchum, R.E. A mass spectrometry method for measuring 15N incorporation into pheophytin. Anal. Biochem. 2002, 307, 219–225. [Google Scholar] [CrossRef]
- Verma, S.K.; White, J.F. Indigenous endophytic seed bacteria promote seedling development and defend against fungal disease in browntop millet (Urochloa ramosa L.). J. Appl. Microbiol. 2018, 124, 764–778. [Google Scholar] [CrossRef]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Jensen, H.L. Jensen, H.L. Nitrogen fixation in leguminous plants. II. Is symbiotic nitrogen fixation influenced by Azotobacter? Proc. Linn. Soc. NSW 1942, 57, 205–212. [Google Scholar]
- Norris, J.R.; Chapman, H.M. Classification of Azotobacter. In Identification Methods for Microbiologists; Gibbs, B.M., Shapton, D.A., Eds.; Academic Press: New York, NY, USA, 1968; pp. 19–27. [Google Scholar]
- White, J.F.; Kingsley, K.L.; Zhang, Q.; Verma, R.; Obi, N.; Dvinskikh, S.; Elmore, M.T.; Verma, S.K.; Gond, S.K.; Kowalski, K.P. Review: Endophytic microbes and their potential applications in crop management. Pest Manag. Sci. 2019, 75, 2558–2565. [Google Scholar] [CrossRef]
- Lagendijk, E.L.; Validov, S.; Lamers, G.E.M.; De Weert, S.; Bloemberg, G.V. Genetic tools for tagging Gram-negative bacteria with mCherry for visualization in vitro and in natural habitats, biofilm and pathogenicity studies. FEMS Microbiol. Lett. 2010, 305, 81–90. [Google Scholar] [CrossRef]
- Martínez-Garciá, E.; Fraile, S.; Rodríguez Espeso, D.; Vecchietti, D.; Bertoni, G.; De Lorenzo, V. Naked Bacterium: Emerging properties of a surfome-streamlined Pseudomonas putida Strain. ACS Synth. Biol. 2020, 9, 2477–2492. [Google Scholar] [CrossRef]
- Benito, P.; Carro, L.; Bacigalupe, R.; Ortuzar, M.; Trullo, M.E. From roots to leaves: The capacity of Micromonospora to colonize different legume tissues. Phytobiomes 2021, 6, 35–44. [Google Scholar] [CrossRef]
- Elmore, M.T.; White, J.F.; Kingsley, K.L.; Diehl, K.H.; Verma, S.K. Pantoea spp. Associated with smooth crabgrass (Digitaria ischaemum) seed inhibit competitor plant species. Microorganisms 2019, 7, 143. [Google Scholar] [CrossRef]
- Soares, M.A.; Li, H.; Bergen, M.S.; Silva, J.F.; Kowalski, K.P.; White, J.F. Functional role of an endophytic Bacillus amyloliquefaciens in enhancing growth and disease protection of invasive English ivy (Hedera helix L.). Plant Soil 2015, 405, 107–123. [Google Scholar] [CrossRef]
- Li, H.; Soares, M.A.; Torres, M.S.; Bergen, M.; White, J.F. Endophytic bacterium, Bacillus amyloliquefaciens, enhances ornamental hosta resistance to diseases and insect pests. J. Plant Interact. 2015, 10, 224–229. [Google Scholar] [CrossRef]
- Sevigny, J.L.; Lloyd, B.; McComish, C.; Ramsey, A.; Koziol, L. Whole-genome sequences of Pantoea agglomerans BL3, Pseudomonas fluorescens BL, and Pseudomonas stutzeri CM14, isolated from hops (Humulus lupulus). Microbiol. Resour. Announc. 2019, 8, e00545-19. [Google Scholar] [CrossRef] [PubMed]
- Schindler, F.; Fragner, L.; Herpell, J.B.; Berger, A.; Brenner, M.; Tischler, S.; Bellaire, A.; Schönenberger, J.; Li, W.; Sun, X.; et al. Dissecting metabolism of leaf nodules in Ardisia crenata and Psychotria punctata. Front. Mol. Biosci. 2021, 8, 683671. [Google Scholar] [CrossRef] [PubMed]
- Vacher, C.; Hampe, A.; Porté, A.J.; Sauer, U.; Compant, S.; Morris, C.E. The phyllosphere: Microbial jungle at the plant–climate interface. Annu. Rev. Ecol. Evol. Syst. 2016, 47, 1–24. [Google Scholar] [CrossRef]
- Ricky, M.; Anderson, R. Effects of nitrogen addition on the invasive grass Phragmites australis and a native competitor Spartina pectinate. J. Appl. Ecol. 2004, 41, 888–896. [Google Scholar] [CrossRef]
- Uddin, M.N.; Robinson, R.W.; Asaeda, T. Nitrogen immobilization may reduce invasibility of nutrient enriched plant community invaded by Phragmites australis. Sci Rep. 2020, 10, 1601. [Google Scholar] [CrossRef]
- Kowalski, K.P.; Bacon, C.; Bickford, W.; Braun, H.; Clay, K.; Leduc-Lapierre, M.; Lillard, E.; McCormick, M.K.; Nelson, E.; Torres, M.; et al. Advancing the science of microbial symbiosis to support invasive species management: A case study on Phragmites in the Great Lakes. Front. Microbiol. 2015, 6, 95. [Google Scholar] [CrossRef]
- Kusstatscher, P.; Wicaksono, W.A.; Bergna, A.; Cernava, T.; Bergau, N.; Tissier, A.; Hause, B.; Berg, G. Trichomes form genotype-specific microbial hotspots in the phyllosphere of tomato. Environ. Microbiome 2020, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Blaskovich, M.A.T.; Kavanagh, A.M.; Elliott, A.G.; Zhang, B.; Ramu, S.; Amado, M.; Lowe, G.J.; Hinton, A.O.; Pham, D.M.T.; Zuegg, J.; et al. The antimicrobial potential of cannabidiol. Commun. Biol. 2021, 4, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Balcke, G.U.; Bennewitz, S.; Bergau, N.; Athmer, B.; Henning, A.; Majovsky, P.; Jiménez-Gómez, J.M.; Hoehenwarter, W.; Tissier, A. multi-omics of tomato glandular trichomes reveals distinct features of central carbon metabolism supporting high productivity of specialized metabolites. Plant Cell 2017, 29, 960–983. [Google Scholar] [CrossRef] [PubMed]
- Pagnani, G.; Pellegrini, M.; Galieni, A.; D’Egidio, S.; Matteucci, F.; Ricci, A.; Stagnari, F.; Sergi, M.; Lo Sterzo, C.; Pisante, M.; et al. Plant growth-promoting rhizobacteria (PGPR) in Cannabis sativa ‘Finola’ cultivation: An alternative fertilization strategy to improve plant growth and quality characteristics. Ind. Crops Prod. 2018, 123, 75–83. [Google Scholar] [CrossRef]
- Ooki, Y.; Banba, M.; Yano, K.; Maruya, J.; Sato, S.; Tabata, S.; Saeki, K.; Hayashi, M.; Kawaguchi, M.; Izui, K.; et al. Characterization of the Lotus japonicus symbiotic mutant lot1 that shows a reduced nodule number and distorted trichomes. Plant Physiol. 2005, 137, 1261–1271. [Google Scholar] [CrossRef][Green Version]
- Guinel, F.C. Ethylene, a hormone at the center-stage of nodulation. Front. Plant Sci. 2015, 6, 1121. [Google Scholar] [CrossRef] [PubMed]
- Iskra, A.E.; Lafontaine, S.R.; Trippe, K.M.; Massie, S.T.; Phillips, C.L.; Twomey, M.C.; Shellhammer, T.H.; Gent, D.H. Influence of nitrogen fertility practices on hop cone quality. Am. Soc. Brew. Chem. 2019, 77, 199–209. [Google Scholar] [CrossRef]
- Tang, K.; Fracasso, A.; Struik, P.C.; Yin, X.; Amaducci, S. Water-and nitrogen-use efficiencies of hemp (Cannabis sativa L.) based on whole-canopy measurements and modeling. Front. Plant Sci. 2018, 9, 951. [Google Scholar] [CrossRef]
- Beltran-Garcia, M.J.; Martinez-Rodriguez, A.; Olmos-Arriaga, I.; Valdez-Salas, B.; Chavez-Castrillon, Y.Y.; Di Mascio, P.; White, J.F. Probiotic endophytes for more sustainable banana production. Microorganisms 2021, 9, 1805. [Google Scholar] [CrossRef]
- De-Polli, H.; Boyer, C.D.; Neyra, C.A. Nitrogenase activity associated with roots and stems of field grown corn (Zea mays L.) plants. Plant Physiol. 1982, 70, 1609–1613. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.F.; Thake, B.; Martin, W.F. Nitrogenase inhibition limited oxygenation of earth’s proterozoic atmosphere. Trends Plant Sci. 2019, 24, 1022–1031. [Google Scholar] [CrossRef] [PubMed]
- Wurm, C.J.; Lindermayr, C. Nitric oxide signaling in the plant nucleus: The function of nitric oxide in chromatin modulation and transcription. J. Exp. Bot. 2021, 72, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Verchot-Lubicz, J.; Goldstein, R.E. Cytoplasmic streaming enables the distribution of molecules and vesicles in large plant cells. Protoplasma 2010, 240, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Woodhouse, F.G.; Goldstein, R.E. Cytoplasmic streaming in plant cells emerges naturally by microfilament self-organization. Proc. Natl. Acad. Sci. USA 2013, 110, 14132–14137. [Google Scholar] [CrossRef]
- Pati, B.R.; Chandra, A.K. Diazotrophic bacterial population and other associated organisms on the phyllosphere of some crop plants. Zentralbl. Mikrobiol. 1993, 148, 392–402. [Google Scholar] [CrossRef]
- Soares, M.A.; Li, H.Y.; Kowalski, K.P.; Bergen, M.; Torres, M.S.; White, J.F. Functional role of bacteria from invasive Phragmites australis in promotion of host growth. Microb. Ecol. 2016, 72, 407–417. [Google Scholar] [CrossRef]
- De Vries, J.; Archibald, J.M. Plant evolution: Landmarks on the path to terrestrial life. New Phytol. 2018, 217, 1428–1434. [Google Scholar] [CrossRef] [PubMed]
- Esau, K. Plant Anatomy, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 1965. [Google Scholar]
- Lee, J.C.; Kundu, J.K.; Hwang, D.M.; Na, H.K.; Surh, Y.J. Humulone inhibits phorbol ester-induced COX-2 expression in mouse skin by blocking activation of NF-κB and AP-1: IκB kinase and c-Jun-N-terminal kinase as respective potential upstream targets. Carcinogenesis 2007, 28, 1491–1498. [Google Scholar] [CrossRef]
- Nuutinen, T. Medicinal properties of terpenes found in Cannabis sativa and Humulus lupulus. Eur. J. Med. Chem. 2018, 157, 198–228. [Google Scholar] [CrossRef]
- Pellati, F.; Borgonetti, V.; Brighenti, V.; Biagi, M.; Benvenuti, S.; Corsi, L. Cannabis sativa L. and nonpsychoactive cannabinoids: Their chemistry and role against oxidative stress, inflammation, and cancer. Biomed Res. Int. 2018, 2018, 1691428. [Google Scholar] [CrossRef]
- Sunda, F.; Arowolo, A. A molecular basis for the anti-inflammatory and anti-fibrosis properties of cannabidiol. FASEB J. 2020, 34, 14083–14092. [Google Scholar] [CrossRef] [PubMed]
| Species | Family | Nuclear Symbiosis | Trichome Type | Ethylene | Reducing Sugars | Superoxide | Hydrogen Peroxide | Nitric Oxide | Nitrate | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F 2 | G 2 | AM | SM | PP | CS | NBT | DAB | FS | DS | |||
| Agave boldinghiana | Asparagaceae | √ 3 | √ | √ | √ | |||||||
| Agave palmeri | Asparagaceae | √ | √ | √ | √ | √ | ||||||
| Ailanthus altissima | Simaroubaceae | √ | √ | √ | √ | √ | √ | √ | √ | √ | ||
| Apocynum cannabinum | Apocynaceae | √ | √ | |||||||||
| Cannabis sativa | Cannabaceae | √ | √ | √ | √ | √ | √ | √ | √ | |||
| Celtis occidentalis | Cannabaceae | √ | √ | √ | √ | √ | ||||||
| Citronella mucronata | Cardiopteridaceae | √ | √ | √ | √ | √ | √ | |||||
| Digitaria sanguinalis | Poaceae | √ | √ | √ | √ | √ | √ | |||||
| Digitaria ischaemum | Poaceae | √ | √ | √ | ||||||||
| Eupatorium altissimum | Asteraceae | √ | √ | √ | √ | |||||||
| Eutrochium maculatum | Euphorbiaceae | √ | √ | √ | √ | √ | √ | |||||
| Festuca rubra | Poaceae | √ | √ | √ | √ | √ | √ | |||||
| Glycine max | Fabaceae | √ | √ | √ | √ | √ | √ | √ | √ | |||
| Helianthus hirsutus | Asteraceae | √ | √ | |||||||||
| Hedera helix | Araliaceae | √ | √ | √ | ||||||||
| Hosta plantaginea | Asparagaceae | √ | √ | √ | √ | √ | √ | √ | ||||
| Humulus lupulus | Cannabaceae | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | |
| Lactuca sativa | Asteraceae | √ | √ | |||||||||
| Lonicera japonica | Caprifoliaceae | √ | √ | √ | √ | √ | √ | √ | √ | |||
| Monotropa hypopitys | Ericaceae | √ | √ | √ | √ | √ | ||||||
| Perilla frutescens | Lamiaceae | √ | √ | √ | √ | √ | ||||||
| Phragmites australis | Poaceae | √ | √ | √ | √ | √ | ||||||
| Rhus glabra | Anacardiaceae | √ | √ | √ | √ | √ | √ | √ | ||||
| Solanum dulcamara | Solanaceae | √ | √ | √ | √ | √ | √ | √ | √ | |||
| Solanum lycopersicum | Solanaceae | √ | √ | √ | √ | √ | √ | √ | ||||
| Solanum nigrum | Solanaceae | √ | √ | √ | √ | √ | √ | √ | ||||
| Solidago canadensis | Asteraceae | √ | √ | √ | √ | √ | ||||||
| Stachys byzantina 4 | Lamiaceae | √ | ||||||||||
| Thespesia populnea | Malvaceae | √ | √ | √ | √ | √ | ||||||
| Vanilla phaeantha | Orchidaceae | √ | √ | √ | √ | √ | √ | |||||
| Verbascum thapsus | Scrophulariaceae | √ | √ | √ | √ | √ | ||||||
| Vigna radiata | Fabaceae | √ | √ | √ | √ | √ | √ | √ | ||||
| Plant Species | Treatment | Plant Organ | Delta 15N vs. Air ± SD (Number of Plants) | p-Value (Two-Tailed)/Group ID 1 |
|---|---|---|---|---|
| Agave boldinghiana | 15N2 enriched | Leaf #1 (youngest) | 13.8 ± 12.6 (N = 3) | N/A 2 |
| Leaf #2 | 28.6 ± 9.8 (N = 3) | N/A | ||
| Leaf #3 | 31.6 ± 5.4 (N = 3) | N/A | ||
| Leaf #4 | 19.3 ± 8.8 (N = 3) | N/A | ||
| Leaf #5 (oldest) | 11.2 (N = 2) | N/A | ||
| Roots | 20.6 ± 28.3 (N = 2) | N/A | ||
| Air control | Leaf #1 (youngest) | 0.6 ± 0.4 (N = 2) | N/A | |
| Leaf #2 | 1.2 ± 0.8 (N = 2) | N/A | ||
| Leaf #3 | 1.1 ± 0.4 (N = 2) | N/A | ||
| Leaf #4 | 0.3 ± 1.0 (N = 2) | N/A | ||
| Leaf #5 (oldest) | 0.8 ± 0.4 (N = 2) | N/A | ||
| Roots | 4.2 ± 0.5 (N = 2) | N/A | ||
| Agave palmeri | 15N2 enriched | Seedling leaves | 74.4 ± 21.3 (N = 3) | N/A |
| Air control | Seeding leaves | 3.9 ± 0.8 (N = 2) | N/A | |
| Ailanthus altissima | 15N2 enriched | Leaflets | 1017.6 ± 3.5 (N = 2) | N/A |
| Leaf rachis | 673.3 (N = 1) | N/A | ||
| Air control | Leaflets | 3.38 (N = 1) | N/A | |
| Leaf rachis | 3.48 (N = 1) | N/A | ||
| Festuca rubra | 15N2 enriched | Seedling shoots (leaf blades and sheaths) | 236.7 ± 102.5 (N = 4) | N/A |
| Air | Seedling shoots (leaf blades and sheaths) | 1.3 ± 1.0 (N = 2) | N/A | |
| Hosta plantaginea | 15N2 enriched | Leaves | 674.5 ± 369.4 (N = 4) | 0.03528 (Leaves in air vs. N15) 3 |
| 0.049734 (Leaves vs. roots in N15) | ||||
| Roots | 87.5 ± 56.7 (N = 4) | 0.052163 (Roots in air vs. N15) | ||
| 0.96609 (Leaves vs. roots in air) | ||||
| Air control | Leaves | −1.4 ± 1.8 (N = 3) | ||
| Roots | −1.3 ± 1.9 (N = 3) | |||
| Humulus lupulus | 15N2 enriched | Inflorescences | 2539.6 ± 1329.7 (N = 3) | 0.6283 (Inflorescences vs. leaves/stems in N15) 3 |
| Leaves and stems | 2206.8 ± 1691.8 (N = 3) | 0.6283 (Inflorescences vs. leaves/stems in N15) | ||
| Air control | Inflorescences | 13.5 (N = 1) | N/A | |
| Leaves and stems | 13.1 (N = 1) | N/A | ||
| Lonicera japonica | 15N2 enriched | Leaves | 52 ± 16.0 (N = 6) | 0.000587 (Leaves in air vs. N15) |
| Air control | Leaves | 1.6 ± 0.1 (N = 3) | ||
| Phragmites australis | 15N2 enriched | Leaves | 23.0 ± 2.0 (N = 3) | No significant differences between plant parts (Leaves/stems/roots in N15) |
| Stems | 21.0 ± 7.1 (N = 3) | |||
| Roots | 15.5 ± 4.7 (N = 3) | |||
| Air control | Leaves | 4.8 (N = 1) | N/A | |
| Stems | 1.8 (N = 1) | N/A | ||
| Roots | 2.1 (N = 1) | N/A | ||
| Thespesia populnea | 15N2 enriched | Leaf #1 (youngest) | 545.2 ± 179.0 (N = 4) | A (Comparison of plant parts in N15) 4 |
| Leaf #2 | 650.2 ± 66.9 (N = 4) | A | ||
| Leaf #3 | 488.1 ± 79.0 (N = 4) | AB | ||
| Leaf #4 (oldest) | 441.1 ± 103.2 (N = 4) | AB | ||
| Roots | 248.9 ± 89.1 (N = 4) | B | ||
| Air control | Leaf #1 (youngest) | 3.9 (N = 1) | N/A | |
| Leaf #2 | 4.4 (N = 1) | N/A | ||
| Leaf #3 | 4.9 (N = 1) | N/A | ||
| Leaf #4 (oldest) | 4.2 (N = 1) | N/A | ||
| Roots | 2.6 (N = 1) | N/A | ||
| Vanilla phaeantha | 15N2 enriched | Leaf #1 (youngest) | 63.9 ± 14.4 (N = 8) | A (Comparison of plant parts in N15) 4,5 |
| Leaf #2 | 40.9 ± 20.8 (N = 8) | AB | ||
| Leaf #3 | 24.9 ± 12.6 (N = 8) | BC | ||
| Leaf #4 (oldest) | 20.2 ± 18.7 (N = 5) | BC | ||
| Roots | 6.8 ± 1.6 (N = 4) | C | ||
| Air control | Leaf #1 (youngest) | 3.9 (N = 1) | N/A | |
| Leaf #2 | 4.2 (N = 1) | N/A | ||
| Leaf #3 | 2.3 (N = 1) | N/A | ||
| Leaf #4 (oldest) | 3.4 (N = 1) | N/A | ||
| Roots | 2.3 (N = 1) | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Micci, A.; Zhang, Q.; Chang, X.; Kingsley, K.; Park, L.; Chiaranunt, P.; Strickland, R.; Velazquez, F.; Lindert, S.; Elmore, M.; et al. Histochemical Evidence for Nitrogen-Transfer Endosymbiosis in Non-Photosynthetic Cells of Leaves and Inflorescence Bracts of Angiosperms. Biology 2022, 11, 876. https://doi.org/10.3390/biology11060876
Micci A, Zhang Q, Chang X, Kingsley K, Park L, Chiaranunt P, Strickland R, Velazquez F, Lindert S, Elmore M, et al. Histochemical Evidence for Nitrogen-Transfer Endosymbiosis in Non-Photosynthetic Cells of Leaves and Inflorescence Bracts of Angiosperms. Biology. 2022; 11(6):876. https://doi.org/10.3390/biology11060876
Chicago/Turabian StyleMicci, April, Qiuwei Zhang, Xiaoqian Chang, Kathryn Kingsley, Linsey Park, Peerapol Chiaranunt, Raquele Strickland, Fernando Velazquez, Sean Lindert, Matthew Elmore, and et al. 2022. "Histochemical Evidence for Nitrogen-Transfer Endosymbiosis in Non-Photosynthetic Cells of Leaves and Inflorescence Bracts of Angiosperms" Biology 11, no. 6: 876. https://doi.org/10.3390/biology11060876
APA StyleMicci, A., Zhang, Q., Chang, X., Kingsley, K., Park, L., Chiaranunt, P., Strickland, R., Velazquez, F., Lindert, S., Elmore, M., Vines, P. L., Crane, S., Irizarry, I., Kowalski, K. P., Johnston-Monje, D., & White, J. F. (2022). Histochemical Evidence for Nitrogen-Transfer Endosymbiosis in Non-Photosynthetic Cells of Leaves and Inflorescence Bracts of Angiosperms. Biology, 11(6), 876. https://doi.org/10.3390/biology11060876








